Abstract
The receptor for activated C kinase 1 (RACK1) serves as an adaptor for a number of proteins along the MAPK, protein kinase C, and Src signaling pathways. The abundance and near ubiquitous expression of RACK1 reflect its role in coordinating signaling molecules for many critical biological processes, from mRNA translation to cell motility to cell survival and death. Complete deficiency of Rack1 is embryonic lethal, but the recent development of genetic Rack1 hypomorphic mice has highlighted the central role that RACK1 plays in cell movement and protein synthesis. This review focuses on the importance of RACK1 in these processes and places the recent work in the larger context of understanding RACK1 function.
Keywords: RACK1, translation, ribosomes, stress response, cell migration, cancer
Background
The 36-kDa receptor for activated C kinase 1 (RACK1) was first identified in a rat brain cDNA library screen designed to isolate anchor proteins that bound protein kinase C (PKC) in the presence of its activators diacylglycerol, calcium, and phosphatidylserine. Inhibition of the interaction was later shown to destabilize PKC and reduce the phosphorylation of its substrates.1-3 However, because RACK1 contained no catalytic domains, it was suggested to function as a scaffold protein that shuttled activated PKC to the sites of its substrates.4 Although the RACK1–PKC interaction was the first to be described, it is now known that RACK1 interacts with numerous proteins in distinct cellular compartments and plays a critical role in many fundamental physiological processes.
RACK1 belongs to the family of WD40 repeat proteins (short ~40-amino acid motifs often terminating in a Trp-Asp [W-D] dipeptide, also known as WD or β-transducin repeats). More than 100 WD repeat proteins are known, and although they play varying roles, they share an ability to function as protein scaffolds.5 WD domains adopt β-propeller structures that are thought to serve as docking sites for interacting proteins.6 RACK1 contains a 7-bladed β-propeller structure and shows high homology to Gβ, the best-characterized WD repeat protein. RACK1 is evolutionarily conserved in yeast to humans, and the crystal structures from 4 species, including humans, have been determined.7-12
Expression of RACK1, encoded by the GNB2L1 (guanine nucleotide binding protein [G protein], β polypeptide 2-like 1) gene, is ubiquitous and tightly regulated.13 Indeed, aberrant expression is associated with numerous pathologies, including cancer and age-related diseases.14-16 More than 100 proteins are known to interact with RACK1, either directly or as part of a complex. Among these 100 proteins, 72 have been experimentally validated as binding partners that are functionally modulated by RACK1 (Table 1). Considering the diverse functions of these proteins, it is not surprising that RACK1 has been implicated in such fundamental processes as cell growth, proliferation, morphology, movement, death, and survival. Table 1 describes the 72 RACK1-interacting proteins segregated into functional clusters.
Table 1.
Putative Functional Binding Partners of RACK1
| Function | Protein | Score | Function | Protein | Score |
|---|---|---|---|---|---|
| Protein kinases | PRKCE | 0.845 | Cell motility/polarity/organization | PDE4D | 0.845 |
| SRC | 0.845 | PTPRM | 0.845 | ||
| PRKCA | 0.619 | STAT3 | 0.825 | ||
| PRKCB | 0.619 | INSR | 0.824 | ||
| PRKCD | 0.559 | IGF1R | 0.821 | ||
| PRKCQ | 0.559 | ITGB1 | 0.808 | ||
| PRKD1 | 0.559 | TUBB2A | 0.619 | ||
| IFN/immune signaling | IFNAR2 | 0.911 | DNM1 | 0.619 | |
| STAT1 | 0.784 | PLEC | 0.619 | ||
| NSMAF | 0.619 | PLCG1 | 0.619 | ||
| CSF2RB | 0.619 | SERBP1 | 0.552 | ||
| SLC9A3R1 | 0.619 | ITGB7 | 0.538 | ||
| IL2RB | 0.569 | ITGB2 | 0.530 | ||
| IL4R | 0.569 | ITGB5 | 0.530 | ||
| JAK1 | 0.569 | Neuronal function/brain | FYN | 0.845 | |
| JAK3 | 0.569 | GABRB3 | 0.619 | ||
| JAK2 | 0.569 | SYT1 | 0.619 | ||
| TYK2 | 0.569 | GRIN2B | 0.619 | ||
| EPOR | 0.569 | SLC6A16 | 0.530 | ||
| IFNAR1 | 0.530 | SLC6A3 | 0.530 | ||
| Ribosome/translation | RPS13 | 0.837 | Others | GNB1 | 0.619 |
| RPS5 | 0.813 | AGTRAP | 0.619 | ||
| HABP4 | 0.792 | AR | 0.619 | ||
| RPS20 | 0.673 | RASA3 | 0.619 | ||
| EIF6 | 0.619 | NFATC1 | 0.619 | ||
| LARP4B | 0.619 | RASA1 | 0.619 | ||
| PABPC1 | 0.619 | SUMO4 | 0.619 | ||
| RPS3 | 0.554 | SREK1 | 0.603 | ||
| RPS15 | 0.548 | SAP25 | 0.591 | ||
| Cell death/survival | TP63 | 0.809 | LRP12 | 0.563 | |
| TNFRSF1A | 0.619 | ST7 | 0.563 | ||
| TP73 | 0.555 | OTUB1 | 0.562 | ||
| BCL2L11 | 0.538 | GRAP2 | 0.543 | ||
| RB1 | 0.530 | DLC1 | 0.538 | ||
| Cell proliferation | ACHE | 0.619 | HIF1A | 0.530 | |
| CCNA1 | 0.562 | SAT1 | 0.530 |
Note: Seventy-two binding partners with functional relevance were suggested using the String 9.0 database (http://string-db.org) (setting: threshold of 0.5; prediction mode: experiment; protein names and string score are listed). Proteins were manually sorted into putative functional clusters.
Here, we focus on the role of RACK1 in the cellular control of kinase activity in the stress response and its implication for cell migration and mRNA translation. For a more comprehensive discussion of RACK1, the reader is referred to a recently published review.5
RACK1 as a Hub for Protein Kinases
The diverse functions of RACK1 are mediated by acting as a scaffold for its binding partners, which may be constitutively associated (e.g., Src17) or bound in a transient stimulus-dependent manner (e.g., PKCβII), often through interactions between RACK1 WD repeats and modular structural domains of the binding partners (i.e., SH2 of Src). The function of RACK1 within these complexes also varies. For example, RACK1 may shuttle proteins to their sites of action (PKC), facilitate cross-talk between distinct signaling pathways (PKC-MAPK), or recruit other signaling proteins into the complexes. Moreover, binding of RACK1 to kinases can result in increased (JNK, MAPKs) or decreased (Src17,18) catalytic activity. At any given time, multiple RACK1 complexes exist that are coordinated in a dynamic stimulus- and compartment-dependent manner. One example of the latter is the function of RACK1 in suppressing Src tyrosine kinase activity at the membrane. PKC activation enhances the Src–RACK1 interaction,18 which can be disrupted upon activation of IGF-1R.19,20 Another example of compartmentalized RACK1 function occurs during regulation of the circadian cycle. Here, PKCα and RACK1 form a nuclear complex with the transcription factor BMAL1. Interestingly, the PKCα–RACK1–BMAL1 complex is formed in a rhythmic manner during the negative feedback phase of the circadian cycle, and complex formation suppresses CLOCK-BMAL1 transcriptional activity. Consequently, disruption of the complex results in a shortened circadian period.21
As mentioned above, RACK1 was initially identified as a PKCβII-anchoring protein that stabilizes the active conformation of the kinase and shuttles it to its substrates. RACK1 also serves as an adaptor to link PKC activation to other signaling pathways, such as the MAPK pathway. We have previously demonstrated that the RACK1–PKC complex binds to JNK in response to several stress stimuli, including ultraviolet (UV) irradiation or treatment with a PKC-activating phorbol ester.22 Once bound, JNK is phosphorylated by PKC at serine 129, and although this is insufficient to activate JNK, S129 phosphorylation augments JNK activation by the canonical upstream kinases MKK4 and MKK7 (Fig. 1).
Figure 1.
RACK1 signaling complexes. RACK1 links PKC signaling to the MAP kinase pathway: RACK1 binding to activated PKC leads to the recruitment of JNK into the complex. PKC-mediated phosphorylation of JNK augments JNK activation by the canonical JNK pathway. JNK activates c-Jun, which in turn regulates the transcription of cyclin D1 and RACK1. RACK1 further stabilizes c-Jun by inhibiting its proteasomal degradation through Fbw7. By enhancing JNK–c-Jun activity, RACK1 is implicated in tumorigenesis. RACK1 regulates apoptosis via MAPK and Src: RACK1 binds to MAP3K and activates stress-induced MAPK signaling, which results in apoptosis. In contrast, sequestration of RACK1 into stress granules attenuates MTK1 activation, which results in the inhibition of apoptosis. Binding of RACK1 to Src inhibits Src kinase activity. Upon apoptotic stimuli, inhibition of Src results in the oligomerization of Bax at the outer mitochondrial membrane, caspase activation, and cell death. RACK1 induces the oligomerization of Bax at the outer mitochondrial membrane, caspase activation, and cell death. RACK1 regulates Src activity during migration: Tight regulation of Src activity is necessary to mediate paxillin dynamics at focal adhesions during migration. Src phosphorylates paxillin, and RACK1 regulates paxillin phosphorylation dynamics by binding and releasing Src at its site of action, thereby enabling migration.
RACK1 can also influence the fate of the cell following exposure to stressors such as UV, H2O2, or sorbitol through regulation of the upstream MAP3K, MTK1. RACK1 binding is essential for the activation of MTK1, which induces apoptosis via the downstream stress-activated p38 and JNK enzymes.23 Interestingly, RACK1 is also able to prevent apoptotic responses to other stressors (e.g., arsenite) by associating with stress granules.24 Recruitment of RACK1 to stress granules significantly reduces its ability to associate with MTK1 and therefore inhibits the MTK1-dependent apoptotic stress response in a seemingly passive manner. On the other hand, sequestration of RACK1 in stress granules might be an active mechanism to inhibit translation, although this remains to be clarified. The function of RACK1 in the stress response is not confined to mammalian cells. In yeast, RACK1 appears to control the stress response by regulating translation of the tyrosine phosphatases that negatively regulate the MAPK pathway.25
Genetic Mouse Models of RACK1 Deficiency
The first mouse models of RACK1 deficiency have been developed only recently.26 Although complete knockout of the gene fails to produce viable embryonic stem cells, a Rack1 hypomorphic allele was successfully constructed by introducing a deletion into intron 2. Crossing of heterozygous (ΔF/+) mice produced wild-type and ΔF/+ animals at the expected Mendelian frequency, but homozygosity at the defective allele (ΔF/ΔF) was lethal at the gastrulation stage. The most obvious phenotype of the ΔF/+ animals was a pigmentation deficit, evident as a white belly spot and hypopigmented tail and paws. Other developmental abnormalities of varying severity were found in young ΔF/+ females but not in male littermates. Notably, these mice also showed defects in protein synthesis. Given the relatively mild downregulation of RACK1 expression in ΔF/+ animals, the observed pigmentation and protein expression phenotypes are consistent with a critical role for RACK1 in controlling cell (melanocyte/melanoblast) migration and mRNA translation.
We have previously described a role for RACK1 in melanoma development and resistance to therapy, but a link between this protein and normal melanocyte biology has not been established. During development, melanocytes are specified from pluripotent neural crest cells of the neural tube, which undergo epithelial-to-mesenchymal transition and migrate as melanocyte precursors (melanoblasts) over large distances to invade and populate the epidermis and hair follicles.27 Melanoma cells retain several key features of melanocytes, including the ability to dissociate from an epithelial microenvironment, invade surrounding tissues, and migrate to distant organs. Lineage-specific factors that regulate melanocyte development, such as the tyrosine kinase receptor KIT and microphthalmia-associated transcription factor (MITF), are reactivated or mutated during melanomagenesis and are important for melanoma progression and metastasis. The pigmentation phenotype observed in the ΔF/+ Rack1 mice might thus result from impaired migration of melanoblasts or reduced numbers of mature melanocytes at the affected sites. Several key questions that remain to be addressed include whether RACK1 affects MITF, the master regulator of melanocytes, or directly affects the migration of melanocytes. The results of such studies will undoubtedly provide insights not only into melanocyte biology but also into the development and progression of melanoma.
RACK1 in the Control of Cell Migration
Cell migration is a dynamic process involving multiple steps: cytoskeletal reorganization to form leading edge protrusions, turnover of focal adhesions, generation of mechanic forces, retraction of the cell tail, and detachment from the surrounding extracellular matrix. There is mounting evidence that RACK1 is involved at several stages of this process.
RACK1 interacts with the cytoplasmic domain of the integrin β1/2 subunit in 293 and JY lymphoblastoid cells, respectively.28 RACK1 has been proposed to integrate IGF-1R and integrin signaling, and its scaffolding functions are necessary to regulate focal adhesion turnover and cell migration in v-Src- or IGF-1R-transformed fibroblasts or epithelial cells.29 RACK1 enhances IGF-1R-mediated migration in breast and prostate cancer cells (MCF7 and DU145) by a mutually exclusive association with phosphatase 2A or β1 integrin.30,31 Tight regulation of focal adhesion turnover, a process associated with cell migration, has also been shown to be regulated by RACK1, in part through its effect on Src activity with a concomitant effect on paxillin phosphorylation at focal adhesion complexes (Fig. 1).32
RACK1 also functions in direction sensing of migrating cells. A complex of RACK1, focal adhesion kinase (FAK), and the cAMP-degrading phosphodiesterase PDE4D5 is recruited to nascent adhesions, where it modulates cell polarity.33 The RACK1–FAK–PDE4D5 complex acts by signaling to its small GTPase target Rap1 via the cAMP downstream target exchange protein activated by cAMP (EPAC, also known as RapGEF3). Disruption of this complex inhibits directional responses such as wound-induced polarization and chemotactic invasion.33,34
RACK1 seems to have a different function in nonadherent cells. For example, binding of chemoattractants to G protein-coupled receptors on Jurkat cells induces the association of RACK1 with Gβγ subunits and recruitment of the complex to the leading edge, where it inhibits chemotaxis. RACK1 is thought to negatively regulate migration by preventing Gβγ stimulation of phosphoinositide 3-kinases and phospholipase C.35,36 Thus, although RACK1 clearly plays a role in fine- tuning the events that control cell adhesion and motility, whether RACK1 enhances or inhibits these processes seems to be cell, context, or stimulus dependent.
RACK1 in Disease
At least some of the importance of RACK1 in disease pathology results from its ability to modulate the innate immune response and activation of interferon (IFN) signaling. Many pathogens, especially viruses, evade the host immune response by interfering with IFN signaling. RACK1 interacts with the IFNα/β receptor and recruits STAT1 into this complex, which is required for IFN signaling.37 The mumps virus protein V and the measles virus accessory proteins C and V bind to RACK1, which induces dissociation of the IFNAR–RACK1–STAT1 complex and inhibits the IFN response.38,39 In addition to its role in innate immunity, RACK1 has been implicated in several psychological disorders and neuronal diseases. These aspects of RACK1 function are discussed in more detail in a recent review.5
Considering the pivotal role of RACK1 in cell growth, migration, and cell death, it is not surprising that it has been implicated not only in the development of cancer but also in the response to therapy. RACK1 has been suggested to be a prognostic marker in breast cancer, and elevated expression of this protein is associated with poor clinical outcome.40,41 Furthermore, RACK1 promotes proliferation, migration, and metastasis of breast cancer cells in vitro and in vivo through activation of the RhoA/Rho kinase pathway.42,43 RACK1 is thought to confer chemoresistance in breast cancer cells by promoting proteasomal degradation of the proapoptotic protein BIMEL.44 RACK1 has also been implicated in hepatocellular carcinoma (HCC) in which its expression is elevated compared to normal liver cells. In HCC, ribosomal RACK1 was shown to confer resistance to chemotherapeutic drugs and tumor growth in vivo and in vitro. Mechanistically, these tumor-promoting effects of RACK1 are likely mediated by its ribosomal localization and the preferential translation of tumor-promoting genes such as cyclin D1, MYC, and Bcl-2.45 In addition to this translational function, RACK1 stimulates MKK7 in HCC, which augments JNK activity, confers resistance to TRAIL- or Fas-induced apoptosis, and promotes tumor growth in vivo.46 In agreement with this study, we have previously demonstrated that RACK1 augments JNK activation in melanoma and renders melanoma cells resistant to stress-induced apoptosis.22,47,48 We also identified a feed-forward mechanism between JNK activation and constitutively active ERK signaling, which is present in the vast majority of melanomas. Constitutive ERK activation increases both the transcription and stability of c-Jun, which targets cyclin D1 and RACK1. In turn, because RACK1 bolsters phosphorylation of JNK and augments its activity, c-Jun activity is further amplified.22,47,48 RACK1 can also promote this feed-forward signaling loop by inhibiting the proteasomal degradation of c-Jun.49
The involvement of RACK1 in the initiation and progression of tumors and in their resistance to therapy has raised the possibility that it could be an appropriate target for the development of therapeutic drugs. However, optimism for this idea is tempered by the findings that RACK1 has several properties consistent with a tumor suppressor function. For example, RACK1 is known to suppress Src activation and has also been reported to have a proapoptotic role in colon cancer cells.50 In addition, RACK1 has been implicated in the downregulation of HIF1α by competing with HSP90-HIF1α binding.51 Given the near ubiquitous expression of RACK1 and its large number of client proteins, these tumor promoter and suppressor roles are likely to vary in different cells and tissues. Understanding the function of RACK1 in activating and inactivating cancer-related pathways could thus lead to novel therapeutic strategies that specifically target RACK1 tumor- promoting activities while sparing its tumor suppressor functions.
RACK1 in the Control of Protein Synthesis
RACK1 associates with the 40S small ribosomal subunit of numerous species, and it is localized on the head of the 40S subunit near the mRNA exit channel (Fig. 2).52 RACK1 is anchored to ribosomal RNA by the universally conserved residues Arg38 and Lys40.8 Importantly, WD repeats 1, 2, and 5 of ribosome-associated RACK1 are obscured by rRNA, which is likely to modify its signaling functions.10
Figure 2.
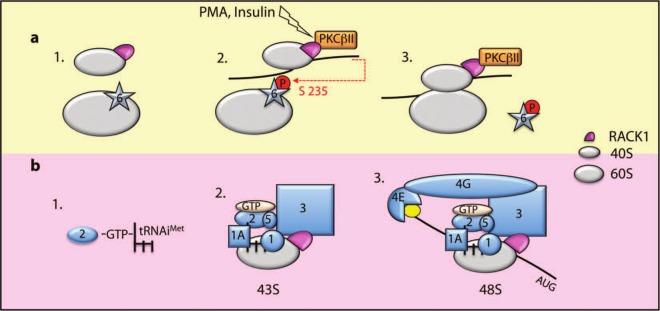
Schematic representation of RACK1’s putative role in translation initiation. (A) RACK1 and the antiassociation factor eIF6 bind 40S and 60S, respectively (1). RACK1 recruits PKCβII to the ribosome, and PKCβII phosphorylates eIF6 on serine 235 (2). Release of phosphorylated eIF6 allows 80S formation on the mRNA (3). (B) The ternary complex (eIF2-GTP-tRNAiMet) (1) is recruited to 40S with a multimeric complex composed of eIF1A, eIF1, eIF5, and eIF3. eIF3 binding to RACK1 stabilizes the ternary complex on the ribosome (2). The 43S complex is then recruited to the mRNA and stabilized through an eIF3-eIF4G interaction (3). The 48S complex scans the mRNA to reach the initiation codon (AUG).
Consistent with its initial identification as a primary RACK1-binding partner,1,53 PKCβII has been identified as an important factor linking ribosomal RACK1 and translational control in mammals. PKCβII binds 40S-associated RACK1 in 2 regions close to WD blades 3 and 4.1,54,55 RACK1 recruits active PKCβII on ribosomes, where it phosphorylates eukaryotic initiation factor 6 (eIF6) on serine 235.55 eIF6 binds to the 60S subunit at the interface with 40S, impeding the association of the 2 subunits.56 Upon phosphorylation, eIF6 is released from the 60S subunit, and the 80S ribosome can be assembled on the mRNA. Indeed, eIF6+/– heterozygous mouse embryonic fibroblasts (MEFs) accumulate monosomes and have defects in PMA- and insulin-stimulated mRNA translation. The defects cannot be rescued by expression of the nonphosphorylatable mutant of eIF6Ser235Ala,56 thus corroborating the importance of the RACK–PKCβII–eIF6 complex in translational control (Fig. 2).
In agreement with a role for RACK1 in the control of protein synthesis, mice heterozygous for RACK1 (ΔF/+) show moderate accumulation of monosomes and impaired protein synthesis, which phenocopies the defects observed in livers and MEFs from eIF6 heterozygous mice. Interestingly, the defects in neural crest–derived melanocyte development observed in RACK1 ΔF/+ mice are also seen in mice heterozygous for the ribosomal protein rpL24, a protein localized on the large ribosomal subunit close to the eIF6 binding site.
The role of ribosomal RACK1 in the control of gene expression at the posttranscriptional level has been extensively studied in yeast, revealing properties of ribosomal RACK1 that would merit further investigation in higher eukaryotes. In yeast, the RACK1 homolog Asc1 associates with the 40S subunit in growing cells but is found predominantly in the ribosome-free fraction of cells entering the stationary phase,57 suggesting that RACK1 may be necessary only on actively translating ribosomes. RACK1 interacts with the C-terminal domain of eIF3c/NIP1 at WD blades 1 to 3, stabilizing the binding between eIF3c and the 40S subunit.58 eIF3c is a component of the multimeric translation initiation factor eIF3, which, together with eIF1 and eIF1A, binds to the free 40S subunit and acts as an acceptor for the ternary complex (eIF2–GTP–tRNAiMet).59 Thus, eIF3c binding to RACK1 may anchor the ternary complex to the 40S subunit. Indeed, loss of RACK1 impairs the recruitment of the ternary complex, suggesting that RACK1 could regulate the initiation phase of translation.58 Intriguingly, eIF3 is released upon 80S assembly, while RACK1 remains associated to translating ribosomes, namely polysomes. This observation suggests that RACK1 could modulate not only translation initiation but also subsequent steps of protein synthesis (e.g., elongation). A previously identified RACK1 interactor on polysomes is the Scp160 protein. Scp160 co-sediments with polysomes60 and associates with specific polyadenylated mRNAs.61 Mutation of the conserved amino acid residues on RACK1 necessary for its binding to the 40S subunit impaired the recruitment of Scp160 on the polysomes.62 The fact that Scp160 associates with specific mRNAs suggests that RACK1 may regulate the translation of a subset of specific mRNAs rather than global translation. In agreement with this hypothesis, RACK1 is not an essential gene in yeast. RACK1 yeast mutants are viable63 and show a slight impairment in the translation of a specific subset of mRNAs64 unless challenged with stress.25,63
RACK1 as a Repressor of Protein Synthesis
The role of RACK1 in translational regulation may be even more complex than the preceding section suggests. In addition to a positive role for ribosomal RACK1 in mRNA translation, a negative role for RACK1 in gene expression at the posttranscriptional level has been reported in yeast. Depletion of RACK1 results in increased ribosomal activity in a cell-free in vitro translation assay and a concomitant increase in protein levels in vivo.65 A more recent finding has linked ribosomal RACK1 with miRNA-dependent repression of translation in Caenorhabditis elegans and humans.66 RACK1 interacts with Ago2 and let-7 miRNA. Analysis on sucrose gradients revealed that let-7 and RACK1 co-sediment on the polysomes, while RACK1 depletion significantly affects the amount of let-7 miRNAs on translating ribosomes (Fig. 3).66
Figure 3.

Schematic representation of RACK1 in miRNA-mediated repression. RACK1 binds translating ribosomes, where it recruits the Ago2–let-7 complex to silence the mRNA translation of let-7 targets.
RACK1 as a Regulator of the Quality Control of Newly Synthesized Polypeptides
Mistakes are commonly made during mRNA transcription that generate aberrant transcripts, including mRNAs with a premature stop codon and mRNAs lacking a poly(A) tail or an in-frame stop codon. In the absence of a stop codon, the ribosomes would reach the poly(A) tail in the translating mRNAs and stall on the lysine-rich sequence, leading to elongation arrest.67,68 This phenomenon is probably a result of the high affinity between the positive charges in the lysine-rich region in the nascent polypeptides and the negative charges of the exit tunnel of the ribosome.69 These aberrant nascent polypeptides must be co-translationally degraded by the proteasome70 to avoid the accumulation of misfolded proteins that may have deleterious effects on cell physiology. In yeast, ribosomes lacking RACK1 are unable to impede the translation of aberrant polypeptides, and their cellular levels increase.71 Thus, ribosomal RACK1 could also participate in the quality control of nascent polypeptides, although it is not yet clear how this might occur. RACK1 binding to the 40S subunit can remodel the 80S ribosome,52 perhaps stabilizing factors that monitor the properties of the nascent polypeptides.
Our recent studies identified RACK1 recruitment of JNK as part of the mechanism underlying the quality control of newly synthesized proteins under stress conditions.72 In response to phorbol ester treatment or UV irradiation, RACK1 acts as a scaffold protein to recruit activated JNK on 40S and on translating ribosomes. Strikingly, disruption of RACK1 binding to 40S impairs ubiquitination and degradation of newly synthesized polypeptides (NSPs), pointing to its pivotal role in the control of NSP stability. Inhibition of JNK upon stress recapitulates this phenomenon, thus revealing a role for the RACK–JNK complex in the degradation of NSPs. Notably, our studies identified that RACK1 recruits JNK to polysomes, resulting in the phosphorylation of the elongation factor eEF1A2 at serine 205 and 358, which in turn promotes its binding to NSPs, with concomitant effects on their ubiquitination and degradation. These findings are consistent with the reported binding of eEF1A2 to damaged NSPs,73,74 which was reported to promote their degradation by the proteasome.75-77
Summary and Perspectives
RACK1 is an abundant adaptor protein, and as such, it is not surprising that it participates in numerous cellular functions. Yet, recent data obtained from genetic RACK1 hypomorphic animals revealed distinct phenotypes associated with only 2 RACK1 functions: namely, its role in melanocyte biogenesis, which may be associated with cellular migration and movement of components of the neural crest lineage, and its clear role in mRNA translation. While this review has focused primarily on these aspects of RACK1 biology, it is apparent that many questions must be answered before we have a complete understanding of the function of RACK1 in mRNA translation, cellular movement, and melanocyte biology. In addition, as a component of signaling complexes that respond to extracellular stimulation, RACK1 regulation and function are expected to be both temporal and spatial. The mechanisms underlying the dynamics of RACK1 complex assembly and localization at subcellular domains will need to be deciphered as our ability to monitor such events at high resolution improves. Tracking the movement of RACK1 along cytoskeletal filaments in response to varying external stimuli may allow us to understand the roadmap used for the diverse functions of RACK1. Whether select RACK1 complexes could be targeted as novel therapeutic modalities remains an important area of research with implications for translation to the clinic.
Acknowledgments
The authors thank Drs. N. Sonenberg and S. Biffo for critical reading of the article and Dr. E. Lau for help with bioinformatics analysis.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: Support from the National Cancer Institute (grant CA051995 to Z.A.R.) and the Canadian Institutes of Health Research (MOP grant to I.T.) is gratefully acknowledged.
References
- 1. Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci U S A. 1994;91:839-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mochly-Rosen D, Smith BL, Chen CH, Disatnik MH, Ron D. Interaction of protein kinase C with RACK1, a receptor for activated C-kinase: a role in beta protein kinase C mediated signal transduction. Biochem Soc Trans. 1995;23:596-600 [DOI] [PubMed] [Google Scholar]
- 3. Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci U S A. 1991;88:3997-4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ron D, Jiang Z, Yao L, Vagts A, Diamond I, Gordon A. Coordinated movement of RACK1 with activated betaIIPKC. J Biol Chem. 1999;274:27039-46 [DOI] [PubMed] [Google Scholar]
- 5. Adams DR, Ron D, Kiely PA. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun Signal. 2011;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261-73 [DOI] [PubMed] [Google Scholar]
- 7. Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol. 2009;29:1626-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goncalves KA, Borges JC, Silva JC, et al. Solution structure of the human signaling protein RACK1. BMC Struct Biol. 2010;10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730-6 [DOI] [PubMed] [Google Scholar]
- 11. Yatime L, Hein KL, Nilsson J, Nissen P. Structure of the RACK1 dimer from Saccharomyces cerevisiae. J Mol Biol. 2011;411:486-98 [DOI] [PubMed] [Google Scholar]
- 12. Ruiz Carrillo D, Chandrasekaran R, Nilsson M, et al. Structure of human Rack1 protein at a resolution of 2.45 A. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:867-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chou YC, Chou CC, Chen YK, et al. Structure and genomic organization of porcine RACK1 gene. Biochim Biophys Acta. 1999;1489:315-22 [DOI] [PubMed] [Google Scholar]
- 14. Battaini F, Pascale A, Paoletti R, Govoni S. The role of anchoring protein RACK1 in PKC activation in the ageing rat brain. Trends Neurosci. 1997;20:410-5 [DOI] [PubMed] [Google Scholar]
- 15. Sanguino E, Roglans N, Alegret M, Sanchez RM, Vazquez-Carrera M, Laguna JC. Prevention of age-related changes in rat cortex transcription factor activator protein-1 by hypolipidemic drugs. Biochem Pharmacol. 2004;68: 1411-21 [DOI] [PubMed] [Google Scholar]
- 16. Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog Neurobiol. 2006;78:117-34 [DOI] [PubMed] [Google Scholar]
- 17. Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang BY, Chiang M, Cartwright CA. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J Biol Chem. 2001;276:20346-56 [DOI] [PubMed] [Google Scholar]
- 19. Kiely PA, Sant A, O’Connor R. RACK1 is an insulin-like growth factor 1 (IGF-1) receptor-interacting protein that can regulate IGF-1-mediated Akt activation and protection from cell death. J Biol Chem. 2002;277:22581-9 [DOI] [PubMed] [Google Scholar]
- 20. Kiely PA, Leahy M, O’Gorman D, O’Connor R. RACK1-mediated integration of adhesion and insulin-like growth factor I (IGF-I) signaling and cell migration are defective in cells expressing an IGF-I receptor mutated at tyrosines 1250 and 1251. J Biol Chem. 2005;280:7624-33 [DOI] [PubMed] [Google Scholar]
- 21. Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327:463-6 [DOI] [PubMed] [Google Scholar]
- 22. Lopez-Bergami P, Habelhah H, Bhoumik A, Zhang W, Wang LH, Ronai Z. RACK1 mediates activation of JNK by protein kinase C [corrected]. Mol Cell. 2005;19:309-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324-32 [DOI] [PubMed] [Google Scholar]
- 24. Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nunez A, Franco A, Madrid M, et al. Role for RACK1 orthologue Cpc2 in the modulation of stress response in fission yeast. Mol Biol Cell. 2009;20:3996-4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volta V, Beugnet A, Gallo S, et al. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell Mol Life Sci. 2013;70:1439-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uong A, Zon LI. Melanocytes in development and cancer. J Cell Physiol. 2010;222:38-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liliental J, Chang DD. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J Biol Chem. 1998;273:2379-83 [DOI] [PubMed] [Google Scholar]
- 29. Hermanto U, Zong CS, Li W, Wang LH. RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol Cell Biol. 2002;22:2345-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiely PA, O’Gorman D, Luong K, Ron D, O’Connor R. Insulin-like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and beta1 integrin to promote cell migration. Mol Cell Biol. 2006;26:4041-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiely PA, Baillie GS, Lynch MJ, Houslay MD, O’Connor R. Tyrosine 302 in RACK1 is essential for insulin-like growth factor-I-mediated competitive binding of PP2A and beta1 integrin and for tumor cell proliferation and migration. J Biol Chem. 2008;283:22952-61 [DOI] [PubMed] [Google Scholar]
- 32. Doan AT, Huttenlocher A. RACK1 regulates Src activity and modulates paxillin dynamics during cell migration. Exp Cell Res. 2007;313:2667-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serrels B, Sandilands E, Serrels A, et al. A complex between FAK, RACK1, and PDE4D5 controls spreading initiation and cancer cell polarity. Curr Biol. 2010;20:1086-92 [DOI] [PubMed] [Google Scholar]
- 34. Serrels B, Sandilands E, Frame MC. Signaling of the direction-sensing FAK/RACK1/PDE4D5 complex to the small GTPase Rap1. Small GTPases. 2011;2:54-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen S, Lin F, Shin ME, Wang F, Shen L, Hamm HE. RACK1 regulates directional cell migration by acting on G betagamma at the interface with its effectors PLC beta and PI3K gamma. Mol Biol Cell. 2008;19:3909-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Runne C, Chen S. WD40-repeat proteins control the flow of Gbetagamma signaling for directional cell migration. Cell Adh Migr. 2013;7:214-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Usacheva A, Smith R, Minshall R, et al. The WD motif-containing protein receptor for activated protein kinase C (RACK1) is required for recruitment and activation of signal transducer and activator of transcription 1 through the type I interferon receptor. J Biol Chem. 2001;276:22948-53 [DOI] [PubMed] [Google Scholar]
- 38. Kubota T, Yokosawa N, Yokota S, Fujii N. Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J Virol. 2002;76:12676-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yokota S, Saito H, Kubota T, Yokosawa N, Amano K, Fujii N. Measles virus suppresses interferon-alpha signaling pathway: suppression of Jak1 phosphorylation and association of viral accessory proteins, C and V, with interferon-alpha receptor complex. Virology. 2003;306:135-46 [DOI] [PubMed] [Google Scholar]
- 40. Al-Reefy S, Mokbel K. The role of RACK1 as an independent prognostic indicator in human breast cancer. Breast Cancer Res Treat. 2010;123:911, author reply 2. [DOI] [PubMed] [Google Scholar]
- 41. Cao XX, Xu JD, Liu XL, et al. RACK1: a superior independent predictor for poor clinical outcome in breast cancer. Int J Cancer. 2010;127:1172-9 [DOI] [PubMed] [Google Scholar]
- 42. Cao XX, Xu JD, Xu JW, et al. RACK1 promotes breast carcinoma proliferation and invasion/metastasis in vitro and in vivo. Breast Cancer Res Treat. 2010;123:375-86 [DOI] [PubMed] [Google Scholar]
- 43. Cao XX, Xu JD, Xu JW, et al. RACK1 promotes breast carcinoma migration/metastasis via activation of the RhoA/Rho kinase pathway. Breast Cancer Res Treat. 2011;126:555-63 [DOI] [PubMed] [Google Scholar]
- 44. Zhang W, Cheng GZ, Gong J, et al. RACK1 and CIS mediate the degradation of BimEL in cancer cells. J Biol Chem. 2008;283:16416-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruan Y, Sun L, Hao Y, et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J Clin Invest. 2012;122:2554-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo Y, Wang W, Wang J, et al. Receptor for activated C kinase 1 promotes hepatocellular carcinoma growth by enhancing mitogen-activated protein kinase kinase 7 activity. Hepatology. 2013;57:140-51 [DOI] [PubMed] [Google Scholar]
- 47. Lopez-Bergami P, Huang C, Goydos JS, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lopez-Bergami P, Ronai Z. Requirements for PKC-augmented JNK activation by MKK4/7. Int J Biochem Cell Biol. 2008;40:1055-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang J, Zhu F, Li X, et al. Rack1 protects N-terminal phosphorylated c-Jun from Fbw7-mediated degradation. Oncogene. 2012;31:1835-44 [DOI] [PubMed] [Google Scholar]
- 50. Mamidipudi V, Cartwright CA. A novel pro-apoptotic function of RACK1: suppression of Src activity in the intrinsic and Akt pathways. Oncogene. 2009;28:4421-33 [DOI] [PubMed] [Google Scholar]
- 51. Liu YV, Semenza GL. RACK1 vs. HSP90: competition for HIF-1 alpha degradation vs. stabilization. Cell Cycle. 2007;6:656-9 [DOI] [PubMed] [Google Scholar]
- 52. Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957-62 [DOI] [PubMed] [Google Scholar]
- 53. Ron D, Luo J, Mochly-Rosen D. C2 region-derived peptides inhibit translocation and function of beta protein kinase C in vivo. J Biol Chem. 1995;270:24180-7 [DOI] [PubMed] [Google Scholar]
- 54. Grosso S, Volta V, Sala LA, et al. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem J. 2008;415:77-85 [DOI] [PubMed] [Google Scholar]
- 55. Sharma G, Pallesen J, Das S, et al. Affinity grid-based cryo-EM of PKC binding to RACK1 on the ribosome. J Struct Biol. 2013;181:190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gandin V, Miluzio A, Barbieri AM, et al. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baum S, Bittins M, Frey S, Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J. 2004;380:823-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kouba T, Rutkai E, Karaskova M, Valasek L. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res. 2012;40:2683-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580-7 [DOI] [PubMed] [Google Scholar]
- 60. Lang BD, Fridovich-Keil JL. Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 2000;28:1576-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li AM, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li AM, Vargas CA, Brykailo MA, Openo KK, Corbett AH, Fridovich-Keil JL. Both KH and non-KH domain sequences are required for polyribosome association of Scp160p in yeast. Nucleic Acids Res. 2004;32:4768-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chantrel Y, Gaisne M, Lions C, Verdiere J. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics. 1998;148:559-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J Biol Chem. 2003;278:49119-28 [DOI] [PubMed] [Google Scholar]
- 65. Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol. 2004;24:8276-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jannot G, Bajan S, Giguere NJ, et al. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011;12:581-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3’-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol. 2008;384:73-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuroha K, Akamatsu M, Dimitrova L, et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010;11:956-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gandin V, Gutierrez GJ, Brill LM, Varsano T, Feng Y, Aza-Blanc, et al. Degradation of newly synthesized polypeptides by ribosome-associated RACK1/JNNK/eEF1A2. Mol. Cell. Biol. In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hotokezaka Y, Tobben U, Hotokezaka H, et al. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J Biol Chem. 2002;277:18545-51 [DOI] [PubMed] [Google Scholar]
- 74. Meriin AB, Zaarur N, Sherman MY. Association of translation factor eEF1A with defective ribosomal products generates a signal for aggresome formation. J Cell Sci. 2012;125:2665-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol. 2005;25:403-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gonen H, Dickman D, Schwartz AL, Ciechanover A. Protein synthesis elongation factor EF-1 alpha is an isopeptidase essential for ubiquitin-dependent degradation of certain proteolytic substrates. Adv Exp Med Biol. 1996;389:209-19 [DOI] [PubMed] [Google Scholar]
- 77. Gonen H, Smith CE, Siegel NR, et al. Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci U S A. 1994;91: 7648-52 [DOI] [PMC free article] [PubMed] [Google Scholar]



