Abstract
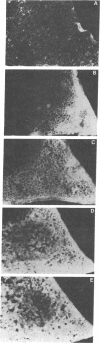
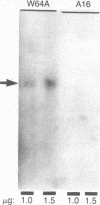
The scutellum of maize is a fully differentiated, nondividing, diploid embryonic tissue. Two distinct structural genes (Cat1 and Cat2) encoding the enzyme catalase (CAT) are differentially expressed in this tissue during postgerminative development. As development proceeds, the expression of Cat1 diminishes, while that of Cat2 is enhanced, leading to the disappearance of the CAT-1 protein and the gradual accumulation of the CAT-2 protein. The present investigation was undertaken to determine whether all scutellar cells may be genetically programmed to activate expression of Cat2 synchronously or whether there is an asynchronous spatial gradient of Cat2 activation. By using immunofluorescence microscopy and anti-CAT-2 IgG, we have found that a gradient of Cat2 activation occurs within the scutellar cell mass during postgerminative development. The gradient of Cat2 activation occurs from the outer perimeter of the tissue inward toward the embryonic axis. To determine a potential site of origin for any putative "triggering signal" for Cat2 activation, we demonstrated that Cat2 is expressed in the single layer of aleurone cells prior to its expression in any other tissue during kernel development. To our knowledge, this is the first observation of a gradient-type spatial pattern of a eukaryote gene activation occurring in a stable, virtually nondividing tissue such as the maize scutellum. The significance of these results with respect to developmental gene regulation is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BEERS R. F., Jr, SIZER I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952 Mar;195(1):133–140. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gierer A. Biological features and physical concepts of pattern formation exemplified by hydra. Curr Top Dev Biol. 1977;11:17–59. doi: 10.1016/s0070-2153(08)60742-5. [DOI] [PubMed] [Google Scholar]
- Hall T. C., Ma Y., Buchbinder B. U., Pyne J. W., Sun S. M., Bliss F. A. Messenger RNA for G1 protein of French bean seeds: Cell-free translation and product characterization. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3196–3200. doi: 10.1073/pnas.75.7.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey B. M., Oaks A. The Hydrolysis of Endosperm Protein in Zea mays. Plant Physiol. 1974 Mar;53(3):453–457. doi: 10.1104/pp.53.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Akazawa T. Enzymic Mechanism of Starch Breakdown in Germinating Rice Seeds: 8. Immunohistochemical Localization of beta-Amylase. Plant Physiol. 1979 Aug;64(2):337–340. doi: 10.1104/pp.64.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik C. M., Storti R. V., Jacobson A. Fractionation and functional analysis of newly synthesized and decaying messenger RNAs from vegetative cells of Dictyostelium discoideum. J Mol Biol. 1979 Mar 5;128(3):371–395. doi: 10.1016/0022-2836(79)90093-7. [DOI] [PubMed] [Google Scholar]
- Quail P. H., Scandalios J. G. Turnover of genetically defined catalase isozymes in maize. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1402–1406. doi: 10.1073/pnas.68.7.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rave N., Crkvenjakov R., Boedtker H. Identification of procollagen mRNAs transferred to diazobenzyloxymethyl paper from formaldehyde agarose gels. Nucleic Acids Res. 1979 Aug 10;6(11):3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios J. G., Chang D. Y., McMillin D. E., Tsaftaris A., Moll R. H. Genetic regulation of the catalase developmental program in maize scutellum: Identification of a temporal regulatory gene. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5360–5364. doi: 10.1073/pnas.77.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios J. G. Genetic control of multiple molecular forms of catalase in maize. Ann N Y Acad Sci. 1968 Jun 14;151(1):274–293. doi: 10.1111/j.1749-6632.1968.tb11896.x. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J. Quantitative ethanol precipitation of nanogram quantities of DNA and RNA. Anal Biochem. 1981 Jan 1;110(1):229–231. doi: 10.1016/0003-2697(81)90139-1. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Tsaftaris A. S., Bosabalidis A. M., Scandalios J. G. Cell-type-specific gene expression and acatalasemic peroxisomes in a null Cat2 catalase mutant of maize. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4455–4459. doi: 10.1073/pnas.80.14.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information and pattern formation. Curr Top Dev Biol. 1971;6(6):183–224. doi: 10.1016/s0070-2153(08)60641-9. [DOI] [PubMed] [Google Scholar]