Abstract
Background
Dengue is an emerging public health problem causing serious morbidity and mortality in tropical developing countries. Early, sensitive and specific diagnosis is paramount for clinical decision making. Currently available diagnostic tests are limited in scope and utility. This study highlights applicability of RT-LAMP in dengue diagnosis.
Methods
100 dengue confirmed cases, 100 dengue negative cases and 79 healthy negative controls from dengue epidemic between Sep 2009 to Jul 2011 were included. Dengue cases were profiled using WHO guidelines 2006, haematological and biochemical parameters evaluated and diagnosed using NS1 antigen, IgM and IgG enzyme immunoassay, RT-PCR and RT-LAMP. Positive cases were serotyped, genotyped and various tests were compared.
Results
Mean haematocrit, PT, PTT, platelet count, activated lymphocytes, serum fibrinogen, transaminases, bilirubin, lactate dehydrogenase, protein and sodium were significantly elevated in DHF/DSS as compared to DF. NS1 antigen, RT-PCR and RT-LAMP were sensitive during 1–3 days while μ-capture IgM EIA was specific after 5–7 days of initial infection. DEN-1 genotype III was predominant.
Conclusion
Deranged haematocrit and liver function tests are indicators of the severity of the disease. RT-LAMP is rapid, cost effective, highly sensitive and specific qualitative and quantitative technique which can detect dengue infection in both early and intermediary stages when NS1 antigen titres are not in the detectable range and the IgM antibody titres have just started to rise. Its superiority over existing techniques, amenability for automation and promising utility in low resource healthcare setups and field conditions raise it as the new gold standard for dengue diagnosis.
Keywords: Dengue, Reverse transcriptase polymerase chain reaction (RT-PCR), Reverse transcription loop-mediated isothermal amplification (RT-LAMP), Gold standard
Introduction
Dengue is a leading public health problem threatening one third of the world population residing in tropical developing countries. Four closely related, but antigenically different serotypes (DEN 1–4) produce either an asymptomatic infection or a moderate febrile illness such as dengue fever (DF) or serious fatal forms, dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS).1
Many outbreaks and epidemics of DF or DHF have been reported during the past few decades. World health organization reported a total of 10,000 laboratory confirmed cases of dengue infection by multiple serotypes in the epidemic of 1996 and 2185 cases in 2003 outbreak.2–4
Dengue exhibits both hyperendemicity and epidemicity. Multiple sequential infections with different serotypes are possible as primary dengue infection induces serotype specific immunity. Seroepidemiological studies have shown that secondary infection is a major risk factor for DHF and DSS through antibody-dependent enhancement.5 Epidemics of DHF/DSS are fret with serious morbidity and mortality and rapidly strain the developing nation health system. In the absence of licensed vaccines or specific antiviral therapy, patient management relies entirely on good supportive care.
Early, sensitive and specific diagnosis is paramount for patient management, prevention of complications, etiologic investigation and disease control. Early diagnosis within 5 days of infection is achieved by NS1 antigen detection, virus isolation and nucleic acid amplification. NS1 antigen detection is a lateral flow chromatographic technique but is unable to identify the serotype. Nucleic acid amplification techniques such as reverse transcriptase polymerase chain reaction (RT-PCR), real-time PCR, multiplex PCR (M-PCR) and nucleic acid sequence based amplification (NASBA) can identify serotypes but are limited by requirement of high-precision instrument for amplification and complicated method for detection of amplified products.6–9 Virus isolation needs sophisticated labs and cannot be used widely. Diagnosis after 5 days is conferred by IgM and IgG based serological techniques such as haemagglutination inhibition, complement fixation, neutralization tests and enzyme immunoassay (EIA). IgM μ-capture enzyme immunoassay is simple and used widely. IgG enzyme immunoassay is nonspecific as it exhibits cross-reactivity among flaviviruses. Both IgM and IgG based immunoassays cannot identify the serotype.6,7
With mankind facing a huge burden of a lethal disease such as dengue, the development of better diagnostic tests is necessitated. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a novel gene amplification technique which has been proven to be highly sensitive, specific and efficient.10 After its introduction by Notomi et al in 2000,10 its applicability was limited by lack of continuous evaluation.
The aim of this study was to develop and evaluate RT-LAMP technique for detection and differentiation of dengue virus serotypes. Dengue was diagnosed using NS1 antigen, IgM and IgG EIA, RT-PCR and RT-LAMP and various tests were compared. The feasibility of the RT-LAMP technology and applicability for clinical diagnosis of dengue virus infection are discussed.
Materials and methods
A total of 279 patients including 100 dengue confirmed cases, 100 dengue negative cases and 79 healthy negative controls between Sep 2009 to Jul 2011 from New Delhi and adjoining areas were included in the study after approval of hospital ethical committee. Cases of all age groups were selected employing the Case Definition Criteria for Dengue Fever as per WHO guidelines 2006. Samples were transported to the laboratory within 6 h of collection and stored at −70 °C until processed. Haematological parameters were performed by coulter machine followed by confirmation under microscope and coagulation profile was done. Liver function, renal function, serum proteins and electrolytes were also assessed.
Samples were subsequently analysed for presence of NS1 antigen, IgM and IgG antibodies and viral detection using RT-PCR, M-PCR and RT-LAMP. NS1 antigen was done by lateral flow rapid test kits. IgM and IgG antibodies were detected by μ-capture enzyme immunoassay kit from National Institute of Virology, Pune, India. IgM values >1.1 NIV units and IgG values >2.2 NIV units were indicative of either an active primary or secondary dengue infection. Nucleic acid amplification was preceded by RNA extraction using the QIAamp viral RNA mini kit (Qiagen, Germany) following the manufacturer's protocol. RT-PCR was done as per the process described by Lanciotti et al with certain modifications.11 cDNA was synthesized from viral RNA using reverse primer (D2) targeting the prM gene, reverse transcriptase of Moloney murine leukaemia virus origin (MMLV-RT) and RNasin® Ribonuclease inhibitor (Promega, USA) following manufacturer's protocol. RT-PCR was carried out using the cDNA as template, forward primer (D1) targeting the capsid gene and thermostable Taq DNA polymerase (Promega, USA) loaded to the thermal cycler (Bio-Rad, USA) following manufacturer's protocol. PCR amplicon was detected by 1% or 2% agarose gel electrophoresis described by Sambrook et al, 1989 followed by visualization in a Gel Documentation System (Bio-Rad, USA) and photographed (Fig. 1). The reference strains were used as positive controls and DEPC treated water as negative control. M-PCR was carried out using the cDNA as template, forward primer (D1), DEN-1–DEN-4 specific reverse primers (TS1–TS4) and thermostable Taq DNA polymerase (Promega, USA) loaded to the thermal cycler (Bio-Rad, USA) following manufacturer's protocol (Fig. 2).
Fig. 1.
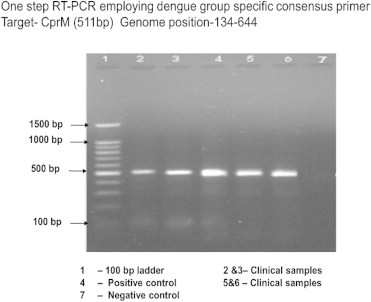
RT-PCR for dengue.
Fig. 2.
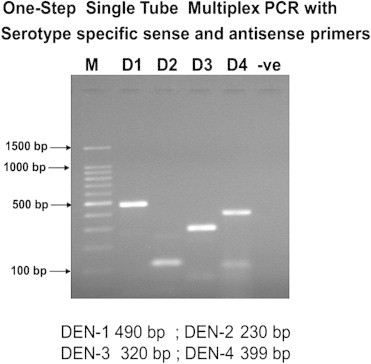
Multiplex PCR for dengue.
RT-LAMP comprised of primer designing, amplification, detection, interpretation and real-time monitoring. A set of six primers comprising two outer, two inner and two loop primers recognizing eight distinct regions on the target sequence were designed employing the LAMP primer designing support software program (Net laboratory, Japan, http://venus.netlaboratory.com). Forward outer primer (F3), backward outer primer (B3), forward inner primer and backward inner primer, and forward loop primer (FLP) and backward loop primer (BLP) were demarcated. Forward inner primer consists of a complementary sequence of F1 and a sense sequence of F2. BIP consists of a complementary sequence of B1 and a sense sequence of B2. Forward inner primer and backward inner primer were high performance liquid chromatography (HPLC) purified primers. The FLP and BLP primers were composed of the sequences that are complementary to the sequence between F1&F2 and B1&B2 regions respectively. The details of the each primer with regards to their positions in the genomic sequences are shown in Table 1 and Fig. 3.
Table 1.
Dengue primer sets for RT-LAMP.
| Name of the primer | Genome position (3' UTR) |
|||
|---|---|---|---|---|
| DEN-1 (199 bp) (10,469–10,667) | DEN-2 (211 bp) (1049–10,659) | DEN-3 (218 bp) (10,289–10,506) | DEN-4 (229 bp) (10,289–10,517) | |
| Forward outer (F3) | 10,469–10,487 | 10,449–10,466 | 10,289–10,307 | 10,289–10,306 |
| Backward outer (B3) | 10,667–10,648 | 10,659–10,642 | 10,506–10,490 | 10,517–10,500 |
| Forward internal primer (FIP) [F1c + TTTT + F2] | F1c 10,552–10,531 | 10,530–10,511 | 10,376–10,355 | 10,375–10,353 |
| F2 10,489–10,506 | 10,470–10,489 | 10,310–10,328 | 10,312–10,329 | |
| Backward Internal primer (BIP) [B1 + TTTT + B2c] | B1 10,557–10,576 | 10,568–10,588 | 10,399–10,418 | 10,424–10,443 |
| B2c 10,629–10,612 | 10,640–10,621 | 10,475–10,457 | 10,491–10,474 | |
| Forward loop primer (FLP) | 10,525–10,508 | 10,509–10,493 | 10,354–10,331 | 10,349–10,333 |
| Backward loop primer (BLP) | 10,583–10,600 | 10,600–10,616 | 10,430–10,446 | 10,445–10,461 |
Fig. 3.
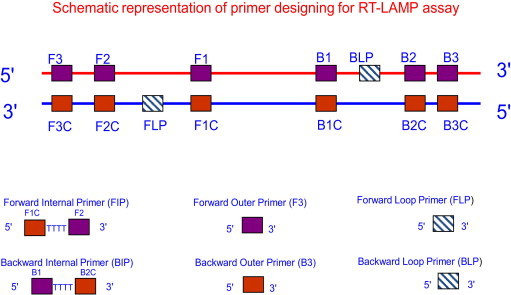
Designing of primers for RT-LAMP.
The RT-LAMP reaction was carried out in a total 25 μl reaction mixture using the Loopamp DNA amplification kit containing 50 pmol each of the primers Forward inner primer and backward inner primer, 5 pmol each of the outer primers F3 and B3, 25 pmol each of loop primers FLP and BLP, 1.4 mM dNTPs, 0.8 M betaine, 0.1% Tween20, 10 mM (NH4)2SO4, 8 mM MgSO4, 10 mM KCl, 20 mM Tris–HCl pH 8.8, 16 units of the Bst DNA polymerase (New England Biolabs), 0.25 units of the AMV Reverse Transcriptase (Invitrogen, USA), and the specified amounts of target RNA were incubated at 63 °C for 60 min in heating block. The analysis of each sample was performed in a set of four tubes each having the primer mix of a particular serotype. Positive and negative controls were included in each run, and all precautions to prevent cross-contamination were observed.
The monitoring of RT-LAMP amplification was carried out both by sophisticated techniques such as real-time spectrophotometry, SYBR Green ultraviolet (UV) transillumination and agarose gel electrophoresis, and field utility techniques such as direct naked eye visualization and SYBR Green colorimetry under natural light or hand held UV torch. Real-time spectrophotometric analysis was done by recording optical density (OD) at 400 nm every 6 s with the help of Loopamp real-time turbidimeter (support from Defence Research and Development Establishment, Gwalior, India). The cut-off value for positivity was determined by checking the time of positivity (Tp) in minutes at which the turbidity increases above the threshold value fixed at 0.1 that is two times more than average turbidity value of the negative controls of several replicates. None of the positive samples tested over multiple times showed positivity in terms of increased turbidity after 30 min. Therefore, a sample having Tp values ≤30 min and turbidity above the threshold value ≥0.1 was considered as positive. SYBR Green monitoring was done by adding 1 μl of SYBR Green I intercalating dye to the reaction tube, checking colour change from yellow/orange to green and bright appearance under UV transillumination as positive result and no colour change as negative result. Agarose gel electrophoresis was done with 10 μl aliquot of RT-LAMP products electrophoresed on 3% NuSieve 3:1 agarose gel (BMA, Rockland, ME, USA) in Tris-borate buffer followed by staining with ethidium bromide and visualization in UV transilluminator at 302 nm. To facilitate filed application of RT-LAMP, naked eye visualization for white turbidity after a pulse spun precipitate deposit and SYBR Green colorimetry by checking orange to green colour change under natural light or under hand held UV light (302 nm) were done. The 85 RT-LAMP positive samples were serotyped by serotype specific primers in a multiplex PCR format and few samples were genotyped by both primer targeted amplification and sequencing of C-prm gene junction.
Descriptive statistics and percentages were worked out. Data was compared using Kappa statistical analysis which included Cohen's un-weighted Kappa, Kappa with lineage weighting, Kappa with quadratic weighting and frequencies and proportion of agreement. Strength of agreement between 0.8 and 1 was taken as perfect, 0.6–0.8 as substantial and 0.4–0.6 as moderate.
Results
Dengue confirmed cases by positive dengue serology or presence of nucleic acid were further classified as DF, DHF and DSS which were 61%, 30% and 9% cases respectively as per WHO case definition.6 92% of cases were seen in the post-monsoon season from September to December in 2009–10. 89% patients were 14 yrs or older. Male:female ratio was 1.7:1.
Mean haemoglobin in DF, DHF and DSS cases was 12.79 g/dl, 12.16 g/dl and 10.35 g/dl respectively. 9.2% DHF patients had severe thrombocytopaenia (less than 20,000/μL) of which four had evidence of spontaneous bleeding. Haematological and biochemical parameters are presented in Table 2.
Table 2.
Haematological and biochemical profile of dengue patients.
| Parameter | DF (N = 61)% | DHF (N = 30)% | DSS (N = 9)% |
|---|---|---|---|
| Haematological profile of dengue patients (N = 100) | |||
| Thrombocytopaenia | 37 (60%) | 30 (100%) | 09 (100%) |
| Neutropenia | 26 (42.6%) | 12 (40%) | 02 (22%) |
| Lymphocytosis | 02 (3.2%) | 02 (6.6%) | 02 (2.2%) |
| HCT>40% | 03 (4.9%) | 17 (56.6%) | 08 (88.8%) |
| Elevated PTT | 01 (1.6%) | 01 (3.3%) | 00 |
| Elevated PT & PTT | 00 | 04 (13.3%) | 06 (66.6%) |
| Decreased fibrinogen | 00 | 03 (10%) | 03 (33.3%) |
| Parameter | DF Mean ± SD | DHF/DSS Mean ± SD | p value |
|---|---|---|---|
| Biochemical profile of dengue patients (N = 100) | |||
| AST (IU/litre) | 70.36 ± 49.96 | 192.62 ± 172.02 | <0.001 |
| ALT (IU/litre) | 79.93 ± 64.06 | 185.36 ± 143.12 | <0.001 |
| S. bilirubin (mg/dl) | 0.74 ± 0.39 | 1.526 ± 1.00 | <0.001 |
| Elevated serum LDH | 356.56 ± 98.68 | 524.46 ± 195.57 | <0.001 |
Age, sex, haemoglobin, leukocyte count parameters were not found to be statistically significant in DHF/DSS as compared to DF. Mean haematocrit, PT, PTT, serum fibrinogen, platelet count, activated lymphocytes were significantly elevated in DHF/DSS as compared to DF (p value <0.001). There was significant correlation between the bleeding manifestation and the degree of thrombocytopaenia (p value <0.001). Levels of ALT, AST, bilirubin, LDH, protein, BUN, sodium were significantly elevated in DHF/DSS as compared to DF (p value <0.001).
Out of a total of 100 dengue infection positive cases, 55% serum samples were positive for NS1 Ag, 56% for IgM and 23% for IgG of which 15% were positive for both NS1 Ag and IgM, 17% samples were positive for both IgM and IgG antibodies and 5% were positive for both NS1 Ag and IgG. In DF cases, IgM serology was positive in 55% patients, IgG in 18% and both IgG and IgM was positive in 14.75% patients. In DHF cases, 66.67% were IgM positive, 30% IgG positive and 26.67% positive for both IgG and IgM. 77% of total serum samples were positive for RT-PCR and 83% for RT-LAMP. 77% samples showed common positivity for RT-PCR and RT-LAMP. Sensitivity, specificity, positive and negative predictive values (PPV and NPV) of NS1 Ag with RT-PCR were 94.54%, 88.84%, 67.53% and 98.51% while that with RT-LAMP was 100%, 87.5%, 66.26% and 100%. Strength of agreement was substantial in both cases. There was low sensitivity, specificity, PPV and NPV when NS1 and RT-PCR were compared to IgM however RT-LAMP showed sensitivity, specificity, PPV and NPV of 49.39%, 92.35%, 73.21% and 81.16%. Strength of agreement was moderate. RT-LAMP compared with RT-PCR showed a perfect agreement with 95% confidence interval of 0.9. 65% of total samples were found to be positive for DEN-1 and 18% were positive for DEN-3. DEN-1 genotype III was the predominant circulating virus in the outbreak studied along with sporadic circulation of DEN-3 genotype III (Fig. 4).
Fig. 4.
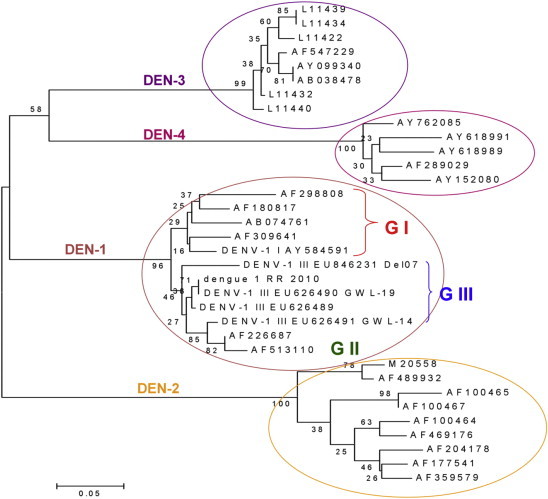
Dengue serotypes and genotypes determined by sequencing.
Discussion
The dramatic resurgence and emergence of epidemic DF and DHF have been attributable to substandard housing, overcrowding, deterioration in water and waste management systems associated with unplanned urbanization resulting in ideal conditions for increased propagation of mosquitoes and subsequent increase in transmission of dengue. Other factors include decay in public health infrastructure and lack of effective mosquito control. Air travel is known to transport the virus to newer locations. Post-monsoon dengue outbreak in Indian subcontinent9 is attributable to increased vector breeding due to conducive temperature, humidity and availability of water reservoirs. Hyperendemicity of dengue virus, as evidenced by this study, plays an important role in the increasing frequency and severity of DHF/DSS.12–14 All four serotypes have been reported.9,15–18 DEN-3 has caused unexpected epidemics of DHF in Sri Lanka, East Africa, Latin America19 and New Delhi.19,20
The study affirmed NS1 Ag as a marker of acute dengue infection, being positive prior to appearance of IgM antibodies. However, it was seen also positive in very early seroconverts where IgM antibody was just in detectable range. Initial high titre of IgG antibodies overlapping with IgM was also seen indicating severe form of dengue developed due to sequential infection by a different serotype. The commonly used antibody μ-capture EIA is superior to indirect EIA as it removes competing antibodies in the capture step. RT-PCR showed a 67.53% and 49.35% sensitivity in acute and late infection respectively. RT-PCR detected 25 NS1 Ag negative, 39 IgM negative and 63 IgG negative cases. Similar studies showed that the RT-PCR was as accurate as 77–100% and is considered to be the gold standard for the diagnosis of dengue in the first 4 days of illness when the dengue serology is negative.21–23
Immunological testing in dengue
The antibody based tests (IgM and IgG) are not helpful in the first 5 days and need to be repeated after 5 days. Diagnosis is achieved by NS1 antigen detection and nucleic acid technology in the first 5 days. NS1 antigen cannot identify serotypes.
Nucleic acid technology in the diagnosis of dengue
Nucleic acid technologies corroborate with viraemia detectable after 24 h of acute dengue infection. Conventional techniques such as RT-PCR, and nested PCR are sensitive and specific, but limited by requirement of infrastructure, cost and expertise. Fully automated real-time amplification is rapid, quantitative, more sensitive and specific and easily standardized but requires precision instruments. Newer isothermal amplification methods such as NASBA and self-sustained sequence replication can detect less than 10 copies within an hour but are compromised in specificity due to low stringency (40 °C) leading to poor target sequence selection. Strand displacement amplification largely overcomes these shortcomings by using four primers but has increased backgrounds due to digestion, irrelevant DNA contained in the sample and the necessity to use costly nucleotides as a substrate. Multiple primers in nested PCR and SDA have improved amplification specificity for the target sequence. The inherent need of expertise and expensive instruments restricts nucleic acid technology to sophisticated labs. RT-LAMP overcomes these restrictions and is of tremendous value in timely medical intervention.
RT-LAMP
RT-LAMP is based on the principle of a strand displacement reaction and stem-loop structure (Fig. 5). In the present study, the RT-LAMP primers were designed from the distal half of the 3 prime noncoding region, as the proximal half has major mismatches and missing sequences. RT-LAMP assay primers are not effective when there are 4–6 mismatches and homology is less than 90%. Buffer, primers, reverse transcriptase and DNA polymerase in a single tube were incubated at 63 °C for 1 h. RT-LAMP has high amplification efficiency, sensitivity and specificity due to continuous isothermal amplification employing six primers recognizing eight distinct target regions.10 The yield of pyrophosphate ion forms a white precipitate of magnesium pyrophosphate and enables naked eye visualization of amplified products. The increased turbidity correlates with the amount of DNA synthesized and enables automated measurement of turbidity real-time,24,25 under SYBR Green UV transillumination. Field visualization techniques such as direct naked eye visualization and SYBR Green colorimetry under natural light or hand held UV torch enable applicability in low resource setups. Colour change is permanent and can be kept as a record (Fig. 6). Further, the entire reaction is carried out within an hour at a fraction of the cost involved, making it faster and cheaper than conventional nucleic acid techniques.
Fig. 5.
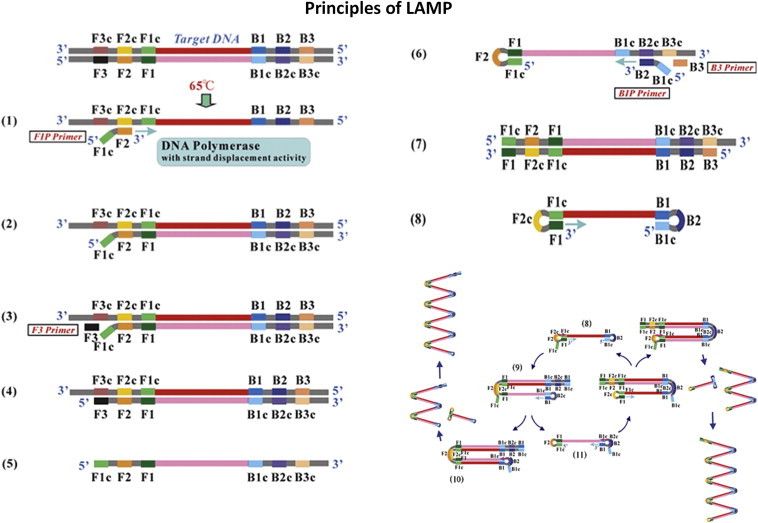
Principle of RT-LAMP.
Fig. 6.
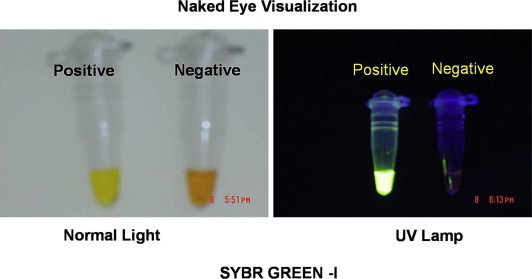
Naked eye visualization of amplification products (can be kept as records).
In our study, RT-LAMP detected dengue in 83 samples as against RT-PCR which detected in 77 samples. The fact that all samples positive by RT-PCR were also positive for RT-LAMP and six samples undetected by RT-PCR were detected by RT-LAMP, establishes the superiority of RT-LAMP against RT-PCR. RT-LAMP scored over RT-PCR in terms of sensitivity and specificity by its ability to detect dengue infection in both early and intermediary stages when NS1 antigen titres are not in the detectable range and the IgM antibody titres have just started to rise.
RT-LAMP scores over conventional and newer amplification techniques as it is highly sensitive, specific, relatively simple and low on infrastructure requirement. Our experience with RT-LAMP emphasized its utility as a rapid, simple, easy, cost effective, single tube, isothermal, highly sensitive and specific field applicable technique for qualitative and quantitative detection of dengue virus serotypes which doesn't have the deficiencies present in existing techniques. Its applicability in tertiary care institutes is emphasized by its ability in viral quantification and evaluation of viraemia in patients. It can suitably be introduced in zonal/peripheral hospitals as an adjunct to existing immunological tests acting as parallel controls. RT-LAMP as the new gold standard for dengue diagnosis may be the panacea of future.
Conclusion
The 2009–2011 dengue epidemic in Delhi and adjoining areas was caused predominantly due to DEN-1 genotype III with some cases of DEN-3 genotype III. NS1 antigen, RT-PCR and RT-LAMP are reliable and sensitive tests for detection during 1–3 days of dengue infection. μ-Capture IgM EIA is reliable and specific test after 5–7 days of initial infection. Deranged haematocrit and liver function tests are indicators of the severity of the disease. RT-LAMP is rapid, cost effective, highly sensitive and specific qualitative and quantitative technique which can detect dengue infection in both early and intermediary stages when NS1 antigen titres are not in the detectable range and the IgM antibody titres have just started to rise. Its amenability for automation and promising utility in low resource healthcare setups and field conditions raise it as the new gold standard for dengue diagnosis.
Intellectual contributions of authors
Study concept: Brig AK Sahni, Col Naveen Grover, Brig Ajay Sharma
Drafting & Manuscript revision: Brig AK Sahni, Maj Inam Danish Khan
Statistical analysis: Brig AK Sahni, Maj Inam Danish Khan, Jugal Kishore
Technical support: Brig AK Sahni, Maj Inam Danish Khan, Jugal Kishore
Study supervision: Brig AK Sahni, Col Naveen Grover, Brig Ajay Sharma
Funding source
This study has been financed by research grants from the O/o DGAFMS, New Delhi.
Conflicts of interest
All authors have none to declare.
References
- 1.Chaturvedi U.C., Agarwal R., Elbishbishi E.A., Mustafa A.S. Cytokine cascade in dengue haemorrhagic fever: implications for pathogenesis. FEMS Immunol Med Microbiol. 2000;28:183–188. doi: 10.1111/j.1574-695X.2000.tb01474.x. [DOI] [PubMed] [Google Scholar]
- 2.Berry N., Chakravarti A., Gur R., Mafthur M.D. Serological investigation of a febrile outbreak in Delhi, India, using a rapid immunochromatographic test. J Clin Microbiol. 1998;36:2795–2796. doi: 10.1128/jcm.36.9.2795-2796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripathi B.K., Gupta B., Sinha R.S., Prasad S., Sharma D.K. Experience in adult population in dengue outbreak in Delhi. J Assoc Physicians India. 1998;46:273–276. [PubMed] [Google Scholar]
- 4.Kumar M., Pasha S.T., Mittal V. Unusual emergence of guate 98 like molecular subtype of Den-3 during 2003 dengue outbreak in Delhi. Dengue Bull. 2004;28:101–167. [Google Scholar]
- 5.Gubler D.J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO . WHO; 1999. Strengthening Implementation of the Global Strategy for Dengue Fever/Dengue Hemorrhagic Fever Prevention and Control. Report on the Informal Consolation. [Google Scholar]
- 7.Vijayakumar T.S., Chandy S., Sathish N., Abraham M., Abraham P., Sridharan G. Is dengue emerging as a major public health problem? Indian J Med Res. 2005;121:100–107. [PubMed] [Google Scholar]
- 8.Vaughn D.W., Kalayanarooj S., Innis B.L. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 9.Shu P.Y., Chang S.F., Kuo Y.C. Development of group and serotype specific one-step SYBR Green I based real-time reverse transcription PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notomi T., Okayama H., Masubuchi H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanciotti R.S., Calisher C.H., Gubler D.J., Chang G.J., Vorndam A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarti A., Kumaria R. Eco-epidemiological analysis of dengue infection during an outbreak of dengue fever, India. Virol J. 2005;2:32. doi: 10.1186/1743-422X-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigau-Perez J.G., Vorndam A.V., Clark G.G. The dengue and dengue hemorrhagic fever epidemic in Puerto Rico, 1994–1995. Am J Trop Med Hyg. 2001;64:67–74. doi: 10.4269/ajtmh.2001.64.67. [DOI] [PubMed] [Google Scholar]
- 14.Vajpayee M., Mohankumar K., Wali J.P., Dar L., Seth P., Broor S. Dengue virus infection during post-epidemic period in Delhi, India. Southeast Asian J Trop Med Public Health. 1999;30:507–510. [PubMed] [Google Scholar]
- 15.Kumar M, Rai A, Pasha ST, Datta KK. Molecular Detection and Typing of Dengue Viruses in Clinical Specimen from Dengue Outbreak in Delhi in 2000. The Proceedings of the International Conference of Indian Public Health Congress. New Delhi; April 2001:14–16.
- 16.Singh U.B., Seth P. Use of nucleotide sequencing of the genomic cDNA fragments of the capsid/premembrane junction region for molecular epidemiology of dengue type 2 viruses. Southeast Asian J Trop Med Public Health. 2001;32(2):326–335. [PubMed] [Google Scholar]
- 17.Das P.K., Parida M.M., Saxena P. Emergence and continued circulation of dengue-2 (genotype IV) virus strains in northern India. J Med Virol. 2004;74(2):314–322. doi: 10.1002/jmv.20166. [DOI] [PubMed] [Google Scholar]
- 18.Monath T.P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci U S A. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libraty D.H., Endy T.P., Houng H.S. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185:1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 20.Corwin A.L., Larasati R.P., Bangs M.J. Epidemic dengue transmission in southern Sumatra, Indonesia. Trans R Soc Trop Med Hyg. 2001;95:257–265. doi: 10.1016/s0035-9203(01)90229-9. [DOI] [PubMed] [Google Scholar]
- 21.Guzman M.G., Kouri G. Advances in dengue diagnosis. Clin Diagn Lab Immunol. 1996;3:621–627. doi: 10.1128/cdli.3.6.621-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callahan J.D., Wu S.J., Dion-Schultz A. Development and evaluation of serotype and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus. J Clin Microbiol. 2001;39:4119–4124. doi: 10.1128/JCM.39.11.4119-4124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houng H.H., Chen R.C.M., Vaughn D.W., Kanesa-thasan N. Development of a fluorogenic RT-PCR system for quantitative identification of dengue virus serotypes 1–4 using conserved and serotype-specific 3' noncoding sequences. J Virol Methods. 2001;95:19–32. doi: 10.1016/s0166-0934(01)00280-4. [DOI] [PubMed] [Google Scholar]
- 24.WHO . WHO Regional Office for South-East Asia Publication; 1999. Guidelines for Treatment of Dengue Fever/Dengue Hemorrhagic Fever in Small Hospitals. [Google Scholar]
- 25.Gubler D.J. Diagnosis of dengue/dengue hemorrhagic fever. Dengue Bull. 1996;20:20–23. [Google Scholar]


