Graphical abstract
Highlights
► Three genera of hematozoa were detected in northern pintails sampled in California. ► Fifteen unique parasite lineages were identified through genetic sequencing. ► Muscle tissue was useful for molecular detection of haemosporidian parasites. ► There was evidence for inter-annual differences in parasite infections in pintails.
Keywords: Hematozoa, Northern pintail, Anas acuta, California, Occupancy modeling
Abstract
Information on the molecular detection of hematozoa from different tissue types and multiple years would be useful to inform sample collection efforts and interpret results of meta-analyses or investigations spanning multiple seasons. In this study, we tested blood and muscle tissue collected from northern pintails (Anas acuta) during autumn and winter of different years to evaluate prevalence and genetic diversity of Leucocytozoon, Haemoproteus, and Plasmodium infections in this abundant waterfowl species of the Central Valley of California. We first compared results for paired blood and wing muscle samples to assess the utility of different tissue types for molecular investigations of haemosporidian parasites. Second, we explored inter-annual variability of hematozoa infection in Central Valley northern pintails and investigated possible effects of age, sex, and sub-region of sample collection on estimated parasite detection probability and prevalence. We found limited evidence for differences between tissue types in detection probability and prevalence of Leucocytozoon, Haemoproteus, and Plasmodium parasites, which supports the utility of both sample types for obtaining information on hematozoan infections. However, we detected 11 haemosporidian mtDNA cyt b haplotypes in blood samples vs. six in wing muscle tissue collected during the same sample year suggesting an advantage to using blood samples for investigations of genetic diversity. Estimated prevalence of Leucocytozoon parasites was greater during 2006–2007 as compared to 2011–2012 and four unique haemosporidian mtDNA cyt b haplotypes were detected in the former sample year but not in the latter. Seven of 15 mtDNA cyt b haplotypes detected in northern pintails had 100% identity with previously reported hematozoa lineages detected in waterfowl (Haemoproteus and Leucocytozoon) or other avian taxa (Plasmodium) providing support for lack of host specificity for some parasite lineages.
1. Introduction
The Central Valley of California provides habitat for an estimated 10–12 million waterfowl annually throughout autumn and winter making it one of the most important areas for migratory waterbirds in the Pacific Americas Flyway (Gilmer et al., 1982, Fleskes, 2012). Anthropogenic impacts to Central Valley wetlands, including urbanization, development of water projects, conversion of lands for agricultural use, and changes in farming practices, have affected the use of habitats by waterfowl in this area (Gilmer et al., 1982, Frayer et al., 1989, Elphick and Oring, 1998, Ackerman et al., 2006). The extent to which these environmental changes have impacted avian health and disease in the Central Valley are unclear. Research on parasitic infections in waterfowl sampled in this area would be of particular value as conversion of land for agricultural use, development of water control projects, and climatic changes may contribute to the emergence and/or proliferation of parasitic disease (Patz et al., 2000). Currently, information on blood parasites in migratory waterfowl sampled in this region is limited.
Northern pintail (Anas acuta) ducks are one of the most abundant avian species inhabiting the Central Valley of California during autumn and throughout winter. Approximately 50–60% of northern pintails wintering in North America and 90% of those in the Pacific Americas Flyway use the Central Valley of California (Bellrose, 1980). In a survey conducted in the 1940s, Herman (1951) found that 2.7% of northern pintails sampled in California (n = 263) were positive for haemosporidian infection based on microscopic examination of thin blood smears. These infections were classified as Leucocytozon simondi (n = 2) or Plasmodium spp. (n = 5). More recently, molecular methods for detection of avian blood parasites have been developed (Feldman et al., 1995, Hellgren et al., 2004) and the sensitivity of PCR-based methods to detect hematozoa infections from avian blood has been reported to be equal to or greater than microscopy (Richard et al., 2002, Valkiūnas et al., 2008). Additionally, molecular screening facilitates genetic characterization of parasites through sequence analysis of phylogenetically informative genes (e.g. cytochrome b). PCR-based methods also provide an opportunity to use non-traditional sample types (i.e. other than blood) that may be made available through biopsy sampling, beached bird surveys, necropsies, or hunter-harvest monitoring to obtain information on haemosporidian infections.
In this study, we tested blood and muscle tissue collected from northern pintails during autumn and winter of different years to evaluate prevalence and genetic diversity of Leucocytozoon, Haemoproteus, and Plasmodium infections in waterfowl from the Central Valley of California. We first compared results for paired blood and wing muscle samples to assess the utility of different tissue types for molecular investigations of haemosporidian parasites. Second, we explored inter-annual variability of hematozoa infections in Central Valley northern pintails and investigated possible effects of age, sex, and sub-region of sample collection on estimated parasite detection probability and prevalence. Results of this study will build upon limited existing data (Herman, 1951) for parasitic infections at an important migratory and wintering area for waterfowl in the Pacific Americas Flyway.
2. Materials and methods
2.1. Sample collection
Tissues were collected from hunter-harvested northern pintails in the Central Valley of California during two sample years: the fall and winter of 2006–2007 and 2011–2012. In 2006–2007, only wing muscle was collected (n = 146) while in 2011–2012, paired blood and wing muscle samples (n = 157) were collected from each individual. Gender and age was determined for each pintail in the field by plumage (Carney, 1992). The location from which northern pintails were harvested was recorded and subsequently assigned to one of two sub-regions according to the watershed of collection: the Sacramento Valley or the San Joaquin Valley (Fig. 1).
Fig. 1.
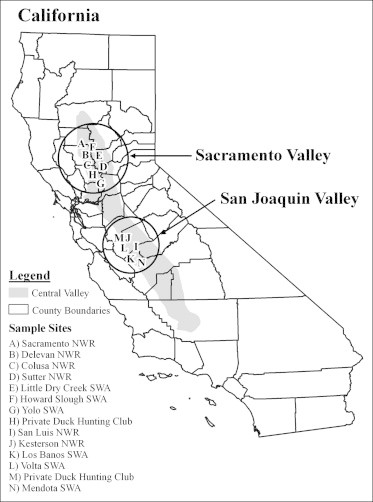
Locations in the Central Valley of California from which northern pintail tissue samples were collected. Samples (n in 2006–2007, n in 2011–2012) were collected in the Sacramento Valley sub-region at: (A) Sacramento National Wildlife Refuge (NWR; 44,92), (B) Delevan NWR (35,7), (C) Colusa NWR (0,2), (D) Sutter NWR (0,1), (E) Little Dry Creek State Wildlife Area (SWA; 0,5), (F) Howard Slough SWA (0,1), (G) Yolo SWA (0,8) and (H) a private duck hunting club (0,1). Samples were collected in the San Joaquin Valley sub-region at: (I) San Luis NWR (7,0), (J) Kesterson NWR (4,0), (K) Los Banos SWA (14,0), (L) Volta SWA (10,0), (M) a private duck hunting club (2,0), and (N) Mendota SWA (30,40).
2.2. Detecting hematozoa infection
DNA was extracted from all samples using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Extracted DNA was screened for presence of hematozoa using a nested PCR as described by Hellgren et al. (2004). A minimum of one negative control every 24 wells was incorporated into each set of PCR reactions. Reactions were conducted in eight well strip tubes with individual caps which remained closed, except while loading template and reagents, to prevent cross contamination. Amplicons were visualized on 0.8% agarose gels stained with Gel Red Nucleic Acid Gel Stain 10,000× in DMSO (Biotium, Hayward, CA). All samples were screened twice for hematozoa by nested PCR.
The target fragment of 479 base pairs (bp) cytochrome b (cyt b) mitochondrial DNA (mtDNA) was sequenced for all samples detected as positive for hematozoa infection to identify parasites by genera, provide information on haplotype diversity, and to prevent misidentification resulting from co-amplification (Cosgrove et al., 2006). PCR products were treated with ExoSap-IT (USB Inc., Cleveland, OH) prior to sequencing. Sequencing was performed with identical primers used for PCR and with BigDye Terminator version 3.1 mix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 3730xl automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequence data was cleaned and edited using Sequencher version 4.7 (Gene Codes Corp., Ann Arbor, MI). Infections were then assigned to three genera (Leucocytozoon, Haemoproteus, or Plasmodium) using the nucleotide BLAST function available through the National Center for Biotechnology Information (NCBI). Assignment was based on the top NCBI BLAST result with a minimum max identity score of 90%. It should be noted that Haemoproteus and Parahaemoproteus parasites were not differentiated in this study as these sub-genera have been demonstrated to be genetically indistinguishable (Ramey et al., 2012) using the molecular methods applied in the current study. Therefore, these two taxa are referred to collectively as Haemoproteus henceforth.
A sample was considered as positive if one or both tests resulted in a double-stranded, target product that was verified through genetic sequencing. Single stranded or otherwise ambiguous products were counted as negative. To confirm the competency of DNA in extracted samples, a 695 bp fragment of the mtDNA cytochrome oxidase I (COI) gene of northern pintail hosts was amplified using PCR protocols as described in Kerr et al. (2007). Infections were further classified as co-infected if the cleaned and sequenced, double-stranded target product contained three or more ambiguous nucleotides (Szymanski and Lovette, 2005). Co-infections were classified as infection by parasites of more than one lineage of (1) Leucocytozoon, (2) Haemoproteus/Plasmodium, or (3) Leucocytozoon and Haemoproteus/Plasmodium based on top NCBI BLAST results. Haemoproteus and Plasmodium were not differentiated for co-infections as both genera were amplified from a single molecular test and samples with large numbers of ambiguities could not be reliably assigned taxonomically. For initial comparisons to assess performance of molecular detection techniques by tissue type, we assumed that the detection probability was equal to one for the sum of all four PCR runs per bird sampled (i.e. two runs per tissue type). For subsequent analyses, we used an occupancy modeling approach to account for imperfect detection to estimate detection probability.
2.3. Assessing prevalence using occupancy modeling
A recent review of statistical modeling approaches for disease investigations highlighted the need to account for imperfect detection for a variety of ecologically important parameters (McClintock et al., 2010). Without explicitly addressing the uncertainty created by imperfect detection, estimates of the parameters of greatest interest may be biased. Therefore, occupancy modeling was used to account for imperfect detection in order to more accurately estimate prevalence (MacKenzie et al., 2006) of Leucocytozoon, Haemoproteus, and Plasmodium parasites. Molecular techniques may have failed to detect hematozoa infections in any given sample for a variety of reasons including strong infection by one parasite masking weak infection by another (Cosgrove et al., 2006), PCR inhibition, or degraded/low quantity template mtDNA. Simple models containing only the prevalence (ψ) and detection parameters (ρ) were used to contrast tissue types for Leucocytozoon, Haemoproteus, and Plasmodium parasites in the 2011–2012 sample collection. More complex models with additional covariates (age, sex, sub-region, year, and associated interactions) were used to explore potential differences in the detection probability and prevalence of haemosporidian parasites between 2006–2007 and 2011–2012 using wing muscle sample collections. Covariates were selected to investigate potential differences in prevalence on account of sex-mediated behavior or immune response (McCurdy et al., 1998), age related differences in vector exposure (Deviche et al., 2001) or recrudescence of infections obtained in prior seasons/years, disparate vector density due to geographic variation (Pagenkopp et al., 2008), and inter-annual differences in parasite transmission (Super and van Riper, 1995).
2.4. Phylogenetic assignment and assessment of haplotype diversity
Phylogenetic analysis was used to confirm assignment of sequence data for parasite infections using the NCBI BLAST function to three genera: Leucocytozoon, Haemoproteus, and Plasmodium. Only sequences resulting from positive samples as previously defined and without ambiguous bases were included in the phylogenetic analysis. A phylogeny was constructed by comparing hematozoa mtDNA cyt b sequence data from California northern pintail samples to the most frequently reported lineages of Leucocytozoon, Haemoproteus, and Plasmodium parasites for ‘America’ (Supplemental Table S1) as reported on the MalAvi public database (accessed 4 October 2012; Bensch et al., 2009). Sequences were aligned using Sequencher version 4.7 and edited to a final contig of 358 nucleotides. MEGA version 4.0.2 (Tamura et al., 2007) was used to create a phylogeny using the maximum composite likelihood model for nucleotide sequences with 10,000 bootstrap replicates to generate neighbor-joining trees.
Haplotype diversity of hematozoa mtDNA cyt b sequences was assessed by summarizing the frequency of unique haplotypes and creating a median-joining minimum spanning network (Bandelt et al., 1999) using Network version 4.6 (available at www.fluxus-engineering.com). Parasite haplotypes detected in northern pintails were also compared to 990 lineages available on the MalAvi public database (accessed 4 October 2012; Bensch et al., 2009) to identify sequences identical to those previously reported for hematozoa infecting wild birds.
3. Results
3.1. Comparison of tissue types for 2011–2012 sample collection
A 695 bp fragment from the mtDNA cytochrome oxidase I gene was successfully amplified from all blood and muscle tissue samples collected from northern pintails used in this study confirming the competency of DNA extractions. Totals of 37 and 38 samples collected in 2011–2012 were positive for haemosporidian infection using wing muscle or blood samples, respectively (Table 1), resulting in 95.5% agreement in detection between the two different tissue types (Table 2). However, the level of agreement was reduced to 82.9% when only positive samples were considered (Table 2). Among individual genera, similar percentages of Leucocytozoon positive samples were identified using blood (84.6%) and muscle tissue (89.7%) when detection was assumed to equal to one (Table 3). However, while 100% of Haemoproteus and Plasmodium positive samples were identified in blood samples when perfect detection was assumed, these parasites were only detected in 67% and 40%, respectively, of wing muscle tissues from birds identified as having an infection with parasites of these genera (Table 3).
Table 1.
Proportion of tissue samples from northern pintails wintering in the Central Valley of California detected as positive for haemosporidian infection using molecular techniques.
| 2006–2007 muscle | 2011–2012 muscle | 2011–2012 blood | |
|---|---|---|---|
| Proportion hematozoa + (%) | 72/146 (49.3%) | 37/157 (23.6%) | 38/157 (24.2%) |
| Proportion Leucocytozoon + (%) | 71/146 (48.6%) | 35/157 (22.3%) | 33/157 (21.0%) |
| Proportion Haemoproteus + (%) | 3/146 (2.1%) | 2/157 (1.3%) | 3/157 (1.9%) |
| Proportion Plasmodium + (%) | 6/146 (4.1%) | 2/157 (1.3%) | 5/157 (3.2%) |
| Proportion co-infected by > one parasite lineage | 25/146 (17.1%) | 8/157 (5.1%) | 10/157 (6.4%) |
| Proportion co-infected by > one Leucocytozoon lineage | 20/146 (13.7%) | 8/157 (5.1%) | 9/157 (5.7%) |
| Proportion co-infected by > one Haemoproteus/Plasmodium lineage | 1/146 (0.7%) | 0/157 (0%) | 0/157 (0%) |
| Proportion co-infected by ⩾ one Leucocytozoon and ⩾ one Haemoproteus/Plasmodiumlineage | 8/146 (5.5%) | 2/157 (1.3%) | 4/157 (2.5%) |
Table 2.
Agreement of molecular detection techniques as applied to double sampled northern pintails wintering in the Central Valley of California 2011–2012.
| All samples | Positive samples only | |
|---|---|---|
| Agreement hematozoa + (%) | 150/157 (95.5%) | 34/41 (82.9%) |
| Agreement Leucocytozoon + (%) | 147/157 (93.6%) | 29/39 (74.4%) |
| Agreement Haemoproteus + (%) | 156/157 (99.4%) | 2/3 (66.7%) |
| Agreement Plasmodium + (%) | 154/157 (98.1%) | 2/5 (40.0%) |
| Agreement co-infected (%) | 145/157 (92.4%) | 3/15 (20.0%) |
| Agreement Leucocytozoon co-infected (%) | 146/157 (93.0%) | 3/14 (21.4%) |
| Agreement Haemoproteus/Plasmodium co-infected (%) | 157/157 (100.0%) | None detected |
| Agreement Leucocytozoon & Haemoproteus/Plasmodium co-infected (%) | 155/157 (98.7%) | 2/4 (50.0%) |
Table 3.
Performance of molecular detection techniques by tissue type as applied to double sampled northern pintails wintering in the Central Valley of California 2011–2012.
| Muscle | Blood | |
|---|---|---|
| Proportion of hematozoa + detected (%) | 37/41 (90.2%) | 38/41 (92.7%) |
| Proportion of Leucocytozoon + detected (%) | 35/39 (89.7%) | 33/39 (84.6%) |
| Proportion of Haemoproteus + detected (%) | 2/3 (66.7%) | 3/3 (100.0%) |
| Proportion of Plasmodium + detected (%) | 2/5 (40.0%) | 5/5 (100.0%) |
| Proportion of co-infected detected (%) | 8/15 (53.3%) | 10/15 (66.7%) |
| Proportion of Leucocytozoon co-infected detected (%) | 8/14 (57.1%) | 9/14 (64.3%) |
| Proportion of Haemoproteus/Plasmodium co-infected detected (%) | None detected | None detected |
| Proportion of Leucocytozoon & Haemoproteus/Plasmodium co-infected detected (%) | 2/4 (50.0%) | 4/4 (100.0%) |
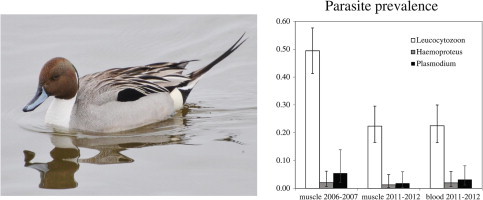
The detection probability for Leucocytoozon was similar for both muscle and blood samples collected from northern pintails in 2011–2012 using occupancy modeling as evidenced by overlapping 95% confidence intervals (Table 4). Detection probabilities for Haemoproteus and Plasmodium parasites were based on few (⩽5) detections (Table 3) and therefore provided limited inference as evidence by large or otherwise uninformative (i.e. undetermined based on ‘perfect’ detection of a small number of samples) confidence intervals (Table 4). Estimates for Leucocytoozon, Haemoproteus, and Plasmodium prevalence using occupancy modeling were equivalent between wing muscle and blood tissue collections based on overlapping confidence intervals (Fig. 2).
Table 4.
Estimates of detection probabilities using molecular techniques for tissue sample collections made from northern pintails wintering in the Central Valley of California through the application of an occupancy modeling approach.
| 2006–2007 muscle | 2011–2012 muscle | 2011–2012 blood | |
|---|---|---|---|
| Leucocytozoon | 0.873 | 0.939 | 0.807 |
| 95% lower confidence limit | 0.798 | 0.845 | 0.671 |
| 95% upper confidence limit | 0.923 | 0.978 | 0.896 |
| Haemoproteus | 1.000 | 1.000 | 0.800 |
| 95% lower confidence limit | 1.000 | 1.000 | 0.266 |
| 95% upper confidence limit | 1.000 | 1.000 | 0.978 |
| Plasmodium | 0.500 | 0.667 | 0.890 |
| 95% lower confidence limit | 0.155 | 0.111 | 0.479 |
| 95% upper confidence limit | 0.845 | 0.970 | 0.986 |
Fig. 2.
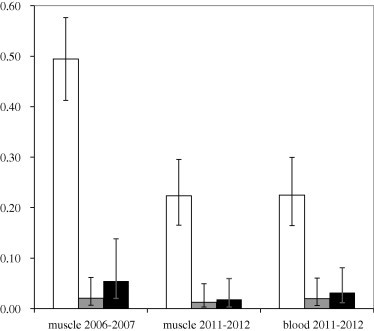
Estimated prevalence of Leucocytozoon (white bars), Haemoproteus (grey bars), and Plasmodium (black bars) parasites in northern pintails sampled in the Central Valley of California in 2006–2007 and 2011–2012 using occupancy modeling. Error bars represent 95% confidence intervals around point estimates.
3.2. Comparison of sample collections between years
Detection probabilities for Leucocytoozon, Haemoproteus, and Plasmodium parasites, as calculated using wing muscle samples, were similar between 2006–2007 and 2011–2012 (Table 4); however, estimates for the latter two parasite genera were limited by few detections (⩽6) per sample collection (Table 1). Leucocytoozon prevalence was estimated to be higher in northern pintails sampled in 2006–2007 (49.4%, 95% confidence interval 41.2–57.6%) than in 2011–2012 (22.4%, 95% confidence interval 16.5–29.6%; Fig. 2). Estimates for Haemoproteus and Plasmodium infection were lower (⩽5.4%) than estimated prevalence rates for Leucocytoozon infection and equivalent between sample years (Fig. 2). Model comparison for 2006–2007 and 2011–2012 wing muscle samples indicated no strong effect of covariates describing age, sex, or Central Valley sub-region on hematozoa prevalence (Table 5). Year effects were incorporated in top models predicting Leucocytoozon and Plasmodium infection, but not in the top model predicting Haemoproteus infection (Table 5).
Table 5.
Most competitive models describing haemosporidian prevalence from wing muscle samples collected from northern pintails in the 2006–2007 and 2011–2012 from the Central Valley of California where psi (ψ) is the estimate of prevalence and rho (ρ) is the estimate of detection probability from occupancy models.
| Model | AICc | ΔAICc | AICc Weights | Model Likelihood | Parameters | Deviance | Estimate | s.e. | Lower c.i. | Upper c.i. | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicting Leucocytozoon infection | |||||||||||
| {ψ Year, ρ hematozoa prevalence} | 505.37 | 0.00 | 0.44 | 1.00 | 3 | 67.85 | Parameters for top Leucocytozoon model | ||||
| {ψ Year + Sub-region, ρ hematozoa prevalence} | 507.27 | 1.90 | 0.17 | 0.39 | 4 | 67.70 | Detection probability | 0.896 | 0.023 | 0.841 | 0.933 |
| {ψ Year, Sub-region, ageSex-no interactions, ρ hematozoa prevalence} | 507.48 | 2.10 | 0.15 | 0.35 | 6 | 63.75 | Prevalence 2006–2007 | 0.492 | 0.042 | 0.410 | 0.573 |
| {ψ Age * Sex * Sub-region * Year, ρ hematozoa prevalence} | 507.87 | 2.50 | 0.13 | 0.29 | 17 | 40.28 | Prevalence 2011–2012 | 0.225 | 0.034 | 0.166 | 0.298 |
| {ψ Age * Sex * Sub-region * Year, ρ year} | 508.11 | 2.74 | 0.11 | 0.25 | 18 | 38.26 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year} | 524.40 | 19.02 | 0.00 | 0.00 | 32 | 21.13 | |||||
| {ψ Age, ρ hematozoa prevalence} | 524.83 | 19.46 | 0.00 | 0.00 | 3 | 87.31 | |||||
| {ψ Year + Age, ρ hematozoa prevalence} | 524.83 | 19.46 | 0.00 | 0.00 | 3 | 87.31 | |||||
| {ψ Sub-region, ρ hematozoa prevalence} | 526.82 | 21.44 | 0.00 | 0.00 | 3 | 89.29 | |||||
| {ψ Sex, ρ hematozoa prevalence} | 528.13 | 22.76 | 0.00 | 0.00 | 3 | 90.61 | |||||
| {ψ Year + Sex, ρ hematozoa prevalence} | 528.13 | 22.76 | 0.00 | 0.00 | 3 | 90.61 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year * time} | 545.96 | 40.59 | 0.00 | 0.00 | 48 | 0.00 | |||||
| Predicting Haemoproteus infection | |||||||||||
| {ψ, ρ hematozoa prevalence} | 52.97 | 0.00 | 0.27 | 1.00 | 1 | 9.02 | Parameters for top Haemoproteus model | ||||
| {ψ Sex, ρ hematozoa prevalence} | 53.96 | 0.99 | 0.16 | 0.61 | 2 | 7.98 | Detection probability | 1.000 | 0.000 | 1.000 | 1.000 |
| {ψ Sub-region, ρ hematozoa prevalence} | 54.43 | 1.46 | 0.13 | 0.48 | 2 | 8.45 | Prevalence (years combined) | 0.017 | 0.007 | 0.007 | 0.039 |
| {ψ Year, ρ hematozoa prevalence} | 54.71 | 1.74 | 0.11 | 0.42 | 2 | 8.74 | |||||
| {ψ Age, ρ hematozoa prevalence} | 54.98 | 2.00 | 0.10 | 0.37 | 2 | 9.00 | |||||
| {ψ Sex + Sub-region, ρ hematozoa prevalence} | 55.32 | 2.35 | 0.08 | 0.31 | 3 | 7.30 | |||||
| {ψ Sex + Year, ρ hematozoa prevalence} | 55.58 | 2.60 | 0.07 | 0.27 | 3 | 7.56 | |||||
| {ψ Sex + Age, ρ hematozoa prevalence} | 55.95 | 2.98 | 0.06 | 0.23 | 3 | 7.93 | |||||
| {ψ Year, Sub-region, AgeSex-no interactions, ρ hematozoa prevalence} | 58.64 | 5.67 | 0.02 | 0.06 | 5 | 6.50 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ hematozoa prevalence} | 78.08 | 25.11 | 0.00 | 0.00 | 17 | 0.00 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ year} | 80.35 | 27.37 | 0.00 | 0.00 | 18 | 0.00 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year} | 87.23 | 34.25 | 0.00 | 0.00 | 21 | 0.00 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year * time} | 99.02 | 46.05 | 0.00 | 0.00 | 26 | 0.00 | |||||
| Predicting Plasmodium infection | |||||||||||
| {ψ Year, ρ hematozoa prevalence} | 95.08 | 0.00 | 0.25 | 1.00 | 3 | 26.94 | Parameters for top Plasmodium model | ||||
| {ψ, ρ hematozoa prevalence} | 95.49 | 0.42 | 0.21 | 0.81 | 2 | 29.40 | Detection probability | 0.545 | 0.181 | 0.223 | 0.834 |
| {ψ Year + Sex, ρ hematozoa prevalence} | 97.02 | 1.95 | 0.10 | 0.38 | 4 | 26.83 | Prevalence 2006–2007 | 0.052 | 0.023 | 0.021 | 0.122 |
| {ψ Year + Age, ρ hematozoa prevalence} | 97.02 | 1.95 | 0.10 | 0.38 | 4 | 26.83 | Prevalence 2011–2012 | 0.016 | 0.012 | 0.004 | 0.066 |
| {ψ Year + Area, ρ hematozoa prevalence} | 97.09 | 2.02 | 0.09 | 0.36 | 4 | 26.90 | |||||
| {ψ Sex, ρ hematozoa prevalence} | 97.28 | 2.20 | 0.08 | 0.33 | 3 | 29.14 | |||||
| {ψ Age, ρ hematozoa prevalence} | 97.38 | 2.30 | 0.08 | 0.32 | 3 | 29.24 | |||||
| {ψ Sub-region, ρ hematozoa prevalence} | 97.52 | 2.44 | 0.08 | 0.30 | 3 | 29.38 | |||||
| {ψ Year, Sub-region, AgeSex-no interactions, ρ hematozoa prevalence} | 101.03 | 5.95 | 0.01 | 0.05 | 6 | 26.69 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ hematozoa prevalence} | 112.95 | 17.87 | 0.00 | 0.00 | 17 | 14.74 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Year} | 115.03 | 19.96 | 0.00 | 0.00 | 18 | 14.57 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year} | 119.09 | 24.01 | 0.00 | 0.00 | 23 | 7.08 | |||||
| {ψ Age * Sex * Sub-region * Year, ρ Age * Sex * Sub-region * Year * time} | 123.98 | 28.91 | 0.00 | 0.00 | 28 | 0.00 | |||||
3.3. Genetic analysis of parasite mtDNA cyt b haplotypes
Fifteen unique haplotypes (Genbank accession numbers: KC409119–KC409133) were observed among 113 genetic sequences for hematozoa mtDNA cyt b derived from Central Valley northern pintail samples. Phylogenetic analysis confirmed correct assignment of all haplotypes to genera using the NCBI BLAST function, although bootstrap support for clades by genera was relatively low (Fig. 3). Ten haplotypes were detected from 2006–2007 wing muscle samples, as compared to six from 2011–2012 wing muscle samples, and 11 from 2011–2012 blood samples (Fig. 4). Four unique haplotypes were detected each from 2006–2007 wing muscle samples and 2011–2012 blood samples (Fig. 4).
Fig. 3.
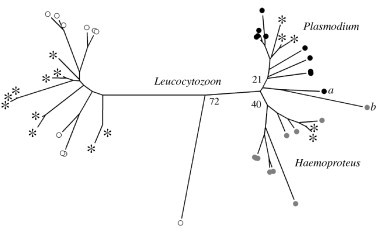
Phylogenetic assignment of hematozoa mitochondrial DNA cytochrome b sequences originating from northern pintails collected from the Central Valley of California (asterisks). Reference sequences for Leucocytozoon (white circles), Haemoproteus (grey circles), and Plasmodium (black circles) parasites were obtained from the National Center for Biotechnology Information. Bootstrap support values for differentiation of broad taxonomic groups are indicated. Values reported for Haemoproteus and Plasmodium show support for phylogenetic differentiation from the mixed subclade formed by reference sequences a and b.
Fig. 4.
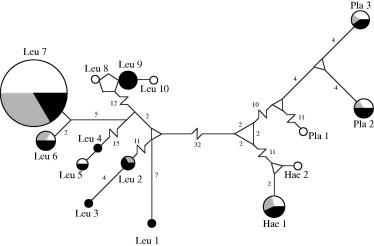
Minimum spanning network for hematozoa mitochondrial DNA cytochrome b haplotypes detected in Central Valley northern pintails. Circles are drawn proportional to the frequency at which haplotypes were observed. Shading represents the sample collection from which haplotypes originated: white (2006–2007 wing muscle), grey (2011–2012 wing muscle), and black (2011–2012 blood). A single mutation separates nodes unless explicitly indicated by number. Lines separating nodes are drawn to scale unless indicated by a break. Parasite taxa have been abbreviated in haplotype names (Leu = Leucocytozoon, Hae = Haemoproteus and Pla = Plasmodium).
Seven of 15 mtDNA cyt b haemosporidian haplotypes detected in northern pintails had 100% identity values when compared to homologous 358 bp sequence of lineages available on the MalAvi database (Bensch et al., 2009). Two Leucocytozoon and Haemoproteus haplotypes detected in northern pintails were identical to lineages reported in other species of waterfowl (Table 6). All three Plasmodium haplotypes detected in northern pintails had 100% identity values when compared to previously reported malarial lineages originating from a wide range of avian hosts (Table 6).
Table 6.
Mitochondrial DNA cytochrome b haplotypes identified from samples collected from northern pintails wintering in the Central Valley of California with 100% identity to previously reported haemosporidian lineages.
| Previously reported avian hosts | Genbank accession numbers | |
|---|---|---|
| Haemoproteus haplotype 1 | Anas platyrhynchos, Cygnus columbianus | AF465591, JQ314226 |
| Haemoproteus haplotype 2 | Anas crecca, Cygnus columbianus | JQ314225 |
| Leucocytozoon haplotype 6 | Cygnus columbianus | JQ314224 |
| Leucocytozoon haplotype 7 | Cygnus columbianus | JQ314222, JQ314223 |
| Plasmodium haplotype 1 | Lamprotornis iris, Strix varia | AY172846, EU627845 |
| Plasmodium haplotype 2 | Accipiter nisus, Accipiter striatus, Aythya marila, Buteo buteo, Buteo lagopus, Calidris melanotos, Carpodacus erythrinus, Cygnus columbianus, Geothlypis trichas, Luscinia svecica, Parus caeruleus, Parus major, Phoenicurus phoenicurus, Pluvialis fulva, Tarsiger cyanurus, Turdus pallidus | AF254978, AY393793, DQ659583, DQ839065, DQ991069, EF380128, EU254539, EU321175, JQ314228 |
| Plasmodium haplotype 3 | Acrocephalus arundinaceus, Acrocephalus schoenobaenus, Calidris melanotos | AF495574 |
4. Discussion
In this study, we assessed the utility of two tissue types collected from northern pintails wintering in the Central Valley of California for the detection of haemosporidian infections. We found limited evidence for differences in detection probability and prevalence of Leucocytozoon, Haemoproteus, and Plasmodium parasites between tissue types, which supports the utility of using tissue other than blood (i.e. muscle) to obtain information on hematozoan infections. However, the molecular detection of 11 haemosporidian mtDNA cyt b haplotypes in blood samples as compared to six from wing muscle tissue collected during the same sample year may indicate an advantage to using blood samples for investigations of genetic diversity. The prevalence of Leucocytozoon parasites was estimated to be more than two times higher during 2006–2007 as compared to 2011–2012, supporting inter-annual differences in parasitic infections in a wintering population of migratory waterfowl. Seven of 15 mtDNA cyt b haplotypes detected in northern pintails had 100% identity with previously reported haemosporidian lineages detected in waterfowl (Haemoproteus and Leucocytozoon) or other avian taxa (Plasmodium) providing evidence that some lineages reported in this study may have been derived from parasites that are not host specific.
The comparable estimates of detection probability for wing muscle and blood indicate that muscle may be equally suitable for identifying Leucocytozoon parasites with molecular techniques. However, the small number of Haemoproteus and Plasmodium positive samples in 2011–2012 precludes clear inference regarding the utility of this tissue for detection of parasites of these two genera. Future work to assess the detection probability of Haemoproteus and Plasmodium from muscle tissue would be useful as differences in detection sensitivity between tissue types could lead to disparate estimates of infection rates in avian populations where prevalence of these parasite genera may be higher. The molecular detection of haemosporidian infections from wing muscle suggests that hunter harvested birds may be a useful resource for obtaining samples; however, consideration should be given to potential biases that may result on account of sampling methodology besides those related to detection probability. For example, hunter-harvested mallards banded in the Mississippi Alluvial Valley were found to be, on average, in poorer condition than non-recovered birds (Hepp et al., 1986). Thus, there could be biases associated with prevalence rates determined using hunter-harvested samples if parasite infection is correlated with body condition in target species.
The only significant inter-annual difference was observed in prevalence of Leucocytozoon parasites, which differed between the two years. Indeed, the most competitive occupancy model describing Leucocytozoon infection for northern pintails from the 2006–2007 and 2011–2012 sample collections included year as a covariate. Differences in Leucocytozoon infection between years may be related to the effect of variable environmental conditions such as temperature, precipitation, and average wind speed on the abundance of blackfly vectors or the ability of vectors to access hosts (Ramey et al., 2012).
Prevalence of haemosporidian infections in northern pintails sampled in California for the current study are comparable to other reports for this species sampled at locations in Canada and analyzed using microscopy (Bennett et al., 1975, Bennett et al., 1982, Williams et al., 1977). However, infection rates are much higher than previously reported for northern pintails sampled in California (Herman, 1951). It remains unclear if these differences in prevalence are due to disparities in detection probability, inter-annul variation, or changes in parasite abundance at transmission areas between sampling periods.
Four unique Leucocytozoon, Haemoproteus, and Plasmodium mtDNA cyt b haplotypes detected in 2006–2007 were not identified in 2011–2012 wing muscle sample collection. Two of these haplotypes (Haemoproteus haplotype 2 and Plasmodium haplotype 1) have been detected previously in wild birds; however two Leucocytozoon lineages (Leucocytozoon haplotypes 8 and 10) had not yet been reported. Haemoproteus haplotype 2 was identified in American green-winged teal (Anas crecca) and tundra swans (Cygnus columbianus) indicating potential transmission of this parasite lineage among sympatric waterfowl. Plasmodium haplotype 1 was previously found in a captive emerald starling (Lamprotornis iris) and barred owl (Strix varia) both sampled in California which is consistent with the previous report of a wide host range for parasites of this genus (Szymanski and Lovette, 2005). Future work could build upon results of this study through the addition of paired thin blood films for microscopial examination with tissue samples for molecular analyses. Thin blood films would allow for verification of the completion of within-host life cycles via the detection of gametocytes in addition to identification of morphospecies from which parasite lineages originated.
Acknowledgments
We thank J. Kohl, D. Skalos, G. Yarris, N. Smith, C. Tierney, and P. Carter of the US Geological Survey (USGS); J. Beam, B. Cook, N. Overton, S. Allen, S. Miyamoto, S. Brueggeman, E. King, G. Gerstenberg, and A. Atkinson of California Department of Fish and Wildlife; R. Wright , J. Laughlin, and D. Loughman of California Waterfowl; C. Hildebrandt of Ducks Unlimited; and numerous other California Wildlife Area and National Wildlife Refuge staff for collecting samples or facilitating sample collection from hunter-shot birds. We are grateful to J. Pearce, T. DeGange, P. Bright, K. Briggs, and S. Gross of USGS and R. Shaffer of the US Fish and Wildlife Service for financial and administrative support. We appreciate reviews provide by J. Pearce, C. Van Hemert (USGS), and two anonymous reviewers. None of the authors have any financial interests or conflict of interest with this article. Any use of trade names is for descriptive purposes only and does not imply endorsement by the US Government.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijppaw.2013.02.001.
Appendix A. Supplementary data
References
- Ackerman J.T., Takekawa J.Y., Orthmeyer D.L., Fleskes J.P., Yee J.L., Kruse K.L. Spatial use by wintering greater white-fronted geese relative to a decade of habitat change in California’s Central Valley. J. Wildl. Manag. 2006;70:965–976. [Google Scholar]
- Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bellrose F.C. Stackpole Books; Harrisburg: 1980. Ducks, Geese, and Swans of North America. [Google Scholar]
- Bennett G.F., Smith A.D., Whitman W., Cameron M. Hematozoa of the Anatidae of the Atlantic flyway II. The maritime provinces of Canada. J. Wildl. Dis. 1975;11:280–289. doi: 10.7589/0090-3558-11.2.280. [DOI] [PubMed] [Google Scholar]
- Bennett G.F., Nieman D.J., Turner B., Kuyt E., Whiteway M., Greiner E.C. Blood parasites of prairie anatids and their implication in waterfowl management in Alberta and Saskatchewan. J. Wildl. Dis. 1982;18:287–296. doi: 10.7589/0090-3558-18.3.287. [DOI] [PubMed] [Google Scholar]
- Bensch S., Hellgren O., Pérez-Tris J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Res. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- Carney S.M. US Department of the Interior; Washington, DC: 1992. Species, Age, and Sex Identification of Ducks using wing Plumage. [Google Scholar]
- Cosgrove C.L., Day K.P., Sheldon B.C. Coamplification of Leucocytozoon by PCR diagnostic tests for avian malaria: a cautionary note. J. Parasitol. 2006;92:1362–1365. doi: 10.1645/GE-879R.1. [DOI] [PubMed] [Google Scholar]
- Deviche P., Greiner E.C., Manteca X. Seasonal and age-related changes in blood parasite prevalence in dark-eyed juncos (Junco hyemalis, Aves, Passeriformes) J. Exp. Zool. 2001;289:456–466. doi: 10.1002/jez.1027. [DOI] [PubMed] [Google Scholar]
- Elphick C.S., Oring L.W. Winter management of Californian rice fields for waterbirds. J. Appl. Ecol. 1998;35:95–108. [Google Scholar]
- Feldman R.A., Freed L.A., Cann R.L. A PCR test for avian malaria in Hawaiian birds. Mol. Ecol. 1995;4:663–674. doi: 10.1111/j.1365-294x.1995.tb00267.x. [DOI] [PubMed] [Google Scholar]
- Fleskes J.P. Wetlands of the Central Valley of California and Klamath Basin. In: Batzer D., Baldwin A., editors. Wetland Habitats of North America: Ecology and Conservation Concerns. University of California Press; Berkeley: 2012. pp. 357–370. [Google Scholar]
- Frayer W.E., Peters D.D., Pywell H.R. US Fish and Wildlife Service; Washington, DC: 1989. Wetlands of the California Central Valley: Status and Trends 1939 to Mid-1980s. [Google Scholar]
- Gilmer D.S., Miller M.R., Bauer R.D., LeDonne J.R. California’s Central Valley wintering waterfowl: concerns and challenges. Trans North American Wildl Nat Resources Conf. 1982;47:441–452. [Google Scholar]
- Hellgren O., Waldenström J., Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J. Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Hepp G.R., Blohm R.J., Reynolds R.E., Hines J.E., Nichols J.D. Physiological condition of autumn-banded mallards and its relationship to hunting vulnerability. J. Wildl. Manag. 1986;50:177–183. [Google Scholar]
- Herman C.M. Blood parasites of California ducks and geese. J. Parasitol. 1951;37:280–282. [PubMed] [Google Scholar]
- Kerr K.C.R., Stoeckle M.Y., Dove C.J., Weigt L.A., Francis C.M., Hebert P.D.N. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. 2007;7:535–543. doi: 10.1111/j.1471-8286.2007.01670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie D.I., Nichols J.D., Royle J.A., Pollock K.H., Bailey L.L., Hines J.E. Academic Press; Burlington: 2006. Occupancy Estimation and Modeling: Inferring Patterns and Dynamics of Species Occurrence. [Google Scholar]
- McClintock B.T., Nichols J.D., Bailey L.L., MacKenzie D.I., Kendall W.L., Franklin A.B. Seeking a second opinion: uncertainty in disease ecology. Ecol. Lett. 2010;13:659–674. doi: 10.1111/j.1461-0248.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- McCurdy D.G., Shutler D., Mullie A., Forbes M.R. Sex-biased parasitism of avian hosts: relations to blood parasite taxon and mating system. Oikos. 1998;82:303–312. [Google Scholar]
- Pagenkopp K.M., Klicka J., Durrant K.L., Garvin J.C., Fleischer R.C. Geographic variation in malarial parasite lineages in the common yellowthroat (Geothlypis trichas) Conserv. Genet. 2008;9:1577–1588. [Google Scholar]
- Patz J.A., Graczyk T.K., Geller N., Vittor A.Y. Effects of environmental change on emerging parasitic diseases. Int. J. Parasitol. 2000:1395–1405. doi: 10.1016/s0020-7519(00)00141-7. [DOI] [PubMed] [Google Scholar]
- Szymanski M.M., Lovette I.J. High lineage diversity and host sharing of malarial parasites in a local avian assemblage. J. Parasitol. 2005;91:768–774. doi: 10.1645/GE-417R1.1. [DOI] [PubMed] [Google Scholar]
- Ramey A.M., Ely C.R., Schmutz J.A., Pearce J.M., Heard D.J. Molecular detection of hematozoa infections in tundra swans relative to migration patterns and ecological conditions at breeding grounds. PLoS One. 2012;7:e45789. doi: 10.1371/journal.pone.0045789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard F.A., Sehgal R.N.M., Jones H.I., Smith T.B. A comparative analysis of PCR-based detection methods for avian malaria. J. Parasitol. 2002;88:819–822. doi: 10.1645/0022-3395(2002)088[0819:ACAOPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Super P.E., van Riper C. A comparison of avian hematozoan epizootiology in two California coastal scrub communities. J. Wildl. Dis. 1995;31:447–461. doi: 10.7589/0090-3558-31.4.447. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G., Iezhova T.A., Križanauskienė A., Palinauskas V., Sehgal R.N.M., Bensch S. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J. Parasitol. 2008;94:1395–1401. doi: 10.1645/GE-1570.1. [DOI] [PubMed] [Google Scholar]
- Williams N.A., Calverley B.K., Mahrt J.L. Blood parasites of mallard and pintail ducks from central Alberta and the MacKenzie Delta, Northwest Territories. J. Wildl. Dis. 1977;13:226–229. doi: 10.7589/0090-3558-13.3.226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


