Abstract
Aim
The main objective of the study was to analyze the structure of data contained in the archives exported from a tomotherapy treatment planning system. An additional aim was to create an application equipped with a user-friendly interface to enable automatic reading of files and data analysis, also using external algorithms. Analyses had to include image registration, dose deformation and summation.
Materials and methods
Files from the archive exported from the tomotherapy treatment planning system (TPS) were analyzed. Two programs were used to analyze the information contained in the archive files: XML Viewer by MindFusion Limited and HxD hex editor by Maël Hora. To create an application enabling loading and analyzing the data, Matlab by MathWorks, version R2009b, was used.
Results
Archive exported from the TPS is a directory with several files. It contains three types of data: .xml, .img and .sin. Tools available in Matlab offer great opportunities for analysis and transformation of loaded information. Proposed application automates the loading of necessary information and simplifies data handling. Furthermore, the application is equipped with a graphical user interface (GUI). The main application window contains buttons for opening the archives and analyzing the loaded data.
Conclusion
The analysis of data contained in the archive exported from the tomotherapy treatment planning system allowed to determine the way and place of saving information of our interest, such as tomography images, structure sets and dose distributions. This enabled us to develop and optimize methods of loading and analyzing this information.
Keywords: Tomotherapy, Non-rigid image registration, Dose distribution, Matlab
1. Background
Tomotherapy is a combination of a therapeutic accelerator with a CT scanner.1 A fan beam is used for irradiation, and the dose is delivered with all possible orientations around the patient. As a result of the combination of rotation of the gantry with the advancing movement of the table, the source of radiation moves round the patient along a spiral track, as in spiral CT.2
The imaging system that was used enables the patient's daily verification and control of the size and location of the tumor. Thanks to that, physicians can observe the evolving tumor and, if necessary, decide to correct the treatment plan. Tomography images obtained this way can be used in the Planned Adaptive module.
The Planned Adaptive module of the treatment planning system allows to create new plans on the basis of verification of doses obtained from previously delivered fractions.3 The system recalculates the dose distribution using MVCT scans collected directly before a session of radiotherapy. The dose distribution obtained this way can be compared with the expected distribution and, if necessary, adjustments in the treatment plan can be made.
Images in the planning system are imposed in linear methods, dose summation is produced by adding a rigid grid of pixels. There is no possibility of including the deformation or change in the volume of various organs. Hence the idea to perform deformation of dose distributions using a non-rigid algorithms that allow local changes in shape and volume, then the summation of doses transformed this way. However, there is no publicly available software for this type of operation; hence the idea to create one.
Image registration is the process of unification of two (or more) images collected at different times, from different viewpoints or with different methods.4 This operation is performed to compare or complement (integrate) data collected in different sets. In medical imaging, image registration algorithms are used to combine images from different imaging systems (e.g. CT–MRI or CT–PET) and comparing images collected at different times.
Image registration algorithms can be divided according to an image transformation method used. The first group consists of algorithms that use linear transformations such as scaling, translation, rotation and other affine transformations. Linear transformations are global in nature and may not take into account local differences between registration images.
The second group consists of algorithms that use flexible transformation (so-called non-rigid). These algorithms may locally deform the target image in order to better match the reference image.
2. Aim
The main objective of this study was to analyze the structure of data contained in the archives exported from the tomotherapy treatment planning system. Specified objectives included: the analysis of the structure of files in the archive, the analysis of data contained in each file, development of a method for reading and correct interpreting of the information contained therein. An additional aim was to create an application equipped with a user-friendly interface to enable automatic reading of files and data analysis, also using external algorithms. Analyses had to include image registration, dose deformation and summation.
3. Material and methods
Files from the archive exported from the tomotherapy treatment planning system have been analyzed. These archives were exported using the Archive patient data option in the planning system5 or through a Data Management System (DMS).6 Such archives are created after patient radiation therapy process to free disk space on the server. They are then stored on storage media, such as DVDs, for future reference.
A tomotherapy archive includes files in three formats, with three different extensions: .xml, .img and .sin. For the analysis of the information contained in .xml files, the program used XML Viewer by MindFusion Limited. It allows to load a file and present its content in a convenient form of a data tree. To preview the contents of .img and .sin files, we used HxD hex editor created by Maël Hora.
Archives exported from the tomotherapy treatment planning system have a specific format. It is not possible to load the information contained therein to other programs or treatment planning systems, as is possible, for example, in the case of DICOM-RT format.
The possibility of loading information contained in the archives allows to perform operations that cannot be done in the tomotherapy treatment planning system, such as dose distribution deformation and non-rigid image registration.
To create an application to enable loading and analyzing data contained in the tomotherapy archive, we used Matlab by MathWorks, version R2009b. It contains Image Processing Toolbox which enables wide analysis of image data.7 Owing to the function enabling the creation of dose–volume histograms, evaluation and comparison of loaded dose distributions is available.8,9
The developed application uses external image fusion algorithms. There are two algorithms written by Dirk-Jan Kroon. The first is the registration with demon (fluid) algorithm,10,11 the second is the registration with a smooth b-spline grid.12 In addition, the second algorithm is used for rigid registration. The application to read xml files uses an external toolbox, written by Marc Molinari (XML Toolbox).13
4. Results
Archive exported from the Thomoterapy treatment planning system is a directory with several files. The directory name contains the patient's name and surname, ID number and date of export. It contains hundreds of files, most of them have names consisting of a sequence of numbers. Archives may have different sizes. Depending on the quantity of images contained, treatment plans, or the quantity of completed fraction, they can vary from 100 megabytes to 1.5 gigabytes.
4.1. Tomotherapy archive structure
Archive contains three types of files:
-
•
files with .xml extension – containing information in the form of a data tree;
-
•
files with .img extension – including graphical information, such as MVCT or kVCT images and dose distribution;
-
•
files with .sin extension – containing information in the form of sinograms.
Files ending with .xml (eXtensible Markup Language) provide information on structured format (data tree). Analysis of the contents of each file using XMLViewer makes it possible to understand the data contained therein and to develop a way to use them.
Four types of .xml files can be distinguished by suffix at the end of the file name. A file ended with _patient is a key one, as it provides information about the patient, the created plans, performed scans, etc. It also contains information about the contents of all the other files, as well as information needed to read and interpret them.
File ended with _machine contains information about the therapeutic machine on which treatment plans have been and/or will be performed. It contains some information about the components of the machinery and used parameters.
Files with the name ending in _imagingequipment contain information about devices used for patient imaging. They provide key information about the file containing the calibration curve, from which the computer system calculates the value of each pixel image on the actual density.
The largest collection of files end with _set. It contains information about structures’ contours. Data are represented by a set of pairs of numerical values which determine the location of points on the CT scans. Each scan corresponds to a branch in the structure of the file. One file stores information on a single structure.
The second group are the files ending in .img. They contain graphic data such as computed tomography images, on which the treatment plan was made, megavolts computed tomography (MVCT) images obtained on the therapeutic machine for correct positioning of the patient, and calculated dose distributions.
These files do not contain any information in the header, so it is impossible to open them in popular graphic programs. Data necessary to read them, such as image size, the number of bits per pixel, or a compression, are stored in a specific branch in a file ended with _patient.xml. All slices are stored in one data file.
In the case of tomographic images, each pixel is described by a nonnegative integer. These values can be easily converted to values expressed in Hounsfield units (HU).
Elements of files with dose distribution are described by nonnegative real numbers. They define explicitly dose value located within an item.
The last group are data files in the form of sinograms. They contain information about how the collimator leaves work (opening time) for each fraction planned. For performed fractions, sinograms contain information collected during patient irradiation through detectors matrix. The files with the .sin extension also hold calibration curves for imaging devices and data from Airscan procedures.
4.2. Loading data
Loading the contents of files with the .xml extension to the Matlab environment is possible by using external package (XML Toolbox). This package reads the file into a structures type variable, maintaining a data tree structure. Another operation on such data, which has been downloaded already, held in the usual way.
The information contained in files with extensions .img and .sin can be loaded into the memory by using Matlab environment's standard commands. Essential in performing this operation correctly is information contained in specific branches of the file ending with _patient.xml (branch called arrayHeader). Information stored there describe: the file name (key binaryFileName), the size of the data matrix (key dimensions) and the precision (key dataType).
Once the contents of files are appropriately loaded into the memory, they become standard variables. In the case of tomography images and dose distribution (.img files) they are three-dimensional arrays, where the third dimension corresponds to the successive slices. For data from .sin files, they are two-dimensional arrays.
4.3. Application
Tools available in Matlab offer great opportunities for analysis and transformation of loaded information. Proposed application automates the loading of necessary information and simplifies data handling. In order to simplify its use, the application is equipped with a graphical user interface (GUI).
The main application window contains buttons for opening the archives and displaying tomography images with dose distribution and sets of structures. The menu bar contains several options to allow execution of functions and analysis of loaded data.
After selecting a plan to display, the program loads into its memory appropriate .img files with CT images and dose distribution and .xml files with a set of structures. All these data are displayed in the preview window (Fig. 1), containing a series of controls for changing the display (choice of projection, slice, change of values and colors of isodoses, change of displayed structures, zoom in and out).
Fig. 1.
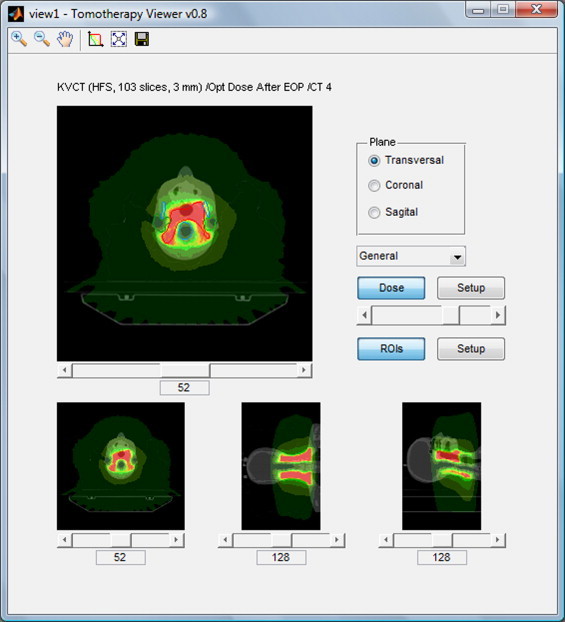
Developed application – preview window.
There are also two buttons in the preview window. The first allows you to save displayed images in an image file (tiff format), the second calls the function of the dose–volume histogram for the currently displayed contours (Fig. 2).
Fig. 2.
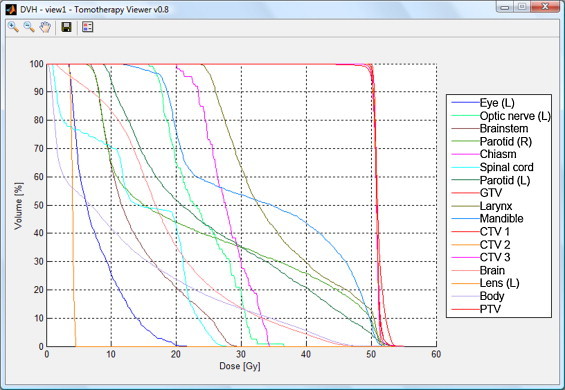
Dose–volume histogram created in the application.
The application menu contains two groups of commands: Register and Dose. Functions called by the command of the first group allow to call image registration window, load the results of previously executed registration and reads and shows the shifts which were accepted during patient positioning verification.
The image registration window contains a set of controls enabling o change the options of image registration (Fig. 3). The user can select the method of registration (rigid, affine, non-rigid demon, b-spline), modes (original or reduced resolution) and way of comparison. Other options allow to create a mask with weights based on the structures set, usage of initial shifts accepted during patient positioning verification or to save a matrix of deformation in a file for further use.
Fig. 3.
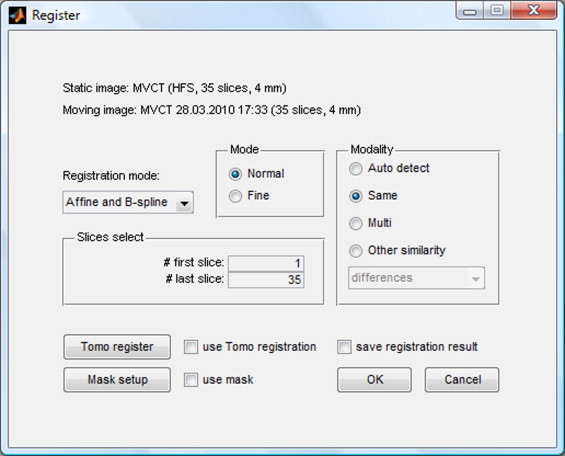
Image registration window.
The developed application was equipped with four methods of image registration. Two rigid type algorithms include linear transformation (shifts and scalar multiplications) and affine transformation. The two types non-rigid algorithms are demon registration algorithm, which is a type of a fast fluid like registration and a b-spline based on a grid of control points.
Once the scans are prepared, the process of registration begins. Then, the preview window displays registered images and calculated average image displacement vectors as well as other statistics for structures set. The result of the algorithm is a matrix of deformation which describes, how to deform an image to fit it to another. Matrix of deformation has a form of three arrays (one for each direction) where each cell corresponds to one voxel and contains information about the transfer. In the case of rigid algorithms, the average value of the array corresponds to a shift the whole image made in that direction.
The second group of commands (Dose) contains features for deformation, loading, summation and comparison of dose distributions. The first function allows the deformation of dose distribution using the previously stored deformation matrix from the image registration. This function deforms the selected dose distribution in the same way, as the moving image was deformed to be fitted to the base image. Deformed dose distribution can be displayed and/or saved in a file.
Another feature allows one to load dose distribution (saved deformed dose distribution or dose distribution from the Planned Adaptive module). The application automatically finds and loads CT scans and the set of ROIs associated with the dose distribution.
The third function allows summing selected dose distributions. Deformed doses as well as doses from standard plans and from the Planned Adaptive module can be summed up. For each dose a multiplier factor can be defined, allowing to assign a different relevance to each dose distribution. Owing to that, it is possible to sum up dose distributions, even if dose distributions are not created for all deformed fractions and some of them have to be multiplied.
The fourth and the last function from the Dose group allows to compare two dose distributions. The result of comparison is presented in two forms: a dose–volume histogram, where a solid line represents the first distribution and a dashed line the second one, and a map of differences imposed on the CT scans (Fig. 4).
Fig. 4.
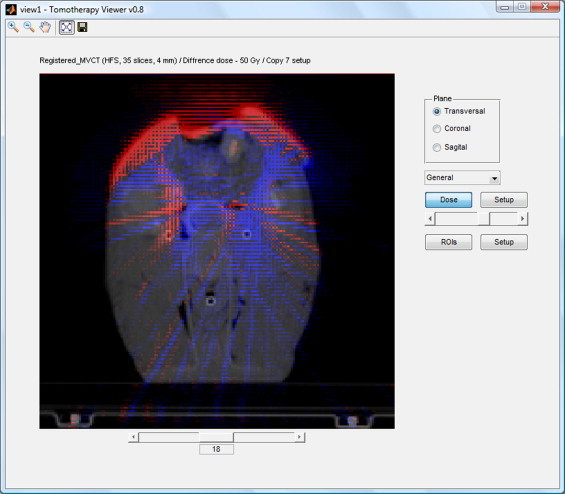
Map of differences in dose distributions imposed on CT scans.
The last feature of the application is the summation of dose distributions from different treatment plans for a single patient (for example, successive stages of treatment). The condition is that these plans were calculated on the same sets of CT scans.
5. Discussion
A key element to understand the structure of archives exported from tomotherapy treatment planning system was the content of .xml file ending with _patient. Analysis of the information contained in it helped to find and understand the information contained in all other files. The information contained in specific branches (called arrayHeader) made it possible to load the contents of files containing the graphical data (extended with .img) and the data in the form of sinograms (extended with .sin) to the Matlab environment.
To be able to properly read the information contained in files with .img and .sin extensions, it was necessary to change the order of reading bits/bytes in the file (machineformat parameter). Default for Windows is a little-endian ordering, while for correct reading of data from the archive it was necessary to use a big-endian ordering, as is common in Unix family systems.
All slices in the set of tomographic scans (dose distributions) are stored in one .img file. Therefore, the loading must be done slice by slice in a loop, where the number of runs is equal to the number of slices. In addition, to display the images in the correct orientation, it is necessary to transpose the matrix with a single slice. By saving all the slices into one three-dimensional matrix, it is possible to achieve the two other tomography projections (coronal and sagital).
The application thus developed automates and simplifies the loading and analysis of the data contained in the archive exported from the tomotherapy treatment planning system. One of the main features is the ability to create dose–volume histograms for a set of loaded structures. The application allows to create a histogram including or not an Overlap Priority parameter.
A single voxel can only belong to one structure. In the case of overlapping structures, a common voxel is assigned to this structure where Overlap Priority parameter is lower. That creates holes in the structures of a higher value of this parameter.
The possibility to create a dose–volume histogram without using an Overlap Priority parameter allows to compare plans created in the tomotherapy treatment planning system with the plans created in other systems. Comparison based on histograms exported from the tomotherapy planning system, particularly in largely overlapping structures, may deliver ambiguous results.
To register images, our application uses external algorithms; therefore, we had no influence on their performance. To speed up the algorithms we had to prepare the input data properly. Double reduction of resolution of scans shortened calculation time, particularly in the case of non-rigid registration. However, in the case of rigid option and a large number of slices, the initial reduction of images takes longer than the time saved by speeding up the algorithm. The initial shift of images based on values accepted during patient positioning verification has also some influence on the shortening of image registration.
6. Conclusions
The analysis of data contained in the archive files exported from the tomotherapy treatment planning system allowed to determine the way and place of saving information of our interest, such as tomography images, structures set and dose distributions.
This enabled the development and optimization of loading of this information. It also allows to continue further analysis, also using external algorithms that require specific input format.
The result is an application that allows loading of treatment plans; display of tomographic images dose distributions and sets of structures; creation of dose–volume histograms; registration of images with non-rigid methods; deformation and summation of dose distributions.
The application has a great potential for development. All it takes is simply to add new functions. The search for the method of verification of results obtained by using the existing features of the application is currently in progress.
Conflict of interest
Authors do not have conflict of interest.
References
- 1.Mackie T.R. History of tomotherapy. Phys Med Biol. 2006;51(13):R427–R453. doi: 10.1088/0031-9155/51/13/R24. [DOI] [PubMed] [Google Scholar]
- 2.Mackie T.R., Holmes T., Swerdloff S., Reckwerdt P., Deasy J.O., Yang J. Tomotherapy: a new concept for the delivery of dynamic conformal radiotherapy. Med Phys. 1993;20(6):1709–1719. doi: 10.1118/1.596958. [DOI] [PubMed] [Google Scholar]
- 3.TomoTherapy Incorporated . 2007. TomoTherapy Hi·Art System® Planned Adaptive Guide 100527 A. [Google Scholar]
- 4.Zitova B., Flusser J. Image registration methods: a survey. Image Vis Comput. 2003;21(11):977–1000. [Google Scholar]
- 5.TomoTherapy Incorporated . 2007. TomoTherapy Hi·Art System® Getting Started Guide 100521 A. [Google Scholar]
- 6.TomoTherapy Incorporated . 2007. TomoTherapy Hi·Art System® Data Management System 100742 A. [Google Scholar]
- 7.Shirazi A., Mahdavi S., Sardari D., Sadri L. Portal image contrast enhancement. Rep Pract Oncol Radiother. 2006;11(1):23–28. [Google Scholar]
- 8.Ślosarek K., Grządziel A., Szlag M., Bystrzycka J. Radiation Planning Index for dose distribution evaluation in stereotactic radiotherapy. Rep Pract Oncol Radiother. 2008;13(4):182–186. [Google Scholar]
- 9.Petrovic B., Grządziel A., Ślosarek K. Quality Assurance of TPS: comparison of dose calculation for stereotactic patients in Eclipse and iPlan RT Dose. Rep Pract Oncol Radiother. 2009;14(1):5–10. [Google Scholar]
- 10.Kroon D. Multimodality non-rigid demon algorithm image registration. Available from: http://www.mathworks.com/matlabcentral/fileexchange/21451-multimodality-non-rigid-demon-algorithm-image-registration [access April 2010].
- 11.Kroon D., Slump C.H. MRI modality transformation in demon registration. Proceedings of the Sixth IEEE international conference on symposium on Biomedical Imaging: from Nano to Macro; Boston, MA, USA, June 28–July 01; Piscataway, NJ: IEEE Press; 2009. pp. 963–966. [Google Scholar]
- 12.Kroon D. B-spline Grid, Image and Point based Registration. Available from: http://www.mathworks.com/matlabcentral/fileexchange/20057-b-spline-grid-image-and-point-based-registration [access April 2010].
- 13.Molinari M. XML Toolbox. Available from: http://www.mathworks.com/matlabcentral/fileexchange/4278-xml-toolbox [access April 2010].


