Abstract
This review focuses on anabolic signaling pathways through which insulin, amino acids, and resistance exercise act to regulate the protein kinase complex referred to as mechanistic target of rapamycin complex (mTORC) 1. Initially, individual pathways through which the 3 anabolic signals act to modulate mTORC1 signaling will be discussed, followed by a summation of evidence showing an additive effect of the regulators. The emphasis will be on mTORC1 signaling in skeletal muscle and its contribution to modulation of rates of protein synthesis. In addition, results from studies using cells in culture will be used to provide a more complete picture of the molecular details of the individual pathways.
INTRODUCTION
Changes in muscle mass occur in response to alterations in the balance between protein synthesis and degradation. An increase in protein synthesis relative to degradation is necessary for increased muscle mass accretion, whereas a decrease in the ratio has the opposite effect and leads to muscle loss. Although changes in protein degradation may contribute to muscle hypertrophy under a variety of conditions, the focus of the present article will be on the role played by the stimulation of protein synthesis in the accretion of muscle mass.
Protein synthesis in skeletal muscle is regulated through a number of signaling pathways that control the individual steps in messenger RNA (mRNA)5 translation. Absolute rates of protein synthesis therefore depend on the integration of the signals generated by the various pathways. The most studied anabolic inputs to muscle protein synthesis include hormones such as insulin and insulin-like growth factor I (IGF-I), amino acids, and exercise. Therefore, the primary emphasis of this review will be on the signaling pathways activated by those inputs, the integration of the pathways, and the step or steps in mRNA translation targeted by the pathways.
MECHANISTIC TARGET OF RAPAMYCIN SIGNALING PATHWAY
The mechanistic target of rapamycin (mTOR; also known as the mammalian target of rapamycin) is a protein kinase that exists in 2 complexes referred to as mTOR complex (mTORC) 1 and mTORC2 (reviewed in reference 1). In addition to mTOR, both complexes contain mammalian lethal with secretory (SEC13) protein 8 (mLST8), but the complexes are distinguished by the presence of the regulatory-associated protein of mTOR, complex 1 (Raptor), in mTORC1 and the Raptor-independent companion of mTOR, complex 2, in mTORC2. Although they may have other roles, one function of Raptor and Raptor-independent companion of mTOR, complex 2, is to engender substrate specificity to the mTOR complexes. For example, mTORC1 phosphorylates proteins such as the 70-kDa ribosomal protein S6 kinase 1 (p70S6K1) and eukaryotic initiation factor (eIF) 4E binding protein (4E-BP) 1, whereas mTORC2 phosphorylates Ak transforming (Akt) and the serum/glucocorticoid regulated kinase 1 (SGK1). As a consequence of their different substrate preferences, the mTOR complexes have distinct functions: mTORC1 regulates cell proliferation and cell cycle progression, whereas mTORC2 modulates cell survival. Of particular relevance to this review, mTORC1 has multiple downstream targets that act to regulate mRNA translation and ribosome biogenesis, and therefore its regulation will be emphasized herein.
Downstream targets of mTORC1
The mTORC1 complex regulates protein synthesis both acutely and chronically (2). Chronically, activation of mTORC1 leads to induced expression of many of the proteins that function in the process of mRNA translation, including a number of initiation and elongation factors. It also promotes ribosome biogenesis, resulting in increased capacity for mRNA translation. Acutely, mTORC1 phosphorylates 4E-BP1 and p70S6K1 and thereby stimulates the binding of eIF4A and eIF4E to eIF4G to form the eIF4F complex. For example, the binding of 4E-BP1 to eIF4E prevents it from associating with eIF4G; phosphorylation of 4E-BP1 by mTORC1 prevents its association with eIF4E, allowing eIF4E to bind to eIF4G (3). In a similar manner, the binding of programmed cell death 4 (PDCD4) to eIF4A blocks its interaction with eIF4G, and phosphorylation of PDCD4 by p70S6K1 results in its release from eIF4A, allowing eIF4A to bind to eIF4G (4). Once assembled, eIF4F mediates the binding of mRNA to the 43S preinitiation complex, resulting in formation of the 48S preinitiation complex. Scanning of the 48S preinitiation complex along the 5′-untranslated region of the mRNA is enhanced by eIF4B, and phosphorylation of eIF4B by p70S6K1 augments its function (5). Thus, activation of mTORC1 promotes both the cap-dependent association of the 43S preinitiation complex with mRNA and scanning of the complex along the 5′-untranslated region to permit its localization at the AUG start codon.
Regulation of mTORC1 signaling
Signaling through mTORC1 is controlled by its association with various small GTPases, such as the Ras homolog enriched in brain (Rheb). Rheb activates mTORC1 when it is bound to GTP but not when it is associated with GDP (6). The GTP/GDP loading status of Rheb is controlled primarily by the GTPase activator protein (GAP) complex consisting of tuberous sclerosis (TSC) proteins 1 and 2 (7) and the ubiquitin specific peptide 6 (Trc-2)/budding uninhibited by benzimidazole/cell division cycle (TBC) domain family member, TBC1D7 (8). Notably, both Rheb and TSC1 localize to intracellular membranes, and the relevance of such localization will be expanded on below. The GAP activity of TSC1/2 is modulated through phosphorylation of both TSC1 and TSC2 by various upstream kinases, including Akt, the extracellular-regulated protein kinases (ERK) 1 and 2, the 90-kDa ribosomal protein S6 kinase (p90RSK), and the AMP-activated protein kinase (AMPK) (9). Phosphorylation by Akt, ERK, or p90RSK represses TSC1/2 GAP activity resulting in accumulation of Rheb in its GTP-bound form, whereas phosphorylation by AMPK has the opposite effect. Thus, by activating the Akt and ERK/p90RSK signaling pathways, hormones such as insulin and IGF-I stimulate mTORC1 signaling, whereas through activation of AMPK, endurance exercise acts to repress it.
Signaling through mTORC1 is also controlled by a GTPase heteroduplex consisting of either Ras-related GTP binding (Rag) A or RagB (referred to as RagA/B subsequently) in association with either RagC or RagD (RagC/D). When RagA/B is bound to GTP and RagC/D is bound to GDP, the heterodimer potently stimulates mTORC1 signaling, but when RagA/B is bound to GDP and RagC/D is bound to GTP, it does not (10). In addition to binding to mTORC1, the RagA/B-RagC/D complex also associates with a heteropentameric protein complex referred to as Ragulator (10, 11). The 5 subunits of Ragulator are encoded by the late endosomal/lysosomal adaptor, mitogen-activated protein kinase and MTOR activator (LAMTOR) genes 1–5. The protein encoded by LAMTOR1, p18, is myristoylated and palmitoylated, and acts to target the complex to late endosomal and/or lysosomal membranes (12). Chromosome 7 open reading frame 59 (C7orf59) and hepatitis B virus X interacting protein (HBXIP), products of the LAMTOR4 and LAMTOR5 genes, respectively, form a subcomplex that permits Ragulator to act as a guanine nucleotide exchange factor (GEF) for RagA/B (11). Amino acids also modulate RagA/B GTP loading through a hetero-octameric complex referred to as GAP activity toward Rags (GATOR) (16). The proteins that form the GATOR complex exist in 2 subcomplexes, GATOR1 and GATOR2. GATOR1 is a GAP for RagA/B and, like Ragulator, localizes to the lysosomal membrane. GATOR2 acts to inhibit GATOR1, and its loss prevents amino acid–induced mTORC1 signaling. Overall, amino acids promote mTORC1 signaling by stimulating Ragulator GEF activity and repressing GATOR1 GAP activity toward RagA/B to increase GTP loading.
The molecular details through which amino acids act to modulate Ragulator and GATOR regulation of mTORC1 are unclear. However, amino acids, and in particular the branched-chain amino acid leucine, stimulate Ragulator GEF activity toward RagA/B through a mechanism involving the vacuolar H+-ATPase, which, like Ragulator and GATOR1, is also localized to late endosomal and/or lysosomal membranes (13). Interestingly, by using a variety of approaches, the vacuolar H+-ATPase was shown to respond to intralysosomal, rather than cytosolic, amino acids (13), suggesting that dietary amino acids might need to be transported into the lysosomal lumen to activate mTORC1. In this regard, several amino acid transporters have been identified on the lysosomal membrane. For example, the proton-assisted amino acid transporter solute carrier family 36 member 1 (SLC36A1) was recently shown to localize, in part, to the lysosomal membrane and to interact with the Rag GTPases (14). However, SLC36A1 is thought to transport amino acids out of the lysosome (15), and exogenous overexpression of the protein inhibits mTORC1 signaling (13). Thus, if it is the accumulation of amino acids within the lumen of the lysosome that is required for mTORC1activation, SLC36A1 transport activity would need to be repressed in order for mTORC1 to be activated by amino acids. Future studies will undoubtedly identify more pieces of this puzzle and provide a clearer picture of how amino acids act to modulate Ragulator and GATOR function.
Compared with the regulation of mTORC1 signaling by insulin/IGF-I and amino acids, much less is known about the pathway or pathways through which resistance exercise stimulates the kinase. Early studies showed that IGF-I expression is enhanced in skeletal muscle in animal models of resistance exercise (eg, reference 17) and that exogenous expression of IGF-I in muscle results in hypertrophy (18). In contrast, the results of a subsequent study showed that exercise-induced muscle hypertrophy occurs through an IGF-I receptor–independent mechanism (19), suggesting that the effect is not mediated by the growth factor. However, even in the absence of IGF-I signaling, exercise still induces Akt phosphorylation, suggesting that it might stimulate mTORC1 signaling through an Akt-dependent signaling pathway. In support of this suggestion, exogenous expression of a constitutively active Akt in muscle promotes hypertrophy (20). Moreover, in primary cultures of myotubes, inhibition of either phosphatidylinositide 3-kinase (PI3K), an upstream activator of Akt, or mTORC1 attenuates stretch-induced hypertrophy (21). However, in a mouse model of resistance exercise, treatment with the PI3K inhibitor wortmannin did not prevent the exercise-induced stimulation of mTORC1 signaling (22), suggesting the involvement of a pathway distinct from PI3K/Akt. Another signaling pathway through which resistance exercise might act to stimulate mTORC1 signaling is the ERK1/2 pathway. Phosphorylation of ERK1/2 is enhanced both in humans after leg extension exercise (23) and in a rodent model of resistance exercise referred to as functional overload or synergist ablation (22). In mice subjected to functional overload, phosphorylation of TSC2 on Ser664, a known target of ERK1/2 (24), is enhanced, suggesting that exercise might stimulate mTORC1 through an ERK/TSC signaling pathway. Arguing against this suggestion is a report showing that stretch-induced stimulation of mTORC1 in C2C12 myoblasts in culture is not prevented by U0126, an inhibitor of the ERK1/2 pathway (25). Instead, in the study by You et al (25), the binding of phosphatidic acid to mTOR was proposed as a possible mechanism to account for stretch-induced stimulation of mTORC1. Whether or not phosphatidic acid might play a role in exercise-induced stimulation of mTORC1 signaling needs to be further explored.
Another possible mechanism through which resistance exercise might act to stimulate mTORC1 signaling is through reduced expression of the mTORC1 repressor proteins referred to as regulated in development and DNA damage (REDD) 1 and 2. Although the mechanism is incompletely defined, both repressors are thought to act by enhancing the GAP activity of TSC1/2 toward Rheb (eg, reference 26). In a number of studies (27–31), the expression of REDD1 and/or REDD2 was shown to be reduced during the recovery period immediately after a bout of resistance exercise. Direct evidence linking decreased REDD1 and REDD2 expression in skeletal muscle after exercise is lacking; however, it is tempting to speculate that such changes contribute to the stimulation of mTORC1 signaling during recovery from exercise.
Effect of rapamycin on nutrient- and exercise-induced muscle protein synthesis
The macrolide immunosuppressant rapamycin is a selective and direct inhibitor of mTORC1. In a complex with FK506 binding protein 12 (FKBP12), rapamycin binds directly to mTOR when the kinase is present in the mTORC1, but not the mTORC2, complex. When administered to fasted rats (32), pigs (33), or humans (34), it attenuates the amino acid– or diet-induced stimulation of muscle protein synthesis. Similarly, if rapamycin is administered to either sedentary rats (35) or humans (23), it prevents the resistance exercise–induced increase in muscle protein synthesis. Together, the results of these studies strongly suggest that activation of mTORC1 is required for both amino acid– and exercise-induced stimulation of muscle protein synthesis.
INTEGRATION OF SIGNALS FROM INSULIN, AMINO ACIDS, AND EXERCISE
Insulin and amino acids
The cooperative effect of insulin and amino acids in the activation of mTORC1 signaling in skeletal muscle has been reported in many studies. For example, oral administration of leucine stimulates mTORC1 signaling in the gastrocnemius muscle of a control rat but not in a rat model of type 1 diabetes in which plasma insulin concentrations are only ∼20% of that observed in a fasted control animal (36). In contrast, the magnitude of the increase in mTORC1 signaling engendered by oral leucine administration in control animals is similar to that in rats treated with somatostatin to clamp insulin at fasting concentrations (37), suggesting that fasting concentrations of the hormone are sufficient for amino acid–induced stimulation of mTORC1 in skeletal muscle. Similarly, in pigs subjected to a pancreatic–amino acid clamp, either increasing insulin concentrations while maintaining amino acid concentrations at fasting values or increasing amino acid concentrations while maintaining the concentration of insulin at fasting values results in enhanced p70S6K1 phosphorylation and enhanced association of eIF4G with eIF4E (38).
The cooperative effect of insulin and amino acids in the activation of mTORC1 signaling is also observed in cells in culture. For example, in FAO hepatoma cells deprived of serum and amino acids, the addition of either insulin or amino acids to the cell culture medium enhances p70S6K1 activity by ∼2-fold, whereas the addition of both insulin and amino acids results in a 3.5-fold increase (39). A later study using Rat2 fibroblasts showed that insulin and leucine act in an additive manner to promote phosphorylation of p70S6K1 on Thr389, a site directly phosphorylated by mTORC1 (40). Combined, the results of a plethora of studies show that insulin and amino acids act through independent signaling pathways to stimulate mTORC1 signaling. Support for this conclusion is provided by a recent study (41) showing that in serum- and leucine-deprived Rat2 fibroblasts expressing a combination of constitutively active (ca) RagB and caRagC, insulin treatment stimulated mTORC1 signaling, but leucine treatment had no effect. In contrast, in serum- and leucine-deprived cells expressing either caAkt or caRheb, mTORC1 signaling was stimulated in response to treatment with leucine but not insulin. Signaling in cells expressing a combination of caRagB/caRagC together with either caAkt or caRheb was completely resistant to the addition of either insulin or leucine. Overall, the results support a model in which insulin signals to mTORC1 through an Akt/Rheb pathway, whereas leucine acts through the Rag pathway to enhance mTORC1 signaling (Figure 1).
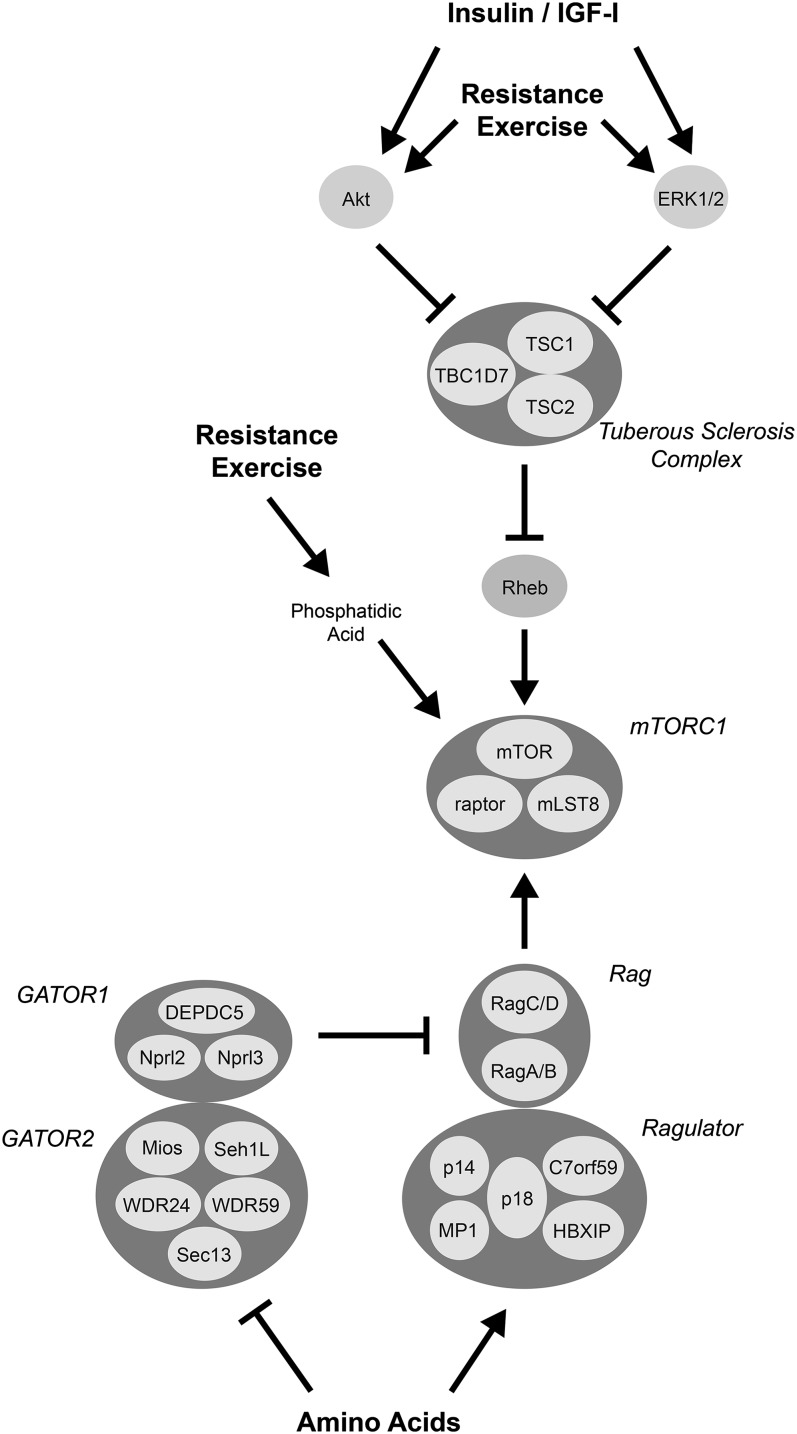
FIGURE 1.
Signaling pathways that modulate amino acid, insulin/IGF-I, and resistance exercise–induced activation of mTORC1 signaling. Hormones such as insulin and IGF-I signal to mTORC1 through Akt- and ERK1/2-dependent inactivation of TSC1/2 GAP activity, leading to enhanced GTP-loading of Rheb at late endosomal/lysosomal membranes. Amino acids promote GTP loading of RagA/B by both inhibiting Ragulator GAP activity and stimulating GATOR GEF activity. Ragulator also acts to localize the Rag complex to the late endosomal/lysosomal membrane. The binding of mTORC1 to the Rag complex allows it to interact with Rheb-GTP and become active. The signaling pathway or pathways through which resistance exercise acts to stimulate mTORC1 signaling is less clear, with Akt, ERK, and phosphatidic acid all playing possible roles. c7orf59, chromosome 7 open reading frame 59; ERK, extracellular-regulated protein kinase; GAP, GTPase activator protein; GATOR, GAP activity toward Rags; GEF, guanine nucleotide exchange factor; IGF-I, insulin-like growth factor I; mLST8, lethal with SEC13 protein 8; mTOR, mechanistic target of rapamycin; mTORC, mTOR complex; Rag, Ras-related GTP binding; raptor, regulatory-associated protein of mTOR, complex 1; Rheb, ras homolog enriched in brain; Sec13, secretory 13; TBC1D7, ubiquitin specific peptide 6 (Trc-2)/budding uninhibited by benzimidazole/cell division cycle; TSC, tuberous sclerosis complex.
Insulin and resistance exercise
Although an increase in plasma insulin concentration may not be required for the exercise-induced increase in muscle protein synthesis, evidence from both rodent and human studies suggests that there is a threshold for the hormone whereby plasma concentrations below a certain value result in protein synthesis being refractory to the stimulatory effect of resistance exercise on muscle protein synthesis. For example, muscle protein synthesis is increased after exercise in rats with moderate type 1 diabetes (fed arterial insulin of ∼180 pmol/L) but not in severely diabetic rats (fed arterial insulin of ∼72 pmol/L) (42, 43). Interestingly, the insulin concentration that is permissive for exercise-induced protein synthesis is at, or below, that observed in a control, fasted animal, suggesting that, in most cases, insulin is not limiting for exercise-induced stimulation of muscle protein synthesis.
Resistance exercise and amino acids
In a variety of studies, amino acids have been shown to enhance the effect of resistance exercise on muscle protein synthesis (reviewed in reference 44). Although the mechanism involved in the effect is poorly characterized, it is tempting to speculate that it might be associated with an exercise-induced stimulation of mTORC1 signaling due to enhanced production of phosphatidic acid. In this regard, the addition of either lysophosphatidic acid (a precursor of phosphatidic acid) or leucine to the culture medium of cells deprived of serum and leucine stimulates mTORC1 signaling ∼2.5-fold, whereas the addition of both lysophosphatidic acid and leucine has an additive effect (40). An alternative, and not mutually exclusive, possibility is that exercise acts to enhance the proportion of Rheb in the GTP-bound form through activation of the Akt and ERK1/2 signaling pathways and/or repression of REDD1/2 expression. All 3 events would lead to inhibition of TSC1/2 GAP activity and an increase in the proportion of Rheb-GTP.
CONCLUSIONS AND FUTURE DIRECTIONS
Although the exact molecular details are incomplete, there exists abundant evidence to support a model in which the intersection of the insulin and amino acid pathways localizes to the late endosome and/or lysosomal membrane. Thus, as depicted in Figure 2, amino acids promote the association of RagA/B with GTP, leading to the localization of mTORC1 to the late endosomal/lysosomal membrane where it associates with, and is activated by, Rheb-GTP. Insulin enhances the effect of amino acids through Akt-dependent inhibition of TSC1/2 GAP activity toward Rheb, resulting in the accumulation of Rheb in the GTP-bound form. Thus, signaling through mTORC1 is ultimately controlled by its association with Rheb-GTP. This idea is supported by the observation that exogenous expression of caRheb at high levels (eg 20- to 50-fold greater than endogenous) leads to both insulin- and amino acid–independent activation of mTORC1 (41). Where input from resistance exercise fits in this model is much less clear, but the available evidence suggests that, similar to insulin, exercise probably stimulates mTORC1 signaling through the Rheb, rather than the Rag, pathway. Future studies should be directed at filling in the missing pieces of the signaling pathways that control mTORC1 signaling. For example, it remains unclear how amino acids act to stimulate the GEF activity of Raptor or repress the GAP activity of GATOR toward RagA/B. Moreover, neither the GAP nor GEF for RagC/D has been characterized. It is also unclear why, in most cells, there is a strong preference for leucine in modulating mTORC1 signaling. The insulin signaling pathway to mTORC1 appears to be more completely defined compared with the amino acid pathway. However, unanswered questions remain, including the mechanism through which Rheb is localized to the late endosomal/lysosomal membrane and the identity of the Rheb GEF, if it exists. Of the signaling pathway and pathways discussed herein, the one through which resistance exercise acts to regulate mTORC1 signaling is the least completely characterized, in part because of the difficulty of mimicking the effect of exercise in vitro. Future studies using genetic approaches such as electroporation of expression plasmids into skeletal muscle, as well as the development of new models using myotubes in culture, will likely greatly expand our knowledge in this important area.
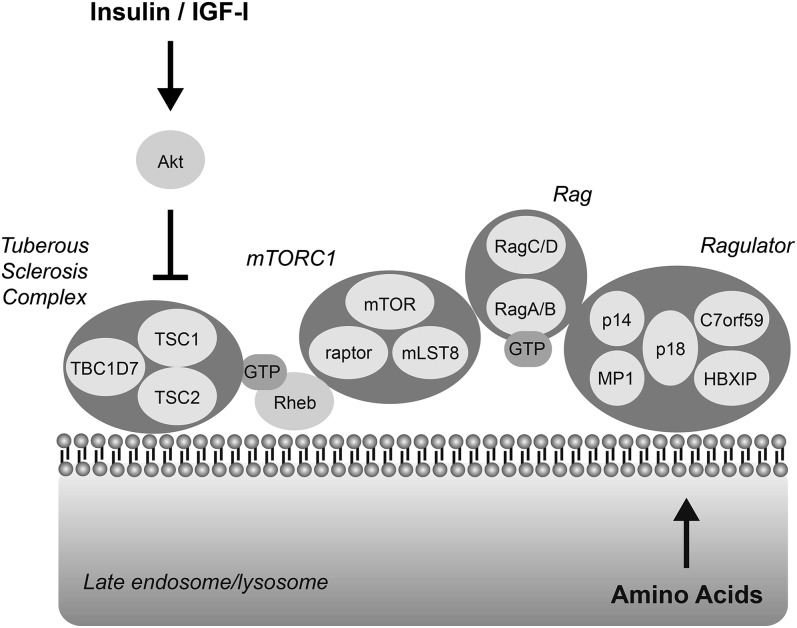
FIGURE 2.
mTORC1 interacts with Rheb and Rag GTPases at the late endosomal/lysosomal membrane. The Ragulator complex is anchored to late endosomal/lysosomal membranes through p18, which is myristoylated and palmitoylated (12). The RagA/B-RagC/D complex binds to Ragulator, and when RagA/B is associated with GTP, it also binds to mTORC1 to recruit it to the membrane where it interacts with Rheb. Rheb is also subject to lipid modification (farnesylation), leading to its localization to intracellular membranes (reviewed in reference 45). The results of studies assessing TSC1/2 subcellular distribution have been inconsistent. However, a portion of the complex has been shown to interact with subcellular membranes (46). It seems likely that TSC1/2 interfaces with Rheb at the late endosomal/lysosomal membrane. c7orf59, chromosome 7 open reading frame 59; IGF-I, insulin-like growth factor I; mLST8, lethal with SEC13 protein 8; mTOR, mechanistic target of rapamycin; mTORC, mTOR complex; Rag, Ras-related GTP binding; raptor, regulatory-associated protein of mTOR, complex 1; Rheb, ras homolog enriched in brain; TBC1D7, ubiquitin specific peptide 6 (Trc-2)/budding uninhibited by benzimidazole/cell division cycle; TSC, tuberous sclerosis complex.
Acknowledgments
I thank Leonard S Jefferson for his careful reading of the manuscript and helpful comments during its preparation.
SRK was solely responsible for the design and content of the material presented in this article and declared no conflicts of interest.
Footnotes
Abbreviations used: Akt, Ak transforming; AMPK, AMP-activated protein kinase; ca, constitutively active; c7orf59, chromosome 7 open reading frame 59; eIF, eukaryotic initiation factor; ERK, extracellular-regulated protein kinase; FKBP12, FK506 binding protein 12; GAP, GTPase activator protein; GATOR, GAP activity toward Rags; GEF, guanine nucleotide exchange factor; HBXIP, hepatitis B virus X interacting protein; IGF-I, insulin-like growth factor I; LAMTOR, late endosomal/lysosomal adaptor, mitogen-activated protein kinase and MTOR activator; mLST8, lethal with secretory (SEC13) protein 8; mRNA, messenger RNA; mTOR, mechanistic target of rapamycin; mTORC, mTOR complex; PDCD4, programmed cell death 4; PI3K, phosphatidylinositide 3-kinase; p70S6K1, 70-kDa ribosomal protein S6 kinase 1; p90RSK, 90-kDa ribosomal protein S6 kinase; Rag, Ras-related GTP binding; Raptor, regulatory-associated protein of mTOR, complex 1; REDD, regulated in development and DNA damage; Rheb, ras homolog enriched in brain; SGK1, serum/glucocorticoid regulated kinase 1; SLC36A1, solute carrier family 36 member 1; TBC, ubiquitin specific peptide 6 (Trc-2)/budding uninhibited by benzimidazole/cell division cycle; TSC, tuberous sclerosis complex; 4E-BP1, eIF4E binding protein 1.
REFERENCES
- 1.Bracho-Valdés I, Moreno-Alvarez P, Valencia-Martinez I, Robles-Molina E, Chavez-Vargas L, Vazquez-Prado J. mTORC1- and mTORC2-interacting proteins keep their multifunctional partners focused. IUBMB Life 2011;63:896–914. [DOI] [PubMed] [Google Scholar]
- 2.Dempsey JM, Mahoney SJ, Blenis J. mTORC1-mediated control of protein translation. In: Fuyuhiko T, Michael NH, eds. The enzymes. Waltham, MA: Academic Press, 2010:1–20. [Google Scholar]
- 3.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 1999;3:707–16. [DOI] [PubMed] [Google Scholar]
- 4.Yang H-S, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 Is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 2003;23:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holz MK, Blenis J. Identification of S6K1 as a novel mTOR-phosphorylating kinase. J Biol Chem 2005;280:26089–93. [DOI] [PubMed] [Google Scholar]
- 6.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol 2005;15:702. [DOI] [PubMed] [Google Scholar]
- 7.Inoki K, Li Y, Xu T, Guan K-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003;17:1829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 2012;47:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 2008;412:179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signalin g to mTORC1. Science 2008;320:1496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012;150:1196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ögmundsdóttir MH, Heublein S, Kazi S, Reynolds B, Visvalingam SM, Shaw MK, Goberdhan DC. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS ONE 2012;7:e36616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagné C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci USA 2001;98:7206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013;340:1100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKoy G, Ashley W, Mander J, Yang SY, Williams N, Russell B, Goldspink G. Expression of insulin growth factor-1 splice variants and structural genes in rabbit skeletal muscle induced by stretch and stimulation. J Physiol 1999;516:583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman ME, DeMayo F, Yin KC, Lee HM, Geske R, Montgomery C, Schwartz RJ. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem 1995;270:12109–16. [DOI] [PubMed] [Google Scholar]
- 19.Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J Physiol 2008;586:283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 2001;3:1014–9. [DOI] [PubMed] [Google Scholar]
- 21.Sasai N, Agata N, Inoue-Miyazu M, Kawakami K, Kobayashi K, Sokabe M, Hayakawa K. Involvement of PI3K/Akt/TOR pathway in stretch-induced hypertrophy of myotubes. Muscle Nerve 2010;41:100–6. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J Physiol 2011;589:1831–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol 2009;587:1535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Teruya-Feldstein J, Bonner P, Bernardi R, Franz DN, Witte D, Cordon-Cardo C, Pandolfi PP. Identification of S664 TSC2 phosphorylation as a marker for extracellular signal-regulated kinase mediated mTOR activation in tuberous sclerosis and human cancer. Cancer Res 2007;67(15):7106–12. [DOI] [PubMed] [Google Scholar]
- 25.You JS, Frey JW, Hornberger TA. Mechanical stimulation induces mTOR signaling via an ERK-independent mechanism: implications for a direct activation of mTOR by phosphatidic acid. PLoS ONE 2012;7:e47258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004;18:2893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond MJ, Fujita S, Abe T, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Med Sci Sports Exerc 2008;40:691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond MJ, Miyazaki M, Dreyer HC, Pennings B, Dhanani S, Volpi E, Esser KA, Rasmussen BB. Expression of growth-related genes in young and older human skeletal muscle following an acute stimulation of protein synthesis. J Appl Physiol 2009;106:1403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry CS, Glynn EL, Drummond MJ, Timmerman KL, Fujita S, Abe T, Dhanani S, Volpi E, Rasmussen BB. Blood flow restriction exercise stimulates mTORC1 signaling and muscle protein synthesis in older men. J Appl Physiol 2010;108:1199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greig CA, Gray C, Rankin D, Young A, Mann V, Noble B, Atherton PJ. Blunting of adaptive responses to resistance exercise training in women over 75y. Exp Gerontol 2011;46:884–90. [DOI] [PubMed] [Google Scholar]
- 31.Liu D, Sartor MA, Nader GA, Gutmann L, Treutelaar MK, Pistilli EE, Iglayreger HB, Burant CF, Hoffman EP, Gordon PM. Skeletal muscle gene expression in response to resistance exercise: sex specific regulation. BMC Genomics 2010;11:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of post-absorptive rats via a rapamycin-sensitive pathway. J Nutr 2000;130:2413–9. [DOI] [PubMed] [Google Scholar]
- 33.Kimball SR, Jefferson LS, Davis TA. Feeding stimulates protein synthesis in muscle and liver of neonatal pigs through an mTOR-dependent process. Am J Physiol Endocrinol Metab 2000;279:E1080–7. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson JM, Fry CS, Drummond MJ, Gundermann DM, Walker DK, Glynn EL, Timmerman KL, Dhanani S, Volpi E, Rasmussen BB. Mammalian target of rapamycin complex 1 activation is required for the stimulation of human skeletal muscle protein synthesis by essential amino acids. J Nutr 2011;141:856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bϵ mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem 2005;280:7570–80. [DOI] [PubMed] [Google Scholar]
- 36.Anthony JC, Reiter AK, Anthony TG, Crozier SJ, Lang CH, MacLean DA, Kimball SR, Jefferson LS. Orally administered leucine enhances protein synthesis in skeletal muscle of diabetic rats in the absence of increases in 4E-BP1 or S6K1 phosphorylation. Diabetes 2002;51:928–36. [DOI] [PubMed] [Google Scholar]
- 37.Anthony JC, Lang CH, Crozier SJ, Anthony TG, MacLean DA, Kimball SR, Jefferson LS. Contribution of insulin to the translational control of protein synthesis in skeletal muscle by leucine. Am J Physiol Endocrinol Metab 2002;282:E1092–101. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor PMJ, Kimball SR, Suryawan A, Bush JA, Nguyen HV, Jefferson LS, Davis TA. Regulation of translation initiation by insulin and amino acids in skeletal muscle of neonatal pigs. Am J Physiol Endocrinol Metab 2003;285:E40–53. [DOI] [PubMed] [Google Scholar]
- 39.Patti M-E, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest 1998;101:1519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter JN, Jefferson LS, Kimball SR. The ERK and Akt signaling pathways function through parallel mechanisms to promote mTORC1 signaling. Am J Physiol Cell Physiol 2011;300:C1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis MD, Baum JI, Kimball SR, Jefferson LS. Mechanisms involved in the coordinate regulation of mTORC1 by insulin and amino acids. J Biol Chem 2011;286:8287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farrell PA, Fedele MJ, Vary TC, Kimball SR, Jefferson LS. Effects of intensity of acute-resistance exercise on rates of protein synthesis in moderately diabetic rats. J Appl Physiol 1998;85:2291–7. [DOI] [PubMed] [Google Scholar]
- 43.Fedele MJ, Hernandez JM, Lang CH, Vary TC, Kimball SR, Jefferson LS, Farrell PA. Severe diabetes prohibits elevations in muscle protein synthesis after acute resistance exercise in rats. J Appl Physiol 2000;88:102–8. [DOI] [PubMed] [Google Scholar]
- 44.Breen L, Phillips SM. Interactions between exercise and nutrition to prevent muscle waste during ageing. Br J Clin Pharmacol 2013;75:708–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aspuria P-J, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal 2004;16:1105–12. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto Y, Jones KA, Mak BC, Muehlenbachs A, Yeung RS. Multicompartmental distribution of the tuberous sclerosis gene products, hamartin and tuberin. Arch Biochem Biophys 2002;404:210–7. [DOI] [PubMed] [Google Scholar]