Abstract
The presence of Cryptococcus spp. has been reported in Mexico’s capital city; however, to our knowledge there are no reports of its presence in the state of Nuevo León located in northeast Mexico. This is presumed to be because the hot and dry climate in this region does not favor cryptococcal proliferation. This study confirmed the presence of C. neoformans and C. albidus in 20% (10/50) of randomly selected fecal samples of pigeons (Columba livia) in the Monterrey metropolitan area. The presence of this yeast in the state of Nuevo León is proof of its adaptation to the typically hot climate of the area and is consistent with recent reviews of cryptococcosis cases in several local hospitals. The two species were identified and characterized through microbiological tests and molecular identification by DNA extraction and PCR amplification of highly conserved 18S ribosomal DNA using ITS1 and ITS2 as target regions. The PCR products were sequenced and compared with those reported in GenBank.
Electronic supplementary material
The online version of this article (doi:10.1186/2193-1801-2-632) contains supplementary material, which is available to authorized users.
Keywords: Cryptococcosis, Neoformans, Albidus, Pigeon (Columba livia), México
Introduction
Thirty-eight yeast species within the Cryptococcus genus have been described, including C. neoformans, C. gatti, C. albidus, and C. laurentii, which have been extensively studied because of their clinical importance as etiologic agents of opportunistic cryptococosis (Rosario et al.2008; Castañón Olivares et al.2000). Cryptococcosis can result in serious meningitis and is acquired by inhalation of infective yeasts present in wood, fruits, rotting vegetables, soil, dairy products, and most frequently fecal material from urban pigeons (Columba livia) (Ellis and Pfeiffer, 1992; Khawcharoenporn et al.2007) that commonly accumulate in metropolitan areas where this species has successfully become established. This kind of mycosis is not considered endemic, mainly because there is only information about its presence in Mexico’s central region. The northeast region of the country has a dry and hot climate, where summer temperatures reach up to 40°C (between 35-40°C) even in the shade (Extreme temperatures and precipitation for Monterrey (OBS) 2000-2010 (in spanish) 2013). It has been suggested that these environmental conditions are not suitable for the growth of Cryptococcus spp. and that most strains cannot survive high temperatures because the majority of isolates reported are from tropical and subtropical countries where the average temperature is 22°C in the hottest month of the year (Lin and Heitman, 2006; Ishaq et al.1968; Ruiz et al.1989; Dromer et al.1996; Mancianti et al.2002; Rivas et al.1999; Quintero et al.2005; Curo et al.2005; Colom Valiente et al.1997; Castañol-Olivares and Lopez-Martinez, 1994; Casali et al.2003). Surprisingly, there have been 190 reports of cryptococcosis in six different state hospitals in Monterrey city from 1995 to 2011 (Casillas 2012). This could be evidence of its successful adaptation to the warm and dry climate present in this region. To investigate this we evaluated 50 random samples of pigeon droppings from three different Monterrey metropolitan locations.
Materials and methods
Fifty fecal samples were collected in the summer between the months of August and September of 2010. We first considered the population density for each of the three municipal zones sampled (Guadalupe, Monterrey, and San Nicolás de los Garza) in the Monterrey metropolitan area in the state of Nuevo León. We obtained one sample for approximately every 45,000 persons reported in the three most densely populated municipal areas according to the Mexican Institute of Statistics and Geography (INEGI) data for 2005 (Table 1) to give a good distribution of samples. Pigeon excreta samples were collected using clean spatulas, transferred to sterile plastic bags, and properly labeled according to site and date. Samples were taken immediately to the laboratory and were processed as previously reported by Shields and Ajello with modifications described by La Pal and Baxter (Shields and Ajello, 1966; Pal and Baxter, 1985). Yeasts were isolated and characterized by phenoloxidase activity determination, capsule formation, presence of urease, sugars, nitrate assimilation, and ability to grow at 37°C (Pérez et al.2003). Species and varieties were identified using creatinine dextrose bromothymol blue thymine medium and l-canavanine glycine bromothymol blue agar. We also evaluated the color, morphology, and colonial growth in CHROMagar Candida media (Irokanulo et al.1994; Kwon-Chung et al.1982; García et al.1998). Cryptococcus neoformans ATCC 66031 and Candida albicans ATCC 14053 were used as microbiological identification controls.
Table 1.
Samples collected for the number of inhabitants
| Municipal zones | Population | Samples | Positive samples (C.n/C.a) |
|---|---|---|---|
| Monterrey | 1.135.550 | 25 | (3/3) |
| Guadalupe | 678.006 | 15 | (1/2) |
| San Nicolás | 443.273 | 10 | (1/0) |
| TOTAL | 2.256.829 | 50 | (5/5) |
Total samples collected in relation to the number of inhabitants in each municipal zones.
Microbiological identification was confirmed by ITS1–ITS2 ribosomal gene sequence determination ( White et al.1990). DNA fragments were obtained by polymerase chain reaction (PCR) amplification from genomic DNA extracted using the Barth and Gaillardin method from isolated yeasts (Barth and Gaillardin, 1996). Sequences were aligned with those deposited in NCBI GenBank. Accession numbers of our nucleotide sequences are JQ794489, JQ794490, JQ794491, JQ794492, JQ794493, JQ794494, JQ794495, JQ794496, JQ794497, and JQ794498 (Table 2).
Table 2.
Sequences characteristics
| Isolate ID | Best alignment sequences (ID) | High similarity specie | Similarity percentage | GenBank access |
|---|---|---|---|---|
| Cs1 | JN939394.1 | C. neoformans | 99% | JQ794489 |
| Cs2 | JN939440.1 | C . neoformans | 95% | JQ794490 |
| Cs3 | HQ231895.1 | C. albidus | 94% | JQ794491 |
| Cs4 | AB032618.1 | C. albidus | 95% | JQ794492 |
| Cs5 | HQ231895.1 | C. albidus | 99% | JQ794493 |
| Cs6 | JN939394.1 | C. neoformans | 98% | JQ794494 |
| Cs7 | AB032617.1 | C. albidus | 96% | JQ794495 |
| Cs8 | AJ560306.1 | C. neoformans | 99% | JQ794496 |
| Cs9 | JN939394.1 | C. neoformans | 99% | JQ794497 |
| Cs10 | AB032617.1 | C. albidus | 99% | JQ794498 |
Percent similarity between the ITS1–ITS2 18s ribosomal gene from genomic DNA amplification from the Cryptococcus spp. isolates in the metropolitan area of Monterrey, Nuevo León, Mexico and those available in GenBank by in silico alignment.
The phylogenetic analysis was performed using MEGA view 5.2 with the Neighbor Joining algorithm (NJ), and included 18 sequences, 8 from Genbank and 10 generated in this research.
We developed a simple regression analysis using correspondence tables to determine whether there was any link between ambient conditions of the sample and the isolation results. The data were analyzed according to the isolation results using the statistics program SPSS version 17.0.
Results
Among the 50 pigeon fecal samples obtained from three different municipal zones in the metropolitan area of Monterrey, Nuevo León, México (Figure 1), 10 Cryptococcus strains were isolated (20% of samples were positive) and identified using different biochemical and microbiological tests (Table 3). For each strain, sequences of approximately 800 bp were PCR amplified from the ITS1–ITS2 genomic DNA (Figure 2). Based on very high similarity between sequences from the present samples and those previously reported in GenBank (Table 2), we identified five C. neoformans and five C. albidus isolates. Most of these (three C. neoformans and three C. albidus) were found in the Monterrey city zone, which was the most populated area (Table 1). Analysis of the cladogram (Figure 3) of the 18S ribosomal sequences of our isolated strains aligned against previously reported sequences in GenBank showed that isolates 1, 9, 6, 8, and 2 are grouped with C. neoformans sequences and 3, 7, 5, 10, and 4 with C. albidus. This cladogram confirms the microbiological identification. Among 10 positive samples, 9 were isolated from shaded sites and simple regression analysis confirmed a statistical correlation (Table 2).
Figure 1.
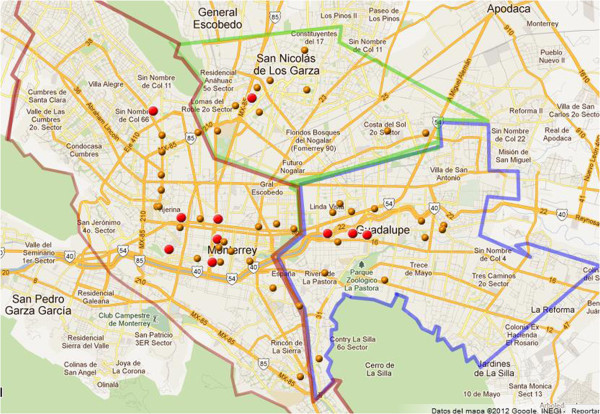
Localization map for collected samples. Dots indicate the location of pigeon fecal samples collected and red dots point to positive samples for Cryptococcus spp.
Table 3.
Isolate’s characteristics
| Isolate identification | Assays | CDBT | CGB | CMa | Microbiologic identification | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | U | I | L | °C | N | C | |||||
| Cs1 | + | + | + | - | + | - | + | - | - | White | C. neoformans var. grubii |
| Cs2 | + | + | + | - | + | - | + | - | - | White | C. neoformans var. grubii |
| Cs3 | - | + | + | + | - | + | + | - | - | Pink lavender | C. albidus |
| Cs4 | - | + | + | + | - | + | + | - | - | Pink lavender | C. albidus |
| Cs5 | - | + | + | - | - | + | + | - | - | Pink lavender | C. albidus |
| Cs6 | + | + | + | - | + | - | + | - | - | White | C. neoformans var. grubii |
| Cs7 | - | + | + | - | - | + | + | - | - | Pink lavender | C. albidus |
| Cs8 | + | + | + | - | + | - | + | - | - | White | C. neoformans var. grubii |
| Cs9 | + | + | + | - | + | - | + | - | - | White | C. neoformans var. grubii |
| Cs10 | - | + | + | + | - | + | + | - | - | Pink lavender | C. albidus |
Cryptococcus species identification using biochemical assays: phenoloxidase activity (P), urease activity (U), inositol (I), lactose (L), Potassium nitrate (N) and Creatinine assimilation (C), growth at 37°C (°C), Color (+) or no color changes (-) observed Creatinine dextrose bromothymol blue thymine (CDBT) agar and Canavanine-Glycine-Bromothymol Blue (CGB) agar and colonial morphotype in CHROMagar plates (CMa). Cryptococcus internal identification for each isolate (Cs1-Cs10).
Figure 2.

PCR results. PCR products obtained from the 10 Cryptococcus isolates. ITS fragments length were approximately 800 bp.
Figure 3.
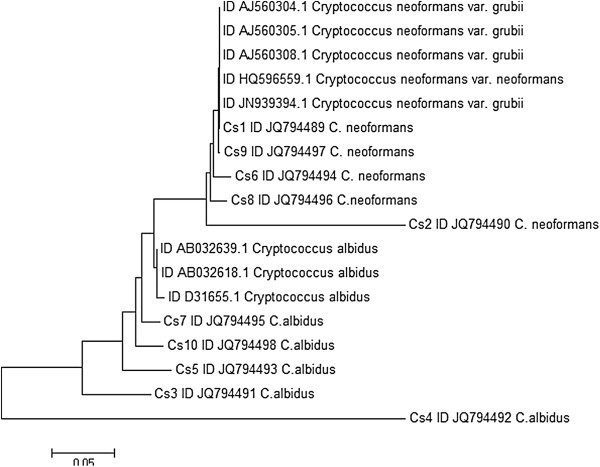
Phylogenetic tree. Phylogenetic relationship between 8 sequences obtained from Genbank and 10 generated in this research.
Discussion
Recent reports have indicated a significant prevalence of cryptococcosis in immunosuppressed patients in Monterrey city (Casillas 2012). The present study adds information about the distribution of this pathogenic yeast in the metropolitan area of Monterrey, a city with a very high population density and extreme temperature conditions in which the probability of Cryptococcus dissemination should be low.
Definitive Cryptococcus spp. identification of isolated yeasts was possible using previously reported biochemical, microbiological, and molecular characterization. From 50 pigeon excreta samples randomly collected from July to September of 2010 we identified five C. neoformans and five C. albidus isolates, representing an incidence of 20%. A similar incidence has been reported in Mexico City and in other countries such as Colombia and Perú ( Castañón Olivares et al. 2000; Ruiz et al. 1989; Rivas et al. 1999; Quintero et al. 2005; Curo et al. 2005; Colom Valiente et al. 1997;Castañol-Olivares and Lopez-Martinez, 1994). The finding that 50% of total Cryptococcus isolates corresponded to C. albidus is not surprising given the fact that 14% and 12% of pigeon excreta isolates in Seoul and Brazil correspond to this species and now it is considered an underestimated pathogen by several researchers ( Jang et al. 2011; Soares et al. 2005). Comparison of 18S RNA sequence against other reported sequences shows that the isolates form clades with their corresponding species according to the microbiological test results. However, two of them group together, mainly because of augmented intra-species sequence variability. This must be probed with a greater number of sequences, therefore it is important to continue to report Criptococcus spp. ITS sequences from other isolates in order to study genetic variation.
The information obtained in this research will inform the medical community about the presence of the etiologic agent of cryptococcosis in this area. Additional epidemiological studies may help set the stage for health campaigns to clean and prevent the accumulation of pigeon droppings in areas of high human traffic, such as subway stations and hospitals.
Acknowledgements
We thank Dr. Laura Raya Garza for support in the standardization of microbiological assays for the characterization of the strains, and Nestor Casillas Vega MD for contributing results obtained from his thesis. This work was developed by the program to support scientific and technological research at the Universidad Autónoma de Nuevo León (PAYCIT SA-3671).
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interest
This Project (No. SA367-10) was financially supported by the Universidad Autónoma de Nuevo León fromthe “Programa de Apoyo a la Investigación Científica y Tecnológica (PAICYT – UANL)”.
Authors’ contributions
YC-G: Took samples, isolated and characterized of Cryptococcus spp. and manuscript writing. RM-H: Revised validation of methodology bystatistic and collaborated with writing. JMA-R: Revised Microbiological characterization of isolates. ETA-C: Developed molecular methodology and revised DNA sequences, revised philogenetic three revised manuscript. All authors read and approved the final manuscript.
Contributor Information
Yolanda Canónico-González, Email: yolis.canonico@hotmail.com.
Juan Manuel Adame-Rodríguez, Email: jmadame@r.uanl.mx.
Roberto Mercado-Hernández, Email: romercad@hotmail.com.
Elva Teresa Aréchiga-Carvajal, Email: elva.arechigacr@uanl.edu.mx.
References
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Non Conventional Yeasts in Biotechnology. Berlin, Heidelberg: Springer Verlag; 1996. pp. 313–388. [Google Scholar]
- Casali AK, Goulart L, Rosa E, Silva LK, Ribeiro AM, Amaral AA, et al. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilianstate Rio Grande do Sul. FEMS Yeast Res. 2003;3:405–415. doi: 10.1016/S1567-1356(03)00038-2. [DOI] [PubMed] [Google Scholar]
- Casillas N. PhD thesis. Universidad Autónoma de Nuevo León: Facultad de Medicina; 2012. Fenotipificación y genotipificación del complejo Cryptococcusneoformans / Cryptococcusgattii en aislamientos clínicos del noreste de México. [Google Scholar]
- Castañol-Olivares LR, Lopez-Martinez R. Isolation of Cryptococcus neoformans from pigeon (Columalivia) droppings in Mexico City. Mycoses. 1994;37:325–327. doi: 10.1111/myc.1994.37.9-10.325. [DOI] [PubMed] [Google Scholar]
- Castañón Olivares L, Arreguin Espinosa R, Ruiz Palacios G, López Martínez A. Frequency of Cryptococcus sp. and varieties in México and their comparison with some Latin American Countries. Rev Latinoam Microbiol. 2000;42:35–40. [PubMed] [Google Scholar]
- Colom Valiente MF, Alberdi M, Meseguer I, Torres-Rodríguez JM. Aislamiento de Cryptococcus neoformans en muestras de medio ambiente de Alicante. Rev Iberoam Micol. 1997;14:63–64. [PubMed] [Google Scholar]
- Curo M, Salinas M, Casquero J. Cryptococcus neoformans in pigeon excreta, soil and air of palomares in the urban perimeter of ICA. Rev Peruana Medicina Experimental y Salud Pública. 2005;22:262–263. [Google Scholar]
- Dromer F, Mathoulin S, Dupont B, Y Laporte A. Epidemiology of cryptoccosis in France: a 9-year survey. Clin Infect Dis. 1996;23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- Ellis D, Pfeiffer T. The ecology of Cryptococcus neoformans. Eur J Epidemiol. 1992;8(3):321–325. doi: 10.1007/BF00158562. [DOI] [PubMed] [Google Scholar]
- Extreme temperatures and precipitation for Monterrey (OBS) 2000-2010 (in spanish) Servicio Meterológico Nacional. 2013. [Google Scholar]
- García P, García R, Hernández J, Marín P, Tallero E, Mira J. Identification of yeasts of clinical interest on CHRO Magar Candida culture medium Rev. Iberoamericana. 1998;15:131–135. [PubMed] [Google Scholar]
- Irokanulo EA, Akueshi CO, Makinde AA. Differentiation of Cryptococcus neoformans serotypes A and D using creatinine dextrose bromothymol blue thymine medium. Br J Biomed Sci. 1994;51(2):100–103. [PubMed] [Google Scholar]
- Ishaq C, y Bulmer GS, Felton FG. An evaluation of various environmental factors affecting the propagation of Cryptococcus neoformans. Mycol Appl. 1968;35:81–90. doi: 10.1007/BF02049570. [DOI] [PubMed] [Google Scholar]
- Jang YH, Lee SJ, Lee JH, Chae HS, Kim SH, Choe NH. Prevalence of yeast-like fungi and evaluation of several virulence factors from feral pigeons in Seoul. Korea Let App Microbiol. 2011;53(4):367–371. doi: 10.1111/j.1472-765X.2011.03009.x. [DOI] [PubMed] [Google Scholar]
- Khawcharoenporn T, Apisamthanarak A, Mundy LM. Non-neoformans cryptococcal infections: a systematic review. Infection. 2007;35:51–58. doi: 10.1007/s15010-007-6142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J ClinMicrobiol. 1982;15(3):535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- Mancianti F, Nardoni S, Ceccherelli R. Occurrence of yeasts in psittacines droppings from captive birds in Italy. Mycopathologia. 2002;153:121–124. doi: 10.1023/A:1014576304894. [DOI] [PubMed] [Google Scholar]
- Pal M, Baxter L. Isolation of Cryptococcus neoformans from the environment using a simplified sunflower seed medium. Proc N Z Microbiol Soc. 1985;29:155–158. [Google Scholar]
- Pérez C, Mata S, Hartung C, Roselló A, Colella MT, Olaizola C, Landaeta M. Preservation of Cryptococcus sp. with the Castellani method. Rev de la SociedadVenezolana de Microbiología. 2003;23:153–157. [Google Scholar]
- Quintero E, Castañeda E, Ruiz A. Environmental distribution of Cryptococcus neoformans in the Department of Cundinamarca Colombia. Rev Ibero Am Micología. 2005;22:93–98. doi: 10.1016/S1130-1406(05)70015-2. [DOI] [PubMed] [Google Scholar]
- Rivas F, De Martin MC, Rojas V. Cryptococcus neoformans isolation from the soil in Panama. Rev Med Panama. 1999;24(1):4–6. [PubMed] [Google Scholar]
- Rosario I, Acosta B, Colom F. Las palomas y otras aves como reservorio de Cryptococcus spp. Rev Ibero Am Micol. 2008;25:S13–S18. doi: 10.1016/S1130-1406(08)70020-2. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Velez D, Fromtling RA. Isolation of saprophytic Cryptococcus neoformans from Puerto Rico: distribution and variety. Mycopathologia. 1989;106:167–170. doi: 10.1007/BF00443058. [DOI] [PubMed] [Google Scholar]
- Shields AB, Ajello L. Medium for selective isolation of Cryptococcus neoformans. Science. 1966;151:218–223. doi: 10.1126/science.151.3707.208. [DOI] [PubMed] [Google Scholar]
- Soares M, Paula C, Dias A, Caseiro M, Costa SO. Environmental strains of Cryptococcus neoformans variety grubii in the city of Santos, SP. Brazil. 2005;47(1):31–36. doi: 10.1590/s0036-46652005000100006. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor S. Amplifications and direct sequencing of fungal ribosomal RNA genes for philogenetics. San Diego, CA: Academic Press, Inc; 1990. PCR protocols: a guide to methods and applications; pp. 315–322. [Google Scholar]


