Abstract
A multitude of different virulence factors as well as the ability to rapidly adapt to adverse environmental conditions are important features for the high pathogenicity of Pseudomonas aeruginosa. Both virulence and adaptive resistance are tightly controlled by a complex regulatory network and respond to external stimuli, such as host signals or antibiotic stress, in a highly specific manner. Here, we demonstrate that physiological concentrations of the human host defense peptide LL-37 promote virulence factor production as well as an adaptive resistance against fluoroquinolone and aminoglycoside antibiotics in P. aeruginosa PAO1. Microarray analyses of P. aeruginosa cells exposed to LL-37 revealed an upregulation of gene clusters involved in the production of quorum sensing molecules and secreted virulence factors (PQS, phenazine, hydrogen cyanide (HCN), elastase and rhamnolipids) and in lipopolysaccharide (LPS) modification as well as an induction of genes encoding multidrug efflux pumps MexCD-OprJ and MexGHI-OpmD. Accordingly, we detected significantly elevated levels of toxic metabolites and proteases in bacterial supernatants after LL-37 treatment. Pre-incubation of bacteria with LL-37 for 2 h led to a decreased susceptibility towards gentamicin and ciprofloxacin. Quantitative Realtime PCR results using a PAO1-pqsE mutant strain present evidence that the quinolone response protein and virulence regulator PqsE may be implicated in the regulation of the observed phenotype in response to LL-37. Further experiments with synthetic cationic antimicrobial peptides IDR-1018, 1037 and HHC-36 showed no induction of pqsE expression, suggesting a new role of PqsE as highly specific host stress sensor.
Introduction
Pseudomonas aeruginosa is a widespread Gram-negative water and soil bacterium, which is, in addition, one of the most important opportunistic human pathogens causing severe infections in immunocompromised persons, such as burn wound, catheter and urinary tract infections or chronic pneumonia in cystic fibrosis (CF) patients [1]. Due to a large arsenal of intrinsic resistance mechanisms such as a low outer membrane permeability, the expression of antibiotic cleaving enzymes, and the existence of multidrug efflux pump systems, P. aeruginosa is inherently resistant to various commonly used antibiotics [2], [3].
Multiple virulence factors have been identified to affect the pathogenicity of P. aeruginosa. These factors comprise the expression of extracellular appendices flagella, type IV pili and type III secretion systems, the production of alginate and lipopolysaccharide (LPS) and the synthesis of secreted exocompounds such as proteases (e.g. elastase) and other enzymes, toxins, phenazines, rhamnolipids, hydrogen cyanide (HCN) and quorum sensing molecules (e.g. 4-quinolone PQS) [4], [5]. P. aeruginosa virulence is controlled by a highly complex and in large parts not fully understood signaling network including the Las, Rhl and PQS quorum sensing systems which induce the expression of various virulence factors e.g. in response to high cell densities or other external stimuli like iron limitation [6].
Adaptive resistance in P. aeruginosa has been reported for aminoglycosides and different cationic antibiotics such as polymyxins and the bovine cationic peptide indolicidin. Although the phenomena of adaptive resistance against the aminoglycoside gentamicin and the polypeptide antibiotic polymyxin B was first mentioned decades ago [7], [8], the underlying signaling pathways and involved defense mechanisms have been elucidated in parts in recent studies [9], [10], [11], [12], [13]. While the two-component systems PhoP-PhoQ and PmrA-PmrB recognize low Mg2+ concentrations and phosphate deprivation in the environment, ParR-ParS and CprR-CprS have been shown to directly sense cationic compounds, such as polymyxin B, colistin, indolicidin and, amongst others, the synthetic antimicrobial peptides HHC-36 and IDR-1018 [12], [13]. Activation of mentioned two-component systems induces the expression of the LPS modifying operon arnBCADTEFugd, leading to a reduced net charge of LPS due to the addition of 4-aminoarabinose to lipid A, which impairs the self-promoted uptake of cationic compounds across the outer membrane and thereby enhances tolerance to these compounds [4], [14].
The increasing occurrence of infections caused by multidrug-resistant bacteria, which tolerate even high concentrations of common antibiotics, calls for the rapid development and clinical application of new anti-infective strategies [15]. Host defense peptides, also termed as antimicrobial peptides (AMPs), have been considered as promising compounds to combat multi-resistant pathogens due to their combinatory actions as antimicrobial, antibiofilm and immunomodulatory agents [15]. The major human host defense peptide, LL-37, is the only cathelicidin class peptide produced in humans and exhibits a modest antibacterial activity against a variety of different pathogens including Staphylococcus epidermidis, Staphylococcus aureus, Streptococcus pneumonia and P. aeruginosa [16]. Additionally, it has been shown to prevent the formation of resistant biofilms and stimulate biofilm dispersal in various bacteria when applied at sublethal concentrations [17]. LL-37 is synthesized by phagocytes, epithelial cells and keratinocytes and has been detected in a large number of different cells, tissues and body fluids at varying concentrations [16]. During infectious diseases, immune cells and epithelial cells secrete a battery of host defense compounds, with either direct antimicrobial or immunomodulatory activities, including cationic peptides [5]. Extracellular LL-37 levels have been observed to be severely increased, reaching local concentrations of 15–20 µg/ml e.g. in the lung fluid of newborns suffering from pulmonary infections [18] and in cystic fibrosis patients [19] – diseases, which are often linked to P. aeruginosa infections [1].
Previous studies demonstrated an influence of human opioids [20], [21], natriuretic peptides [22], INF-γ [23] and the polypeptide antibiotic colistin [24] on virulence and quorum sensing in P. aeruginosa. However, this has not been investigated for human cationic host defense peptides so far. In this study, we elucidated the response of P. aeruginosa towards physiological concentrations of LL-37 by global transcriptional studies and metabolite analyses and observed a strong induction of virulence factor production as well as an increase in efflux pump expression during incubation with LL-37. Further experiments revealed an involvement of the quinolone signal response protein PqsE in the regulation of this LL-37 stimulated enhanced virulence factor production and adaptive resistance in P. aeruginosa.
Materials and Methods
Bacterial Strains, Media and Antimicrobial Peptides
Bacterial strains used in this study are listed in Table 1. Transposon mutants PAO1-pqsE and PA14-mexH were confirmed by PCR (data not shown). All experiments were performed in Mueller Hinton (MH) broth (Merck, Darmstadt, Germany). Bacteria were routinely grown at 37°C with shaking at 170 rpm. Antimicrobial peptides were kindly provided by Prof. Robert Hancock (University of British Columbia, Vancouver, Canada) or purchased from Anaspec (Fremont, CA, USA). The amino acid sequences of antimicrobial peptides used and their minimal inhibitory concentrations (MIC) against PAO1 WT are shown in Table 2. Peptide stock solutions of 2 mg/ml were prepared in sterile ultra pure DI water and stored at −20°C until needed.
Table 1. P. aeruginosa strains used in this study.
| Strain | Description | Reference |
| PAO1 WT | H103 (PAO1 wild-type strain) | [61] |
| PAO1- pqsE | Transposon insertion in pqsE (PA1000), Mutant ID PAO1_lux_76:C11, TcR | [38] |
| K767 | Wild-type of strains K1521, K1536, K1523, K1455, K1525, K2415, K2896 | [57] |
| K1521 | K767ΔmexCD | [57] |
| K1536 | K767ΔnfxB | [57] |
| K1523 | K767ΔmexB | [57] |
| K1455 | K767ΔnalB | [62] |
| K1525 | K767ΔmexXY | [63] |
| K2415 | K767ΔmexZ | [63] |
| K2896 | K767 ΔmexBΔmexCDΔmexXY | [57] |
| K2153 | Clinical isolate, wild-type of efflux mutants K2892 and K2376 | [64] |
| K2892 | K2153ΔmexF | [64] |
| K2376 | K2153ΔmexS | [64] |
| PA14 WT | PA14 wild-type strain | [65] |
| PA14- mexH | Transposon insertion in mexH (PA4206), Mutant ID: PAMr_nr_mas_09_3:C7, GmR | [65] |
GmR: gentamicin resistance, TcR: tetracycline resistance.
Table 2. Antimicrobial peptides used in this study.
| Peptide | Sequence | Source/Reference | MIC [µg/ml] against PAO1 WTa |
| LL-37 | LLGDFFRKSKEKIFKEFKRIVQRIKDFLRNLVPRTES | [66] | 16 |
| HHC-36 | KRWWKWWRR | [47] | 16 |
| IDR-1018 | VRLIVAVRIWRR | [45] | 16 |
| 1037 | KRFRIRVRV | [46] | 16 |
a Minimal inhibitory concentrations (MIC) of PAO1 WT against different antimicrobial peptides were determined in MH broth using a standard two-fold serial dilution protocol for microtiter plates. Data represent mode MIC values of three independent experiments for each strain.
MIC (Minimal Inhibitory Concentration) Determination
MIC values were determined using a standard broth microdilution protocol as described previously [25]. Growth in MH medium in presence or absence of antibiotics or antimicrobial peptides was monitored after 18 h of incubation at 37°C. In case of experiments with cationic peptides, 96-well polypropylene microtiter plates (Eppendorf, Hamburg, Germany) were used in order to prevent high MIC values due to the binding of cationic peptides to polystyrene. MIC values against antibiotics ciprofloxacin and gentamicin were determined in 96-well polystyrene microtiter plates (Nunc, Thermo Fisher Scientific, St. Leon-Rot, Germany).
RNA Extraction, cDNA Synthesis and Microarray Analysis
For global gene expression studies, three independent mid-log phase cultures of P. aeruginosa PAO1 were challenged with LL-37 (20 µg/ml) for 2 h. Untreated bacterial cultures served as negative controls. To ensure homogenous gene expression profiles within treated and untreated groups enabling a precise analysis of transcriptional changes, we used bacteria from the exponential growth phase. Due to the higher cell number in this experiment (∼5×108 cells/ml) in comparison to the MIC assay (5×105 cells/ml), used LL-37 concentrations of 20 µg/ml did not affect bacterial growth during the incubation time. This was confirmed by measuring the optical density of bacterial cultures at 600 nm (OD600), resulting in comparable OD600 values in the range of 0.7–1.2 in treated samples and untreated controls after 2 h of incubation. Following peptide treatment, total RNA was extracted using RNA protect reagent and RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Remaining contaminating DNA was removed from the samples in an off-column DNAse digestion procedure using Ambion® DNA-free™ Kit (Life Technologies GmbH, Darmstadt, Germany). RNA quantity and quality was checked photometrically.
First strand cDNA synthesis from 10 µg total RNA as well as cDNA fragmentation into 50–200 bp fragments, biotin-labeling and subsequent hybridization to Affymetrix GeneChip DNA Microarrays Pae_G1a was carried out according to the manufacturer’s standard protocol (Affymetrix UK Ltd, Freiburg, Germany). Each sample was hybridized to at least two microarray chips as technical repeat. Only genes which showed more than 1.5-fold changes in gene expression between LL-37-treated bacteria and untreated controls were included in further analyses.
Quantitative Real Time PCR (qRT-PCR)
Quantitative Realtime PCR (qRT-PCR) experiments were performed in order to verify microarray results of specific dysregulated genes. To this aim, P. aeruginosa cultures were grown until mid-exponential phase following incubation with peptides LL-37, IDR-1018, 1037 or HHC-36 (20 µg/ml each; MIC values: 16 µg/ml (Table 2)) for 2 h as described for microarray analysis. Isolation of total RNA was carried out using RNA protect reagent and the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Remaining contaminating DNA was removed from the samples in an off-column DNAse digestion procedure using Ambion® DNA-free™ Kit (Life Technologies GmbH, Darmstadt, Germany). RNA quantity and quality was checked photometrically. RNA was converted into first strand cDNA using random hexamers and Maxima Reverse Transcriptase (Thermo Fisher Scientific, St. Leon-Rot, Germany) in a standard PCR protocol which was provided by the manufacturer. cDNA was diluted to a concentration of 4 ng/µl and directly utilized as template for qRT-PCR reactions using the KAPA SYBR Fast Universal qPCR MasterMix (Peqlab Biotechnologie GmbH, Erlangen, Germany) in an Abi 7300 Real Time PCR System (Applied Biosystems Deutschland, Darmstadt, Germany) as described previously [26]. Analysis of melting curves of PCR products ensured specificity of the PCR reactions. Primers used for determination of P. aeruginosa gene expression were designed with Primer Express (Applied Biosystems Deutschland, Darmstadt, Germany). Primer sequences are shown in Table S1. Obtained ct-values were normalized to the expression of housekeeping gene rpoD which was not affected by LL-37 treatment as shown by microarray analysis. Samples were assayed at least three times in duplicate (n≥6).
Time-killing Curves
PAO1 WT cultures were grown to mid-log phase and then incubated with LL-37 (20 µg/ml) or without peptide as controls for 2 h at 37°C in MH broth. Following dilution of bacterial cultures to 107 cells/ml in MH broth, antibiotics ciprofloxacin (0.18 µg/ml) or gentamicin (1.5 µg/ml) were added at 3-fold MIC concentrations. Samples were serially diluted and plated out on LB agar after 0, 2, 5, 7, 10, 15, 20, 30, 60 and 90 min of incubation using an optimized drop plate method [27]. Experiments were conducted in triplicates, each with an independent bacterial culture.
Measurement of Pyocyanin and Elastase Activity
For detection and quantification of virulence-associated metabolites and enzymes, PAO1 WT mid-log phase cultures were incubated with 20 µg/ml LL-37 for 21 h. Untreated cultures served as negative controls. Following peptide treatment, bacteria were spun down by centrifugation (30 min, 9000×g, 4°C) and the supernatants were passed through a 0.22 µm syringe filter (Sarstedt, Nümbrecht, Germany).
Pyocyanin was extracted from 2 ml supernatant by adding 2 ml chloroform following re-extraction with 2 ml 0.2 M HCl and subsequent measurement of absorption at 520 nm (A520) as described previously [28]. A520 was normalized against OD600 of the bacterial culture.
Elastase activity in supernatants was determined by an elastin congo red (ECR) assay which was previously described by Pearson et al. [29]. Elastolytic levels were normalized against the cell density of each sample (OD600).
Pyocyanin and elastase activity analyses were performed at least with six independent bacterial cultures. All data was statistically analyzed using the non-parametric Mann-Whitney test.
PQS Measurement by Liquid Chromatography/Tandem Mass Spectrometry (LC-MS/MS)
Supernatants of PAO1 WT bacteria grown for 21 h in the presence or absence of LL-37 were obtained as described above for pyocyanin and elastase analyses. Supernatants were mixed with 2 volumes dichloromethane and vortexed for 1 min. After centrifugation (10 min, 9000×g, 4°C), the lower organic layer containing PQS was transferred into a new reaction tube following evaporation under nitrogen gas at room temperature. The pellet was then resuspended in pure methanol following quantification by LC-MS/MS. To this aim, a quaternary HPLC pump and an autosampler of the series 200 from Perkin Elmer (Überlingen, Germany) were used. The protocol was adapted from two methods described previously [30], [31] with the following modifications. The separation was performed on a Zorbax Eclipse XCB-C8 5 µm, 150×3.6 mm HPLC column (Agilent, USA). The mobile phase consisted of acetonitrile - water 80∶20 (v/v) with 100 µM EDTA and 0.1% acetic acid at a flow rate of 0.40 ml/min. The injection volume was set to 10 µl per sample. Electro-Spray-Ionisation (ESI)-MS was performed on an API 365 triple quadrupole mass spectrometer (PE Sciex, Toronto, Canada) using a turbo ion spray interface used in positive mode. Single MS experiments (Q1 scan), MS/MS experiments (product ion scan, PIC) and multiple reaction monitoring (MRM) were performed using nitrogen as curtain gas, nebulizer gas, heater gas and collision gas. Instrumental parameters were optimized by infusion experiments with PQS standard solution (10 µg/mL; Sigma-Aldrich, Seelze, Germany) infused into the mass spectrometer using a syringe pump (Harvard Apparatus Inc. South Natick, MA, USA) at a flow rate of 10 µl/min. To quantify PQS with high selectivity and sensitivity, MRM experiments were performed using the transitions from precursor ion to fragment ion: 260/175 (quantifier), 260/146, 260/147 and 260/188 (qualifier). An external calibration was performed using PQS standard solutions with concentrations ranging from 10 ng/ml up to 1000 ng/ml.
PQS concentrations were normalized against the cell density of each sample (OD600). Analyses were performed with six independent bacterial cultures and data was statistically analyzed using the non-parametric Mann-Whitney test.
HCN/CN− Quantification
In order to determine levels of toxic HCN in response to LL-37, P. aeruginosa cultures were grown until mid-log phase followed by 2 h of incubation with LL-37 (20 µg/ml) or without peptide as negative control. Subsequently, cells were sedimented by centrifugation (30 min, 9000×g, 4°C) and supernatants were passed through a 0.22 µm syringe filter (Sarstedt, Germany). Quantification of HCN/CN− production was carried out using a polarographic approach delevoped by Blier et al. [32]. Since HCN/CN− production in P. aeruginosa mainly occurs during the exponential growth phase with a peak after 5 h post-inoculation [32], samples for HCN quantification were taken 2 h after LL-37 addition, which corresponds to the late log phase of bacterial growth. Mean values and pooled standard deviations were calculated from three independent experiments, each measured in triplicate, and normalized to OD600 values. Statistical significance was verified by a two-sided t-test for independent samples.
Microarray Data Accession Number
Complete microarray data is deposited in ArrayExpress under accession number E-MEXP-3970.
Results
LL-37 Induces the Expression of Virulence Factor Synthesis and Multidrug Efflux Pump Genes in P. aeruginosa PAO1
In order to get a detailed insight into the global response of P. aeruginosa to physiological concentrations of the human host defense peptide LL-37, we performed gene expression studies using DNA microarray technology. To this aim, mid-log phase P. aeruginosa cells were exposed to 20 µg/ml LL-37 for 2 h following RNA extraction and transcriptional analysis. Untreated cultures served as negative controls. Determined OD600 values were comparable in treated P. aeruginosa samples and control cultures, ranging between 0.7 and 1.2 after the incubation time of 2 h, which confirmed that growth was not inhibited by applied LL-37 concentrations. Additional CFU counts showed furthermore that the relation between OD600 and cell counts was not altered by the applied peptide concentrations. An OD600-value of 1.0 corresponded to 0.8±0.2×109 CFU/ml in control cultures and to 1.1±0.3×109 CFU/ml in LL-37-treated samples after 2 h of incubation.
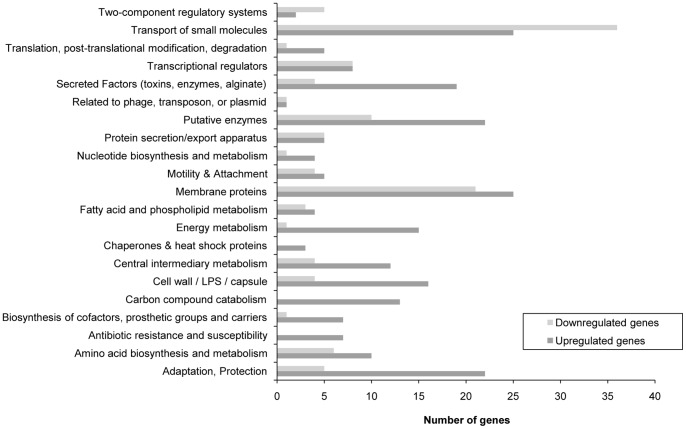
Regarding microarray results, comparison of LL-37-treated bacteria with untreated controls revealed a total number of 420 dysregulated genes (cut-off: 1.5-fold up- or downregulation), of which 280 genes were upregulated and 140 genes were downregulated (Tables 3, S2 and S3). Figure 1 summarizes the functions of dysregulated genes in response to LL-37 and illustrates the diversity of P. aeruginosa stress response to LL-37, since 21 out of the 25 defined gene function classes [33] were affected by the cationic peptide. Quantitative RT-PCR experiments on selected genes were performed in order to confirm microarray data (Table 4).
Table 3. Microarray results of selected dysregulated genes of P. aeruginosa PAO1 WT in response to 2 h of incubation with LL-37 (20 µg/ml) compared to untreated controls.
| PA number | Gene name | Gene product | Fold change in gene expression |
| PQS biosynthesis and response (quorum sensing, virulence factor) | |||
| PA0996 | pqsA | Probable coenzyme A ligase | +2.1 |
| PA0997 | pqsB | PqsB | +1.9 |
| PA0998 | pqsC | PqsC | +1.6 |
| PA0999 | pqsD | 3-oxoacyl-[acyl-carrier-protein] synthase III | +1.7 |
| PA1000 | pqsE | Quinolone signal response protein | +2.1 |
| Pyocyanin biosynthesis (virulence factor) | |||
| PA0051 | phzH | Potential phenazine-modifying enzyme | +1.6 |
| PA1001 | phnA | Phenazine biosynthesis protein PhnA | +2.0 |
| PA1002 | phnB | Phenazine biosynthesis protein PhnB | +2.2 |
| PA1901 | phzC2 | Phenazine biosynthesis protein PhzC | +4.7 |
| PA1902 | phzD2 | Phenazine biosynthesis protein PhzD | +5.8 |
| PA1903 | phzE2 | Phenazine biosynthesis protein PhzE | +6.1 |
| PA1904 | phzF2 | Probable phenazine biosynthesis protein | +6.0 |
| PA1905 | phzG2 | Probable pyridoxamine 5′-phosphate oxidase | +6.3 |
| PA4209 | phzM | Probable phenazine-specific methyltransferase | +5.3 |
| PA4210 | phzA1 | Probable phenazine biosynthesis protein | +7.8 |
| PA4211 | phzB1 | Probable phenazine biosynthesis protein | +4.5 |
| PA4217 | phzS | Flavin-containing monooxygenase | +5.3 |
| Elastase biosynthesis (virulence factor) | |||
| PA1871 | lasA | LasA protease precursor | +1.8 |
| PA3724 | lasB | Elastase LasB | +2.1 |
| Hydrogen cyanide (HCN) production (virulence factor) | |||
| PA2193 | hcnA | Hydrogen cyanide synthase HcnA | +2.4 |
| PA2194 | hcnB | Hydrogen cyanide synthase HcnB | +2.6 |
| PA2195 | hcnC | Hydrogen cyanide synthase HcnC | +2.1 |
| Rhamnolipid production | |||
| PA1130 | rhlC | Rhamnosyltransferase 2 | +2.2 |
| PA3478 | rhlB | Rhamnosyltransferase chain B | +2.0 |
| PA3479 | rhlA | Rhamnosyltransferase chain A | +1.5 |
| Porins, efflux pumps | |||
| PA3279 | oprP | Phosphate-specific outer membrane porin OprP precursor | −5.5 |
| PA3280 | oprO | Pyrophosphate-specific outer membrane porin OprO precursor | −8.2 |
| PA4205 | mexG | Hypothetical protein | +10.2 |
| PA4206 | mexH | Probable RND efflux membrane fusion protein precursor | +4.9 |
| PA4207 | mexI | Probable RND efflux transporter | +2.5 |
| PA4208 | opmD | Probable outer membrane protein precursor | +3.1 |
| PA4597 | oprJ | Multidrug efflux outer membrane protein OprJ precursor | +3.7 |
| PA4598 | mexD | RND multidrug efflux transporter MexD | +4.4 |
| PA4599 | mexC | RND multidrug efflux membrane fusion protein MexC precursor | +9.1 |
| PA4600 | nfxB | Transcriptional regulator NfxB | +1.9 |
| Lipopolysaccharide (LPS) modification | |||
| PA3552 | arnB | ArnB | +1.6 |
| PA3553 | arnC | ArnC | +1.6 |
| PA3555 | arnD | ArnD | +1.5 |
| PA3556 | arnT | Inner membrane L-Ara4N transferase ArnT | +2.0 |
| PA3557 | arnE | ArnE | +2.0 |
| PA3558 | arnF | ArnF | +2.2 |
| PA3559 | ugd | Probable nucleotide sugar dehydrogenase | +2.8 |
| Two-component system PmrA-PmrB | |||
| PA4773 | Hypothetical protein | +4.9 | |
| PA4774 | Hypothetical protein | +3.2 | |
| PA4775 | Hypothetical protein | +2.2 | |
| PA4776 | pmrA | Two-component regulator system response regulator PmrA | +1.9 |
Figure 1. Summarized microarray data of dysregulated P. aeruginosa genes in response to LL-37.
Mid-log phase cultures of P. aeruginosa PAO1 were grown in MH broth containing either 20 µg/ml LL-37 or no LL-37 for 2 h at 37°C following RNA extraction and microarray analysis. The graph shows functions of more than 1.5-fold up- or downregulated genes according to the Pseudomonas Genome Database [28]. Hypothetical genes are not shown.
Table 4. qRT-PCR analysis of P. aeruginosa PAO1 WT and PAO1-pqsE mutant gene expression in response to LL-37 (20 µg/ml)a.
| Gene | PAO1 WT | PAO1-pqsE |
| PA4598 (mexD) | 1.8±0.1b | 1.9±0.2 |
| PA4206 (mexH) | 7.5±0.4 | 1.0±0.1 |
| PA1000 (pqsE) | 1.7±0.3 | n.d. |
| PA3724 (lasB) | 2.6±0.5 | 0.8±0.2 |
| PA2194 (hcnB) | 1.8±0.3 | 0.8±0.1 |
| PA1901 (phzC2) | 2.8±0.4 | 1.0±0.3 |
| PA4776 (pmrA) | 1.6±0.2 | 0.9±0.2 |
| PA3556 (arnT) | 1.5±0.2 | 1.8±0.2 |
P. aeruginosa PAO1 WT or PAO1-pqsE were grown in MH broth containing either 20 µg/ml LL-37 or no LL-37 (control) for 2 h at 37°C following RNA isolation and qRT-PCR analysis.a Mid-log phase cultures of
≥6). ct values were normalized against expression of housekeeping gene rpoD. Fold changes in gene expression of LL-37-treated cells compared to untreated controls were calculated using the ΔΔct method [67]. n.d.: not determined.b Mean averages and standard deviations of three independent experiment, each analyzed at least in duplicate (n
Most strikingly, the microarray data showed an upregulation by 2–8-fold of gene clusters involved in quorum sensing molecule and virulence factor synthesis. Among these were genes coding for the Pseudomonas quinolone signal (PQS) – pqsABCD (PA0996–0999) - and the production of secreted toxic metabolites phenazine (PA0051, PA1001/1002, PA1901–1905, PA4209–4211, PA4217), HCN (PA2193–2195), elastase (PA1871, PA3724) and rhamnolipids (PA3478/3479, PA1130) (Table 3). PqsE (PA1000), which is involved in the regulation of virulence factor expression, influencing e.g. pyocyanin, rhamnolipid and HCN production [6], was also 2-fold upregulated during LL-37 incubation. Although expression of rhamnolipid biosynthesis genes rhlA (PA3479), rhlB (PA3478) and rhlC (PA1130) was enhanced by LL-37 contact, the two main regulators of rhamnolipid production, rhlI (PA3476) and rhlR (PA3477) [34] were not induced (see Tables S2 and S3). Additional qRT-PCR experiments indicated rather a downregulation of major quorum sensing regulators rhlR (fold change −2.0±0.3) and lasR (fold change −1.9±0.5) in response to LL-37. In general, only a few genes (in total 9) encoding transcriptional regulators were more than 1.5-fold upregulated by LL-37 (see Table S2).
In accordance with previous studies [9], [12], we observed an induction of the arnBCADTEFugd LPS modification operon (PA3552-3559) and the two-component regulator pmrA (PA4776). In addition, our microarray data demonstrated an upregulation of Resistance Nodulation Division (RND) efflux pumps genes mexCD-oprJ (PA4597-4599) and mexGHI-opmD (PA4205-4208), which are also involved in multidrug resistance of P. aeruginosa by exporting antibiotics and other toxic compounds [3], [35], [36], [37], whereas genes encoding porin proteins were downregulated by LL-37 (Table 3).
Susceptibility of Different P. aeruginosa Efflux Mutant Strains to LL-37
Since we observed an upregulation of two RND efflux pump systems in response to LL-37, we performed LL-37 susceptibility tests with respective efflux pump mutants PA14-mexH and K1521 (K767ΔmexCD-oprJ) in comparison to the corresponding wild-type strains PA14 and K767. Additional experiments included efflux mutants K1523 (K767ΔmexB), K1525 (K767ΔmexXY) and K2892 (K2153ΔmexF) and K2153, the wild-type strain of K2892. All tested efflux mutants and corresponding wild-type strains showed identical MIC values for LL-37 (16 µg/ml for PA14-mexH, K1521, K1523, K1525 and wild-types PA14 WT and K767; 32 µg/ml for K2892 and wild-type K2153), indicating no impact of a single pump knockout of either MexAB-OprM, MexCD-OprJ, MexXY-OprM, MexEF-OprN or MexGHI-OpmD on susceptibility to LL-37. Since export of many antibiotics is not restricted to one individual efflux pump [3], in case of a single pump knockout, other efflux systems could eventually take over functions of missing efflux pumps, resulting in unaffected MIC values. To test whether a multiple knockout of main P. aeruginosa RND efflux pumps MexAB-OprM, MexXY-OprM and MexCD-OprJ affects susceptibility to LL-37, MIC values were determined for triple mutant K2896 (K767ΔmexBΔmexCDΔmexXY) and wild-type K767, both exhibiting identical MIC values of 16 µg/ml. Next, we could show that susceptibility to LL-37 was not influenced by overexpression of efflux pumps MexAB-OprM (K1455), MexCD-OprJ (K1536), MexXY-OprM (K2415) or MexEF-OprN (K2376) which also resulted in equal MIC values compared to corresponding wild-types K767 (16 µg/ml) or K2153 (32 µg/ml).
In summary, we conclude that susceptibility of P. aeruginosa PAO1 to LL-37 is independent of the tested efflux systems, although expression of RND efflux pump MexCD-OprJ was upregulated in response to LL-37 in our gene expression studies.
LL-37 Enhances Antibiotic Resistance of P. aeruginosa PAO1 Towards Fluoroquinolone and Aminoglycoside Antibiotics
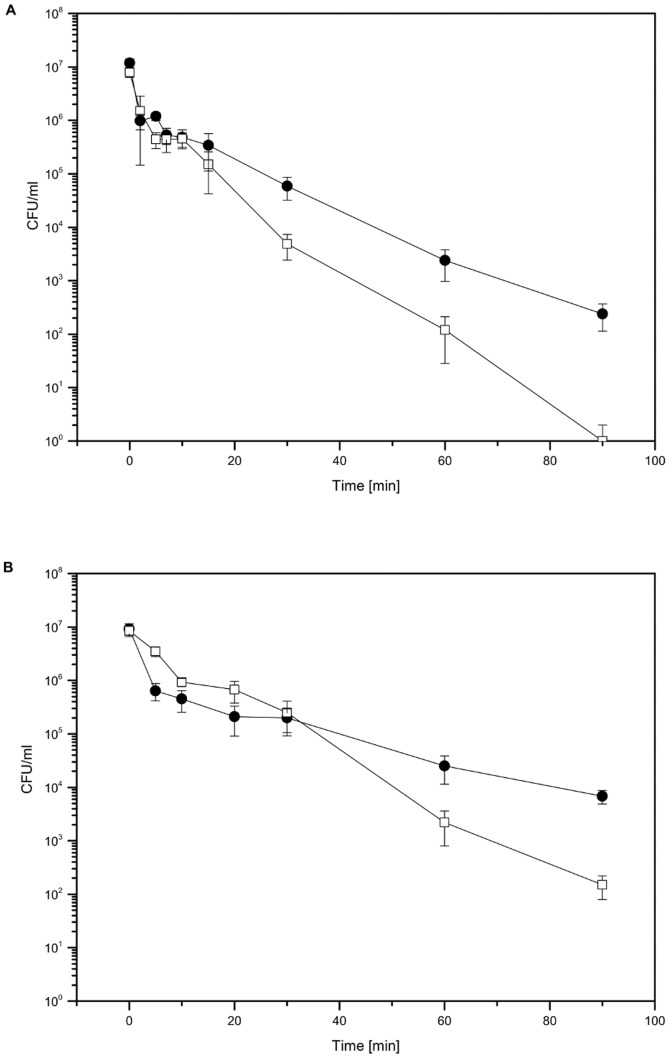
To investigate whether LL-37 was able to trigger adaptive resistance mechanisms in P. aeruginosa towards different antibiotics, time dependent killing of P. aeruginosa PAO1 WT by the fluoroquinolone ciprofloxacin and the aminoglycoside gentamicin was monitored after a 2 h pre-incubation period with LL-37. The peptide itself did not affect bacterial growth as confirmed by determination of OD600. Colony forming units (CFUs) were counted after indicated time points during incubation with antibiotics and compared to cell numbers of control cultures without LL-37 pre-incubation. For ciprofloxacin, we observed an increased resistance of LL-37-treated bacteria compared to the untreated P. aeruginosa cultures already after 30 min of incubation. At this time point, CFU counts revealed only a 2-fold log reduction for LL-37-treated bacteria, but a more than 3-fold log reduction for controls without peptide treatment. After 90 min of incubation, no surviving bacteria in the control group could be detected, whereas P. aeruginosa cultures, which were pre-grown with LL-37, still showed cell numbers of approximately 220 CFU/ml (Figure 2A). Similar results were obtained with gentamicin, however, killing of bacteria was slower and less efficient compared to ciprofloxacin. During the first 30 min of incubation, both LL-37-treated and control bacteria showed a comparable 2-fold log reduction of culturable cells. CFU counts after 60 min demonstrated a beginning resistance of LL-37-treated cells (2×104 CFU/ml) compared to untreated controls (2×103 CFU/ml). After 90 min control cultures contained only 150 CFU/ml, whereas pre-incubation with LL-37 significantly increased cell numbers up to 6800 CFU/ml (Figure 2B). Statistical significance of differences between LL-37-treated bacteria and untreated controls at the end point of the experiment after 90 min of incubation with antibiotics was confirmed by a two-sided t-test for independent samples (p-value <0.001 for both antibiotics). Taken together, we could show that P. aeruginosa resistance to both fluoroquinolone and aminoglycoside antibiotics was enhanced by pre-incubation with the human cathelicidin LL-37.
Figure 2. Time-killing of P. aeruginosa PAO1 by antibiotics ciprofloxacin (A) or gentamicin (B) in the absence or presence of LL-37.
Mid-log phase bacterial cultures were incubated with either 20 µg/ml LL-37 (filled circles) or without LL-37 (open squares) for 2 h. Following dilution of bacterial cultures to 107 cells/ml and addition of 3-fold MIC concentrations of antibiotics ciprofloxacin (0.18 µg/ml) or gentamicin (1.5 µg/ml), colony forming units at indicated time points were determined using the optimized drop plate method [27]. Experiments were performed in triplicate. The figure shows representative results of one experiment. Error bars indicate standard deviations of 10 spots per sample plated out on two different agar plates (n = 10).
Virulence-associated Metabolite Production is Increased during LL-37 Treatment
As shown by our microarray analyses, LL-37 treatment of P. aeruginosa PAO1 induced the expression of several gene clusters which are known to be involved in the production of virulence-associated metabolites such as las, pqs, phz and hcn genes (Table 3).
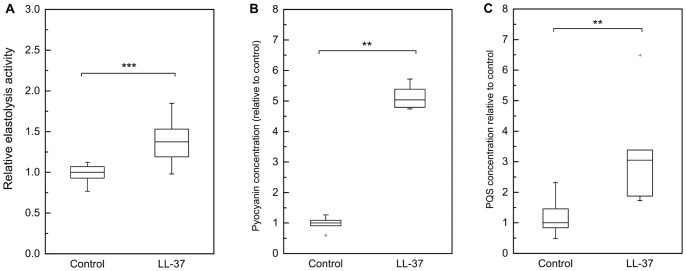
To examine whether this upregulation of gene expression directly leads to an enhanced secretion of virulence factors, levels of pyocyanin, PQS, elastase and HCN were quantified in the bacterial supernatant. For elastase, pyocyanin and PQS determination, supernatants were analyzed after 21 h of incubation with LL-37 in order to ensure an accumulation of adequate amounts of metabolites for subsequent measurements. OD600 values after 21 h of incubation were only marginally decreased in LL-37-treated cultures compared to control cultures (p = 0.14; difference not statistically significant) and therefore LL-37 independent effects of divergent cell densities on quorum sensing and virulence factor levels could be excluded. Photometric determination of elastase expression and pyocyanin synthesis revealed significantly increased elastase (+1.4-fold) and pyocyanin (+5-fold) levels during LL-37 incubation compared to untreated controls (Figure 3A and 3B). Moreover, the LL-37-treated bacterial cultures in contrast to control cultures appeared intensely green (Figure S1), which was most likely due to the elevated levels of the green-blue fluorophore pyocyanin [6]. PQS content in bacterial supernatants was measured using LC-MS/MS and showed 3-fold higher levels in response to LL-37 as well (Figure 3C). HCN quantification also demonstrated increased HCN/CN− levels in the supernatants of LL-37-treated bacteria compared to the untreated controls (Table 5). In conclusion, we could show, that the cathelicidin LL-37 not only affected the expression of various genes which are involved in quorum sensing cascades and virulence phenotype of P. aeruginosa, but was also able to directly enhance the secretion of toxic metabolites pyocyanin, elastase, PQS and HCN.
Figure 3. Quantification of metabolites elastase (A), pyocyanin (B) and PQS (C) in PAO1 WT supernatants after 21 h incubation without or with LL-37.
Mid-log phase cultures of PAO1 WT were grown in MH broth containing either 20 µg/ml LL-37 or no LL-37 (control) for 21 h at 37°C. OD600 values after 21 h were comparable in treated samples and controls, indicating no growth inhibition by LL-37. Elastase activity (A) and pyocyanin concentration (B) in bacterial supernatants were determined photometrically. PQS levels (C) were quantified by LC-MS/MS. Boxes include median (black line), 25th and 75th percentiles of normalized data (n≥6). Statistical significance was calculated by Mann-Whitney-Test (**: p≤0.01, ***: p≤0.001).
Table 5. HCN/CN− concentrations in PAO1 WT supernatantsa.
| Sample | HCN/CN− [µg/l]b |
| PAO1+ LL-37 | 899±31 |
| PAO1 control | 475±18 |
µg/ml LL-37 or no LL-37 (control) for 2 h at 37°C. Cell densities after 2 h peptide treatment were comparable in treated samples and controls, indicating no growth inhibition by LL-37. Supernatants were prepared by centrifugation following polarographic determination of HCN/CN− content.a Mid-log phase cultures of PAO1 WT were grown in MH broth containing either 20
= 9). Statistical significance of differences between mean values was confirmed by a two-sided t-test for independent samples (p<0.001).b Mean averages and pooled standard deviations of three experiments, each measured in triplicate (n
Involvement of pqsE in the Response of P. aeruginosa PAO1 to LL-37
Microarray analysis of P. aeruginosa PAO1 cells treated with LL-37 (20 µg/ml) indicated a dysregulation of 20 genes, of which 9 genes were more than 1.5-fold upregulated, whose gene products exhibit potential roles as transcriptional regulators (see Tables S2 and S3). In addition, we observed an increased expression of virulence regulator PA1000 (pqsE) (Table 3), the fifth gene of the PQS biosynthesis operon pqsABCDE. In order to analyze whether the increased biosynthesis of secreted virulence factors and the enhanced expression of efflux pumps MexCD-OprJ and MexGHI-OpmD in response to LL-37 was influenced by pqsE expression, we performed qRT-PCR experiments using a pqsE transposon insertion mutant (Mutant ID PAO1_lux_76:C11) of the PAO1mini-Tn5 lux transposon mutant library [38], which was either grown for 2 h in the presence or absence of LL-37. Whereas expression of efflux pump gene mexD and one gene of the LPS modification operon, arnT, still remained induced in response to LL-37 in the PAO1-pqsE mutant, our results demonstrated no alterations in the expression of genes mexH, hcnB, lasB and phzC in the PAO1-pqsE mutant treated with LL-37 compared to untreated PAO1-pqsE (Table 4). These findings suggest an involvement of pqsE in the regulation of our observed LL-37 induced adaptive resistance and virulence factor production in P. aeruginosa. To further investigate whether pqsE induction could represent a general response of P. aeruginosa to cationic peptides, we quantified pqsE expression after 2 h of incubation with the synthetic peptides IDR-1018, 1037 and HHC-36 (20 µg/ml each), but observed no changes in pqsE transcriptional levels in case of IDR-1018 (fold change +1.2±0.1) or rather a downregulation in case of 1037 (fold change −2.7±0.7) and HHC-36 (fold change: −3.0±0.7) in comparison to untreated bacteria.
Discussion
The notable repertoire of virulence factors and the ability to rapidly develop adaptive resistances against antibiotics are two crucial factors for the great success of P. aeruginosa as an opportunistic human pathogen [2], [6]. Here we demonstrate that both, virulence factor production as well as the adaptive resistance against fluoroquinolone and aminoglycoside antibiotics, are considerably stimulated by the host defense peptide LL-37, when applied at concentrations that are comparable to the high LL-37 levels found in body fluids at sites of inflammation. Microarray data of LL-37-treated P. aeruginosa cells revealed an upregulation of quorum sensing genes pqsABCDE and significantly increased PQS levels in bacterial supernatants. PQS functions as a signaling molecule in cell-to-cell communication of P. aeruginosa and affects various cellular processes such as virulence, biofilm formation, swarming motility, antibiotic susceptibility and iron binding in an autoinduction mechanism which is dependent on a threshold concentration of PQS [6]. Since cell densities of LL-37-treated cultures and untreated controls were comparable after 2 h as well as after 21 h of incubation, growth effects as a factor influencing the level of quorum sensing signaling molecules and virulence factor production could be ruled out.
In contrast to PAO1 WT, expression of virulence factor genes and of efflux operon mexGHI-opmD was not enhanced in the PAO1-pqsE mutant during LL-37 incubation. These results indicate a regulatory function of pqsE in the adaptation to LL-37, which is comparable to the response to human peptide neuromodulator dynorphin [21] and its synthetic equivalent U50,488 in P. aeruginosa [20]. PqsE (PA1000), although located in one operon together with pqsABCD, is not implicated in PQS biosynthesis. Instead, it has been shown to influence the expression of more than 600 different genes, thus controlling e.g. the production of virulence factors phenazine, rhamnolipids, elastase and HCN and is required for full virulence of P. aeruginosa in mice [39], [40]. Although the recently solved crystal structure of PqsE and amino acid sequence analyses predict a hydrolase activity, there is still a controversy in the literature concerning the precise protein function [41]. Several studies showed that the inducing effect of PqsE on phenazine biosynthesis is controlled by the transcriptional regulator PqsR (MvfR) [40], [42], [43], whereas Farrow et al. observed a RhlR dependent stimulation of virulence factor production by PqsE also in the absence of PqsR [44]. Interestingly, our microarray analysis indicated no induction of major quorum sensing regulators lasR, lasI, rhlI, rhlR or mvfR. In accord with this, these genes were either unaffected or downregulated by U50,488 and the described induction of virulence and adaptive resistance genes was proposed to be regulated by pqsE alone in a yet unknown mechanism [20]. Cummins et al. reported an enhanced expression of pqsB, pqsE, phzF and rhlB in P. aeruginosa PAO1, in conjunction with an increased virulence against Lactobacillus rhamnosus in response to sublethal concentrations of the cationic antibiotic colistin [24].
In the present study, only LL-37, but none of the synthetic cationic peptides IDR-1018, 1037 and HHC-36 were able to induce pqsE expression, although they all target the outer cell membrane of Gram-negative bacteria in order to evolve their antibacterial actions [9]. Hence, the activation of pqsE expression and downstream effects appear to be dependent on other factors such as peptide structure or chemical properties. IDR-1018, 1037 and HHC-36 are small synthetic, 9 to 12 amino acid containing cationic peptides, based on the linear peptide Bac2A [45], [46], [47]. Studies on IDR-1018 structure revealed a β-turn conformation [45], whereas the 37 residue peptide LL-37 forms an α-helix during interaction with lipid bilayers [48]. In contrast to these linear peptides, polypeptide colistin (polymyxin E) exhibits a cyclic structure [24]. One main difference between pqsE affecting agents (LL-37, colistin, dynorphin and U50,488) and the synthetic peptides which showed no influence on pqsE expression (IDR-1018, 1037 and HHC-36), is the molecular mass of the agents, but whether this attribute is critical for the demonstrated induction of pqsE signal pathways, requires further investigation.
Moreover, our microarray data revealed an increased expression of the efflux operon mexGHI-opmD, which is implicated in the resistance against antibiotics norfloxacin [49] and vanadium [36]. In addition, mexGHI-opmD functions as a regulator of PQS and AHL synthesis and promotes quorum sensing and virulence in P. aeruginosa, presumably by exporting toxic quinolone intermediates [37]. Conversely, mexGHI-opmD expression is under the control of PQS and PqsE [39], [50] and has been recently shown to be directly regulated by pyocyanin in a SoxR dependent pathway [51]. Interestingly, only phenazine and PQS gene expression, but not the expression of transcriptional regulator SoxR was upregulated in response to LL-37 in our microarrays, suggesting the involvement of an alternative regulator in the observed induction of mexGHI-opmD. In contrast to our results, Cummins et al. [24] did not observe an upregulation of mexGHI-opmD operon by colistin - although pqsE and phzF expression was induced - emphasizing, again, the complex regulation of virulence and adaptive resistance in P. aeruginosa in response to different environmental stimuli.
Additionally to the enhanced production of toxic, virulence-associated compounds, we noticed an adaptive resistance of P. aeruginosa against antibiotic ciprofloxacin after LL-37 treatment. The response of P. aeruginosa against fluoroquinolone ciprofloxacin and its resistome has been extensively studied and reveals a complex regulation and an involvement of hundreds of different genes [52], [53], including RND efflux pumps MexAB-OprM, MexCD-OprJ and MexEF-OprN [3]. Since MexCD-OprJ was upregulated in reponse to LL-37 in our transcriptional analyses, the adaptive resistance against ciprofloxacin could be due, at least in parts, to this efflux pump induction. MexCD-OprJ efflux system is not expressed in wild-type P. aeruginosa under normal growth conditions, but can be initiated by mutations of repressor nfxB. In previous studies, this activation caused adaptive resistances to various substrates of MexCD-OprJ including macrolides, chloramphenicol, tetracycline and fluoroquinolones [54], [55] and led to a strain-specific induction of virulence [55]. It has been shown recently, that P. aeruginosa mexCD-oprJ expression is also stimulated by cationic biocides benzalkonium chloride and chlorhexidine gluconate [56], [57] and by other components causing membrane damage and envelope stress such as ethanol, SDS, polymyxin B and the antimicrobial peptides V8 and V681 in an algU-dependent pathway [57]. However, LL-37, although it is known to act as a cell membrane-damaging agent [58], did not cause alterations in algU sigma factor expression (see Tables S2 and S3) and the main regulator of MexCD-OprJ, the repressor nfxB, was rather upregulated than downregulated. These findings suggest the existence of an alternative peptide inducible regulator of MexCD-OprJ expression. Although previous experiments from other groups demonstrate that PqsE acts as positive regulator of mexCD-oprJ expression [20], [39], our qRT-PCR results indicate that other regulators may override the lack of PqsE in reponse to LL-37 leading to equally increased mexD levels in the PAO1-pqsE mutant as in the wild-type bacteria.
In addition to the increased ciprofloxacin resistance, susceptibility of P. aeruginosa to aminoglycoside gentamicin was also reduced following LL-37 treatment. Similar results have been reported for Streptococcus pneumonaiae and erythromycin [59]. Since aminoglycosides are mainly exported by the inducible efflux system MexXY-OprM [3], which was not affected by LL-37 in this study, the observed gentamicin resistance has to be caused by other factors. One possible explanation refers to the enhanced expression of the LPS modifying operon arnBCDTEFugd which mediates resistance to aminoglycosides and other cationic antibiotics [4]. In accord with a previous study [9], we observed a 1.9-fold increase in pmrA expression by LL-37, which is involved in arn regulation, whereas other two-component systems controlling arn expression, ParR-ParS [12], PhoP-PhoQ [10] and the recently identified system CprR-CprS [13], were not affected. Since arnT expression, but not pmrA expression, were stimulated in the PAO1-pqsE mutant by LL-37, the observed induction of arnBCDTEFugd expression seems to be independent of both, pmrA and pqsE.
Our observations that virulence factor production and adaptive resistance in response to LL-37, are in parts influenced by PqsE in a yet unknown manner emphasize the crucial role of quorum sensing in P. aeruginosa infections and the high potential of quorum sensing inhibitors as promising agents against infections caused by multi-resistant bacteria, as mentioned previously [60]. Since several cationic peptides, including LL-37, exhibit a potent antibiofilm activity, in several cases in combination with a direct antimicrobial activity against various bacterial species, their prospective medical application is widely discussed [15]. The possibility, that cationic compounds structurally related to LL-37 could be able to affect virulence and adaptive resistance in P. aeruginosa in a similar way, may limit their use as sole drug during infectious diseases and should be considered in further investigations.
Supporting Information
P. aeruginosa PAO1 cultures after 21 h of incubation with LL-37 (left) or without LL-37 (right) in MH medium under shaking conditions at 37°C.
(TIF)
qRT-PCR primers for detection of P. aeruginosa PAO1 gene expression.
(PDF)
Full list of upregulated genes (fold change≥1.5) in response to 20 µg/ml LL-37 compared to non-treated P. aeruginosa PAO1.
(PDF)
Full list of downregulated genes (fold change≥1.5) in response to 20 µg/ml LL-37 compared to non-treated P. aeruginosa PAO1.
(PDF)
Acknowledgments
We would like to thank Bob Hancock for kindly providing antimicrobial peptides, Keith Poole for kindly providing strains used in this study and Manjeet Bains for technical assistance.
Funding Statement
The authors gratefully acknowledge financial support by the BioInterfaces (BIF) Program of Karlsruhe Institute of Technology (KIT) in the Helmholtz Association, the “Concept for the Future” of KIT within the German Excellence Initiative, the Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of KIT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- 2. Hancock RE, Speert DP (2000) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat 3: 247–255. [DOI] [PubMed] [Google Scholar]
- 3. Poole K, Srikumar R (2001) Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr Top Med Chem 1: 59–71. [DOI] [PubMed] [Google Scholar]
- 4. Gooderham WJ, Hancock RE (2009) Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa . FEMS Microbiol Rev 33: 279–294. [DOI] [PubMed] [Google Scholar]
- 5. Gellatly SL, Hancock RE (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67: 159–173. [DOI] [PubMed] [Google Scholar]
- 6. Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, et al. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa . Microbiol Mol Biol Rev 76: 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilleland HE Jr, Murray RG (1976) Ultrastructural study of polymyxin-resistant isolates of Pseudomonas aeruginosa . J Bacteriol 125: 267–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nicas TI, Hancock RE (1980) Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J Bacteriol 143: 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McPhee JB, Lewenza S, Hancock REW (2003) Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa . Mol Microbiol 50: 205–217. [DOI] [PubMed] [Google Scholar]
- 10. Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW (1999) PhoP–PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol 34: 305–316. [DOI] [PubMed] [Google Scholar]
- 11. Macfarlane ELA, Kwasnicka A, Hancock REW (2000) Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146: 2543–2554. [DOI] [PubMed] [Google Scholar]
- 12. Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, et al. (2010) Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54: 3372–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernández L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, et al. (2012) The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother 56: 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, et al. (1999) Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa . Science 286: 1561–1565. [DOI] [PubMed] [Google Scholar]
- 15. Yeung AY, Gellatly S, Hancock RW (2011) Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci 68: 2161–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dürr UHN, Sudheendra US, Ramamoorthy A (2006) LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta 1758: 1408–1425. [DOI] [PubMed] [Google Scholar]
- 17. Jacobsen AS, Jenssen H (2012) Human cathelicidin LL-37 prevents bacterial biofilm formation. Future Med Chem 4: 1587–1599. [DOI] [PubMed] [Google Scholar]
- 18. Schaller-Bals S, Schulze A, Bals R (2002) Increased levels of antimicrobial peptides in tracheal aspirates of newborn infants during infection. Am J Respir Crit Care Med 165: 992–995. [DOI] [PubMed] [Google Scholar]
- 19. Chen CIU, Schaller-Bals S, Paul KP, Wahn U, Bals R (2004) β-defensins and LL-37 in bronchoalveolar lavage fluid of patients with cystic fibrosis. J Cyst Fibros 3: 45–50. [DOI] [PubMed] [Google Scholar]
- 20. Zaborin A, Gerdes S, Holbrook C, Liu DC, Zaborina OY, et al. (2012) Pseudomonas aeruginosa Overrides the Virulence Inducing Effect of Opioids When It Senses an Abundance of Phosphate. PLoS ONE 7: e34883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, et al. (2007) Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa . PLoS Pathog 3: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blier AS, Veron W, Bazire A, Gerault E, Taupin L, et al. (2011) C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa . Microbiology 157: 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu L, Estrada O, Zaborina O, Bains M, Shen L, et al. (2005) Recognition of host immune activation by Pseudomonas aeruginosa . Science 309: 774–777. [DOI] [PubMed] [Google Scholar]
- 24. Cummins J, Reen FJ, Baysse C, Mooij MJ, O’Gara F (2009) Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa . Microbiology 155: 2826–2837. [DOI] [PubMed] [Google Scholar]
- 25. Wiegand I, Hilpert K, Hancock RE (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3: 163–175. [DOI] [PubMed] [Google Scholar]
- 26. Neidig A, Yeung A, Rosay T, Tettmann B, Strempel N, et al. (2013) TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa . BMC Microbiol 13: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herigstad B, Hamilton M, Heersink J (2001) How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods 44: 121–129. [DOI] [PubMed] [Google Scholar]
- 28. Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, et al. (2009) The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa . Microbiology 155: 699–711. [DOI] [PubMed] [Google Scholar]
- 29. Pearson JP, Pesci EC, Iglewski BH (1997) Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179: 5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lepine F, Milot S, Deziel E, He J, Rahme LG (2004) Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa . J Am Soc Mass Spectrom 15: 862–869. [DOI] [PubMed] [Google Scholar]
- 31. Ortori CA, Dubern JF, Chhabra SR, Camara M, Hardie K, et al. (2011) Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal Bioanal Chem 399: 839–850. [DOI] [PubMed] [Google Scholar]
- 32. Blier A-S, Vieillard J, Gerault E, Dagorn A, Varacavoudin T, et al. (2012) Quantification of Pseudomonas aeruginosa hydrogen cyanide production by a polarographic approach. J Microbiol Methods 90: 20–24. [DOI] [PubMed] [Google Scholar]
- 33. Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, et al. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39: D596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reis RS, Pereira AG, Neves BC, Freire DMG (2011) Gene regulation of rhamnolipid production in Pseudomonas aeruginosa – A review. Bioresour Technol 102: 6377–6384. [DOI] [PubMed] [Google Scholar]
- 35. Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V (2009) The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A 106: 7173–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aendekerk S, Ghysels B, Cornelis P, Baysse C (2002) Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148: 2371–2381. [DOI] [PubMed] [Google Scholar]
- 37. Aendekerk S, Diggle SP, Song Z, Høiby N, Cornelis P, et al. (2005) The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151: 1113–1125. [DOI] [PubMed] [Google Scholar]
- 38. Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, et al. (2005) Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: A tool for identifying differentially regulated genes. Genome Res 15: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hazan R, He J, Xiao G, Dekimpe V, Apidianakis Y, et al. (2010) Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog 6: e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deziel E, Lepine F, Milot S, He J, Mindrinos MN, et al. (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu S, Jensen V, Seeliger J, Feldmann I, Weber S, et al. (2009) Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas Quinolone Signal (PQS) response protein. Biochemistry 48: 10298–10307. [DOI] [PubMed] [Google Scholar]
- 42. Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, et al. (2003) The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 50: 29–43. [DOI] [PubMed] [Google Scholar]
- 43. Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, et al. (2001) A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98: 14613–14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Farrow JM 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, et al. (2008) PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190: 7043–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wieczorek M, Jenssen H, Kindrachuk J, Scott WRP, Elliott M, et al. (2010) Structural Studies of a Peptide with Immune Modulating and Direct Antimicrobial Activity. Chem Biol 17: 970–980. [DOI] [PubMed] [Google Scholar]
- 46. de la Fuente-Núñez C, Korolik V, Bains M, Nguyen U, Breidenstein EBM, et al. (2012) Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob Agents Chemother 56: 2696–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cherkasov A, Hilpert K, Jenssen H, Fjell CD, Waldbrook M, et al. (2008) Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem Biol 4: 65–74. [DOI] [PubMed] [Google Scholar]
- 48. Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, et al. (2008) NMR Structure of the Cathelicidin-Derived Human Antimicrobial Peptide LL-37 in Dodecylphosphocholine Micelles. Biochemistry 47: 5565–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sekiya H, Mima T, Morita Y, Kuroda T, Mizushima T, et al. (2003) Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob Agents Chemother 47: 2990–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rampioni G, Pustelny C, Fletcher MP, Wright VJ, Bruce M, et al. (2010) Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ Microbiol 12: 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK (2006) The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa . Mol Microbiol 61: 1308–1321. [DOI] [PubMed] [Google Scholar]
- 52. Brazas MD, Hancock RE (2005) Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa . Antimicrob Agents Chemother 49: 3222–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Breidenstein EB, Khaira BK, Wiegand I, Overhage J, Hancock RE (2008) Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother 52: 4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, et al. (1996) Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa . Mol Microbiol 21: 713–724. [DOI] [PubMed] [Google Scholar]
- 55. Jeannot K, Elsen S, Kohler T, Attree I, van Delden C, et al. (2008) Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52: 2455–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morita Y, Murata T, Mima T, Shiota S, Kuroda T, et al. (2003) Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother 51: 991–994. [DOI] [PubMed] [Google Scholar]
- 57. Fraud S, Campigotto AJ, Chen Z, Poole K (2008) MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother 52: 4478–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sochacki KA, Barns KJ, Bucki R, Weisshaar JC (2011) Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A 108: E77–E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zahner D, Zhou X, Chancey ST, Pohl J, Shafer WM, et al. (2010) Human antimicrobial peptide LL-37 induces MefE/Mel-mediated macrolide resistance in Streptococcus pneumoniae . Antimicrob Agents Chemother 54: 3516–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bhardwaj AK, Vinothkumar K, Rajpara N (2013) Bacterial quorum sensing inhibitors: attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat Antiinfect Drug Discov 8: 68–83. [DOI] [PubMed] [Google Scholar]
- 61. Breidenstein EB, Janot L, Strehmel J, Fernandez L, Taylor PK, et al. (2012) The Lon protease is essential for full virulence in Pseudomonas aeruginosa . PLoS One 7: e49123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Srikumar R, Paul CJ, Poole K (2000) Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-oprM multidrug efflux system of Pseudomonas aeruginosa . J Bacteriol 182: 1410–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morita Y, Sobel ML, Poole K (2006) Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J Bacteriol 188: 1847–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, et al. (2011) mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother 55: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103: 2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B, et al. (1996) The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur J Biochem 238: 325–332. [DOI] [PubMed] [Google Scholar]
- 67. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P. aeruginosa PAO1 cultures after 21 h of incubation with LL-37 (left) or without LL-37 (right) in MH medium under shaking conditions at 37°C.
(TIF)
qRT-PCR primers for detection of P. aeruginosa PAO1 gene expression.
(PDF)
Full list of upregulated genes (fold change≥1.5) in response to 20 µg/ml LL-37 compared to non-treated P. aeruginosa PAO1.
(PDF)
Full list of downregulated genes (fold change≥1.5) in response to 20 µg/ml LL-37 compared to non-treated P. aeruginosa PAO1.
(PDF)