Abstract
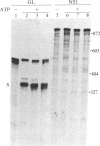
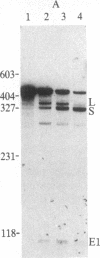
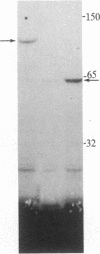
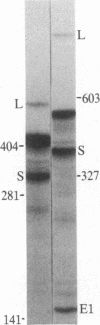
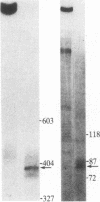
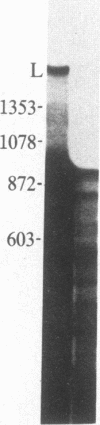
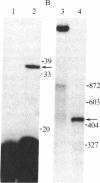
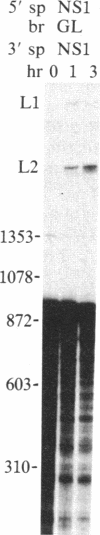
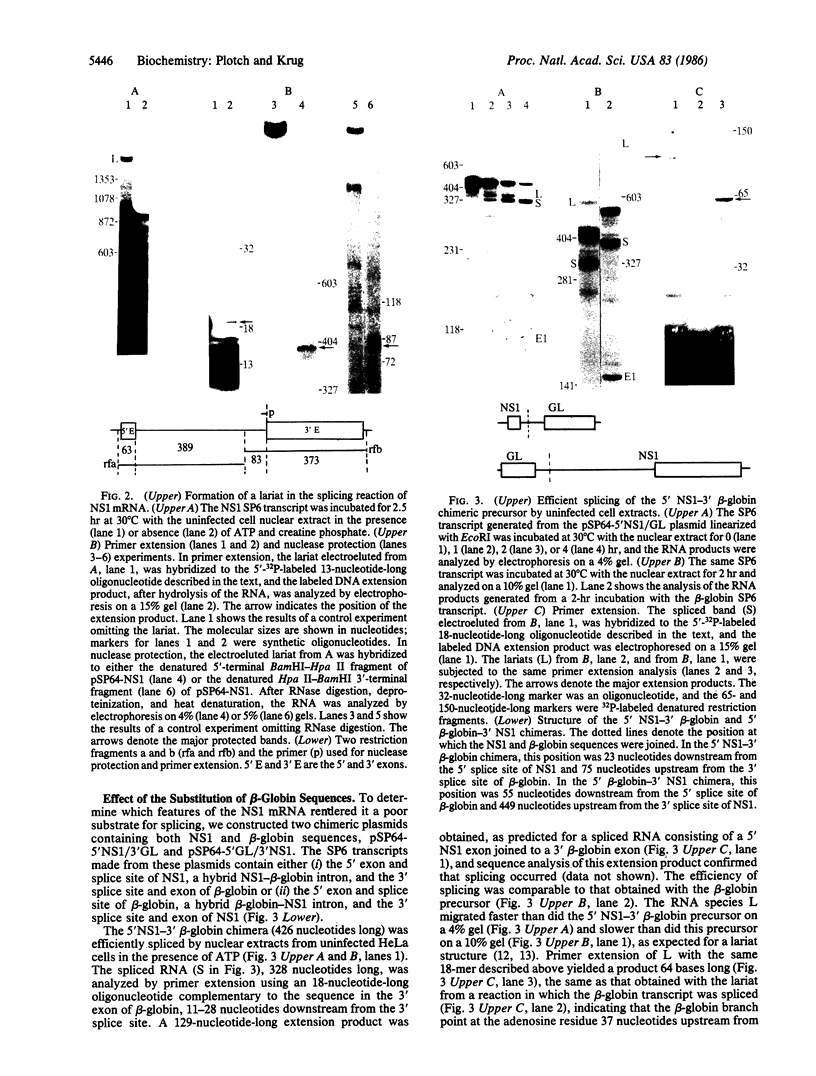
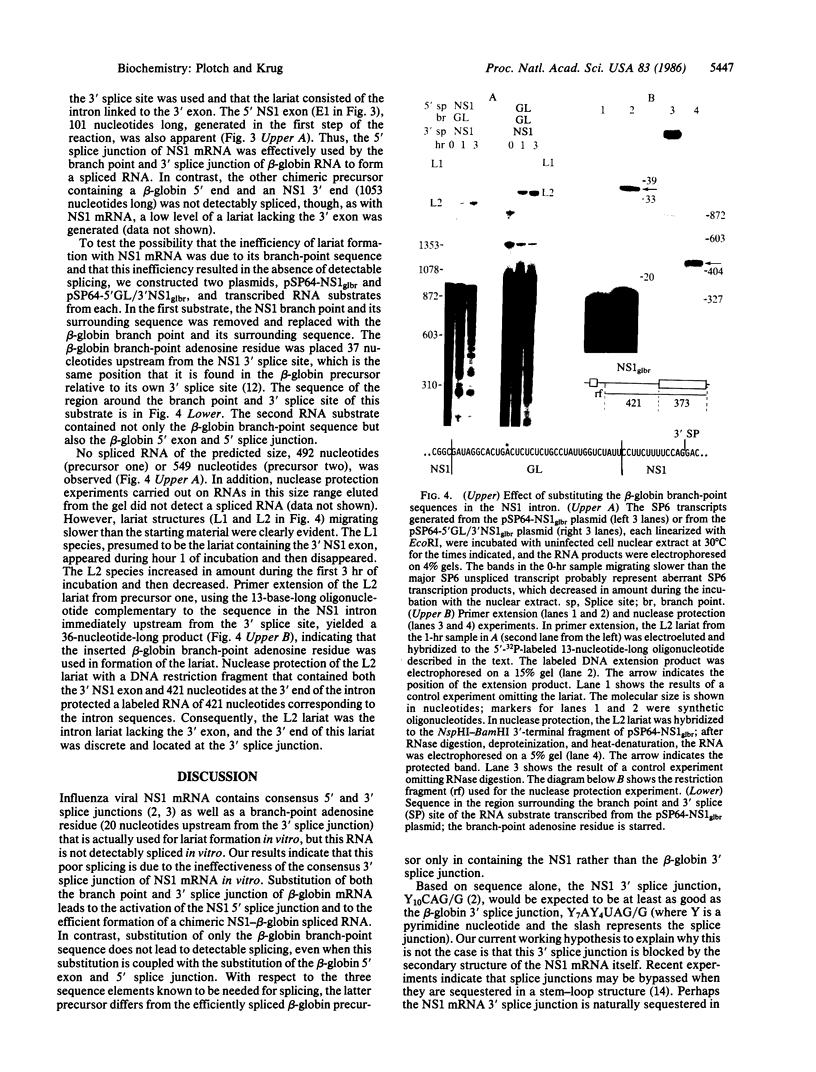
In influenza virus-infected cells, the splicing of the viral NS1 mRNA catalyzed by host nuclear enzymes is controlled so that the steady-state amount of the spliced NS2 mRNA is only 5-10% of that of the unspliced NS1 mRNA. Here we examine the splicing of NS1 mRNA in vitro, using nuclear extracts from HeLa cells. We show that in addition to its consensus 5' and 3' splice sites, NS1 mRNA has an intron branch-point adenosine residue that was functional in lariat formation. Nonetheless, this RNA was not detectably spliced in vitro under conditions in which a human beta-globin precursor was efficiently spliced. Using chimeric RNA precursors containing both NS1 and beta-globin sequences, we show that the NS1 5' splice site was effectively utilized by the beta-globin branch-point sequence and 3' splice site to form a spliced RNA, whereas the NS1 3' splice site did not function in detectable splicing in vitro, even in the presence of the beta-globin branch-point sequence or in the presence of both the branch-point sequence and 5' exon and splice site from beta-globin. With the chimeric precursors that were not detectably spliced, as with NS1 mRNA itself, a low level of a lariat structure containing only intron and not 3' exon sequences was formed. The inability of the consensus 3' splice site of NS1 mRNA to function effectively in in vitro splicing suggests that this site is structurally inaccessible to components of the splicing machinery. Based on these results, we propose two mechanisms whereby NS1 mRNA splicing in infected cells is controlled via the accessibility of its 3' splice site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Lai C. J., Dhar R., Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H., Leder P. Splicing and the formation of stable RNA. Cell. 1979 Dec;18(4):1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- Herz C., Stavnezer E., Krug R., Gurney T., Jr Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell. 1981 Nov;26(3 Pt 1):391–400. doi: 10.1016/0092-8674(81)90208-7. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krug R. M. Priming of influenza viral RNA transcription by capped heterologous RNAs. Curr Top Microbiol Immunol. 1981;93:125–149. doi: 10.1007/978-3-642-68123-3_6. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Khoury G. Deletion mutants of simian virus 40 defective in biosynthesis of late viral mRNA. Proc Natl Acad Sci U S A. 1979 Jan;76(1):71–75. doi: 10.1073/pnas.76.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Choppin P. W., Chanock R. M., Lai C. J. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1857–1861. doi: 10.1073/pnas.77.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J., Choppin P. W. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology. 1984 May;135(1):139–147. doi: 10.1016/0042-6822(84)90124-7. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980 Sep;21(2):475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J. Spliced and unspliced messenger RNAs synthesized from cloned influenza virus M DNA in an SV40 vector: expression of the influenza virus membrane protein (M1). Virology. 1982 Dec;123(2):237–256. doi: 10.1016/0042-6822(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Martinez H. M. An RNA folding rule. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):323–334. doi: 10.1093/nar/12.1part1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Inglis S. C. Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 1985 Sep;4(9):2313–2319. doi: 10.1002/j.1460-2075.1985.tb03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]