SUMMARY
DNA damage is normally detrimental to living organisms. Here we show that it can also serve as a signal to promote immune responses in plants. We found that the plant immune hormone salicylic acid (SA) can trigger DNA damage in the absence of a genotoxic agent. The DNA damage sensor proteins, RAD17 and ATR, are required for effective immune responses. These sensor proteins are negatively regulated by a key immune regulator SNI1 (suppressor of npr1-1, inducible 1), which we discovered as a missing subunit of the Structural Maintenance of Chromosome (SMC) 5/6 complex required for controlling DNA damage. Elevated DNA damage caused by the sni1 mutation or treatment with a DNA-damaging agent markedly enhances SA-mediated defense gene expression. Our study suggests that activation of DNA damage responses is an intrinsic component of the plant immune responses.
INTRODUCTION
Organisms are constantly assaulted by endogenous and environmental agents that could potentially result in tens of thousands of DNA lesions per cell per day (Jackson and Bartek, 2009). These lesions are serious threats to the faithful transmission of genetic information. To maintain genome integrity, the DNA damage responses (DDR) are triggered to carry out repair. There are two major pathways to repair double-strand DNA breaks (DSBs), one through homologous recombination (HR) involving components such as BRCA2 and RAD51 and the other through nonhomologous end joining (NHEJ) requiring components such as Ku70 and Ku80 (Chapman et al., 2012).
Besides abiotic stresses, all organisms are also threatened by various pathogens. To combat infection, organisms rely on their immune systems. Similar to animals, plants have innate immune mechanisms, which can be triggered through detection of pathogen-associated molecular patterns (PAMPs) by cell surface pattern recognition receptors (Jones and Dangl, 2006; Spoel and Dong, 2012). This PAMP-triggered immunity (PTI) is effective in protection against most microorganisms. However, some successful pathogens can overcome PTI by directly delivering effectors into plant cells through the type III secretion system. In response, plants have evolved intracellular receptors, structurally similar to the NLR (nucleotide-binding domain, (ETI). Moreover, ETI at the site of infection can further induce systemic acquired resistance (SAR) resulting in enhancement of general disease resistance throughout the organism (Jones and Dangl, 2006; Spoel and Dong, 2012).
Even though both DDR and immune responses have been studied in depth separately, whether and how they are connected are largely unknown. There are commonalities between these two stress responses; both involve transcriptional reprogramming, cell cycle perturbation, or even cell death (Ciccia and Elledge, 2010; Spoel and Dong, 2012). Interestingly, treating plants with salicylic acid (SA), a necessary and sufficient signal for SAR (Gaffney et al., 1993), not only induces expression of defense genes, but also increases the rate of HR, one of the DNA repair pathways (Kovalchuk et al., 2003; Lucht et al., 2002). However, the biological significance of this increase and the underlying molecular mechanism has yet to be revealed. Through genetic screens, NPR1 (nonexpresser of PR genes 1) and SNI1 (suppressor of npr1-1, inducible 1) have been identified as master regulators of SA-mediated defense responses (Cao et al., 1997; Li et al., 1999). In the npr1 mutant, the SA-induced expression of defense genes, such as pathogenesis-related 1 (PR1) and PR2, is abolished, indicating that NPR1 is a positive regulator of defense (Cao et al., 1997). SNI1 was identified in an npr1 suppressor screen and functions as a negative regulator of defense. The sni1 mutant shows heightened basal level of PR gene expression and sensitivity to SA induction and has increased HR rate (Durrant et al., 2007; Li et al., 1999). Therefore, elucidating the molecular function of SNI1 holds the key to our understanding of the crosstalk between plant immune responses and DDR. However, progress has been significantly hindered because SNI1 is an unknown protein without significant sequence similarity to any other known proteins or domains, even though it is highly conserved in plants (Mosher et al., 2006).
In this study, we purified and characterized the SNI1 complex and found that it is a subunit of the Structural Maintenance of Chromosome (SMC) 5/6 complex involved in DDR. Mutations in SNI1 or treatment of wild-type (WT) plants with SA could both induce DNA damage as well as facilitate defense gene expression. These phenotypes of sni1 were suppressed by mutations in RAD17 and ATR, two DNA damage sensor proteins. Our study therefore suggests that activation of DDR is an intrinsic mechanism during immune responses in plants.
RESULTS
SNI1 Is a Subunit of the SMC5/6 Complex
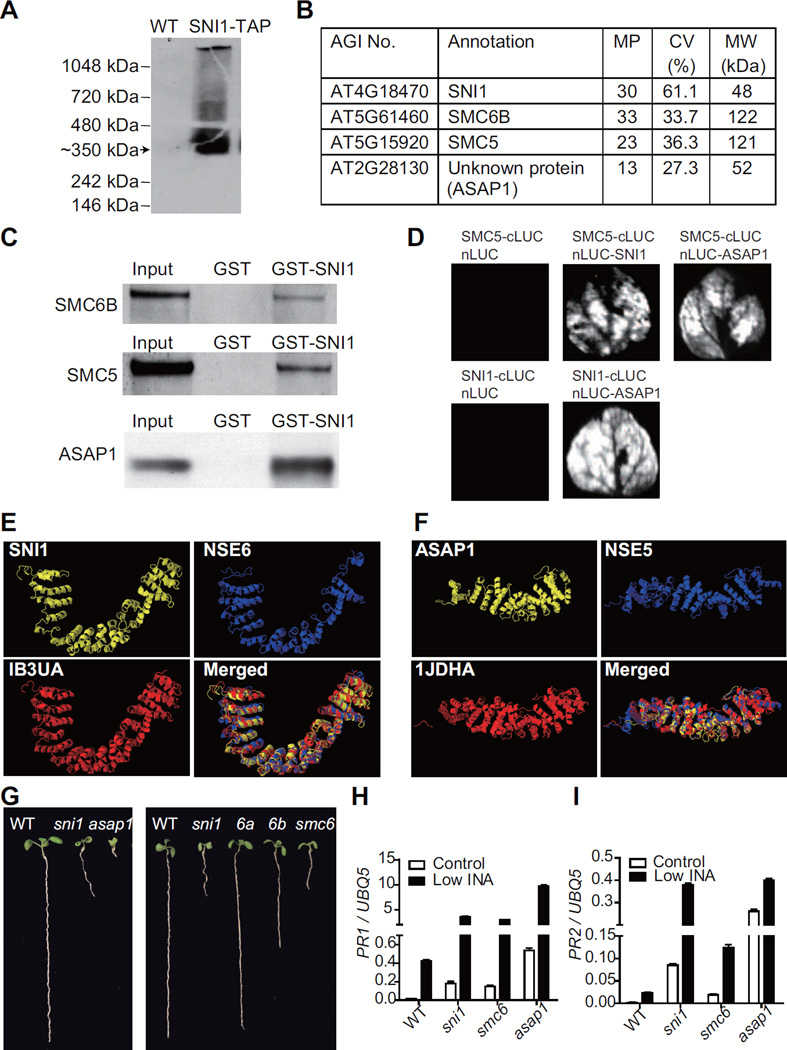
To directly determine the molecular function of SNI1, we purified the SNI1 complex using the tandem affinity purification (TAP) strategy (Rubio et al., 2005). SNI1-TAP driven by the native promoter of SNI1 was shown to be biologically active as it fully complemented the sni1 mutation when transformed into the mutant plants (Song et al., 2011). While the SNI1-TAP fusion protein has a molecular weight (MW) of ∼80 kDa, the major SNI-containing band detected on a Blue-Native PAGE gel was at ∼350 kDa (Figure 1A), indicating that SNI1 is associated with other proteins. After two rounds of purification (for details, see Experimental Procedures and Supplemental Experimental Procedures), the SNI1 complex was subjected to liquid chromatography–tandem mass spectrometry (LC–MS/MS). The top-ranked proteins included SNI1, Structural Maintenance of Chromosome (SMC) 6B, SMC5 and an unknown protein, which we named Arabidopsis SNI1 Associated Protein 1 (ASAP1) (Figure. 1B). Each protein was matched by multiple peptides, with 27.3% to 61.1% coverage of the whole protein (Table S3, available online). The combined MW for these four proteins adds up to about 350 kDa, the size of the SNI1 complex estimated through the Blue-Native PAGE. To confirm the interactions between SMC5, SMC6B, ASAP1 and SNI1, we carried out pulldown assays. As shown in Figure 1C, the MYC-tagged SMC5, SMC6B and ASAP1 proteins could all be pulled down by the glutathione S-transferase (GST)-tagged SNI1 protein, but not the GST control. To further validate the interactions in planta, we performed the split luciferase assay in Nicotiana benthamiana by co-expressing fusion proteins with either the C-terminal half of luciferase (cLUC) or the N-terminal half of the enzyme (nLUC). An interaction between two proteins could bring the two halves of the luciferase together, leading to enzymatic activity. Except SMC6B, which failed to express in N. benthamiana, the other three proteins in the complex could interact with each other (Figure 1D).
Figure 1. SNI1 is a Subunit of the SMC5/6 Complex.
(A) SNI1 forms a complex in plants. Total protein from the SNI1-TAP transgenic line was separated on a Blue-Native PAGE gel and detected with an anti-TAP antibody. The major SNI1-TAP band (arrowed) was ∼350 kDa. The untransformed wild-type (WT) plant (left lane) was used as a negative control.
(B) Proteins identified in the SNI1 complex. MP, the number of matched peptides; CV, the percentage of sequence coverage; MW, molecular weight.
(C) In vitro pull-down assays. GST and GST-SNI1 were expressed in E. coli and purified. SMC5-MYC, SMC6B-MYC and ASAP1-MYC were in vitro translated. The blots were detected with an anti-MYC antibody.
(D) Split luciferase assays. The proteins were fused to either the C- or N-terminal half of luciferase (cLUC or nLUC) and transiently expressed in N. benthamiana. The luciferase activities were monitored by a CCD camera.
(E and F) The 3D structures of SNI1, NSE6, ASAP1 and NSE5 predicted by the I-TASSER server. SNI1 and NSE6 were similar to 1B3UA; ASAP1 and NSE5 were similar to 1JDHA in the PDB database.
(G) The short-root phenotype observed in sni1, asap1, smc6a (6a), smc6b (6b), and smc6a/SMC6A smc6b (smc6) seedlings in comparison to wild-type (WT).
(H and I) The expression of PR1 (H) and PR2 (I) measured by qRT-PCR. Plants were grown on medium with 10 μM INA (Low INA) for 9 days. The expression level was normalized to ubiquitin 5 (UBQ5). The data are presented as mean ± SD (n = 3). See also Figure S1, Tables S1 and S3.
SMC5 and SMC6 are known to interact to form the SMC5/6 complex, which plays a critical role in DDR (Potts, 2009). The structure of this complex has been well-studied in yeast. In addition to SMC5 and SMC6, it contains 6 non-SMC element (NSE) proteins (Hazbun et al., 2003; Pebernard et al., 2006). While NSE1, 2, 3 and 4 are highly conserved in eukaryotes and their homologs have been identified in Arabidopsis (Watanabe et al., 2009), NSE5 and NSE6 have only been identified in budding yeast and fission yeast (Hazbun et al., 2003; Pebernard et al., 2006). Given the conserved function of the SMC5/6 complex, it is believed that other organisms also contain NSE5 and NSE6. However, their primary sequences are not conserved, making them difficult to identify. Based on our complex purification and protein interaction data, we hypothesized that ASAP1 and SNI1 are the functional counterparts of NSE5 and NSE6 in Arabidopsis.
To test our hypothesis, we submitted the amino acid sequences of Arabidopsis SNI1, ASAP1, and fission yeast NSE5, NSE6 to the I-TASSER server (Zhang, 2008) to predict their three-dimensional (3D) protein structures. The models with the best scores are shown in Figure 1E and 1F. Surprisingly, despite limited sequence similarities (Figure S1), both SNI1 and NSE6 showed high structural similarities to the PR65/A subunit of protein phosphatase 2A (PDB ID: 1B3UA) (Groves et al., 1999). ASAP1 and NSE5, on the other hand, were found to be structurally similar to beta-catenin (PDB ID: 1JDHA) (Graham et al., 2001). Structural alignment using TM-align (Zhang and Skolnick, 2005) showed TM-scores of > 0.91 for the SNI1/NSE6-1B3UA pair and > 0.86 for the ASAP1/NSE5-1JDHA pair (Table S1); a TM-score of > 0.5 indicates similar folding of the proteins. Therefore, we propose that ASAP1 and SNI1 are the first NSE5 and NSE6 found in a multicellular organism.
If SNI1, ASAP1, SMC6B and SMC5 are in the same complex, the corresponding mutants should exhibit similar phenotypes. Arabidopsis has one SMC5 homolog and two SMC6 homologs, namely SMC6A and SMC6B (Watanabe et al., 2009). SMC6B, but not SMC6A, was found in the SNI1 complex, consistent with the finding that SMC6B is the major form of SMC6 in Arabidopsis. While the smc5 knockout mutant is embryonic lethal in Arabidopsis, both the asap1 and the smc6b mutants are defective in root development, similar to the sni1 mutant (Figure 1G). Moreover, this smc6b mutant phenotype is exacerbated in the smc6a/SMC6A heterozygous background, while the smc6a smc6b double mutation causes lethality. Another phenotype of the sni1 mutant is the enhancement of defense responses, including elevated basal defense gene expression in the absence of an inducer and increased sensitivity to SA and its synthetic analog 2, 6-dichloroisonicotinic acid (INA) (Li et al., 1999). We tested the expression of two defense marker genes, PR1 and PR2, in the asap1 and smc6a/SMC6A smc6b mutants (smc6) and found higher levels of basal and induced expression in the mutants than in WT (Figures 1H and 1I). Thus, the sni1 phenotypes were recapitulated in the asap1 and smc6 mutants. Together, these data strongly support our hypothesis that SNI1 and ASAP1 are subunits of the SMC5/6 complex.
Mutation in SNI1 or SA Treatment Induces DNA Damage
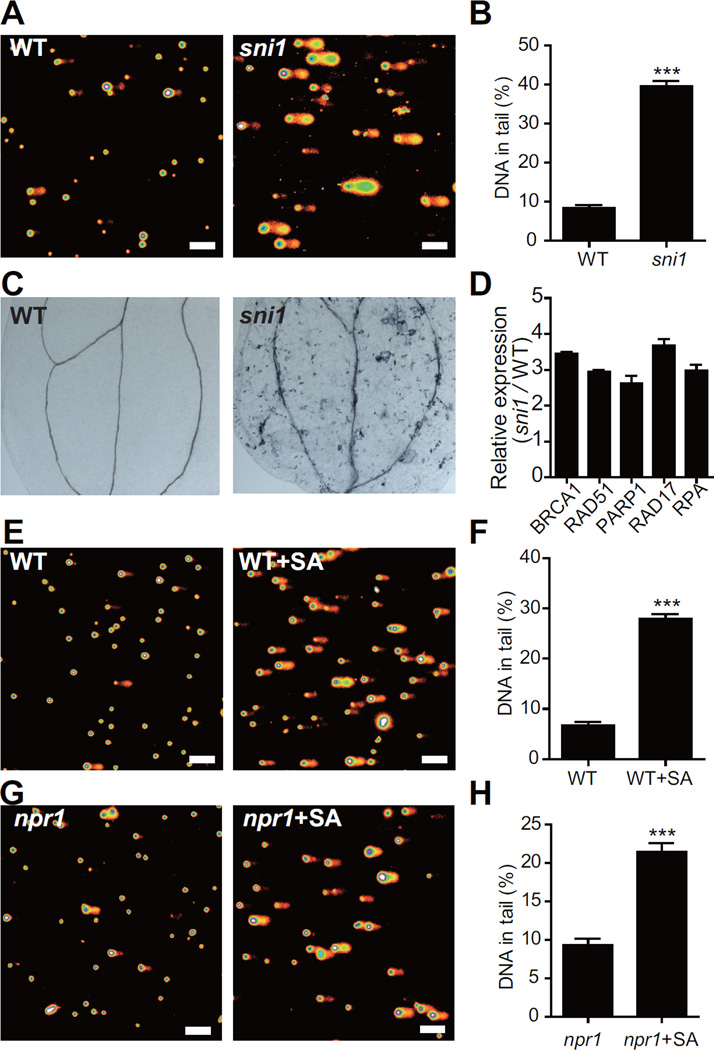
Since the SMC5/6 complex is known to be involved in DDR (Potts, 2009), we tested the sni1 plants for DDR phenotypes. As observed in a comet assay, the nuclei isolated from the sni1 mutant contained significantly more DNA damage than those isolated from the WT (Figures 2A and 2B). Consequently, spontaneous cell death was detected in the sni1 mutant by trypan blue staining (Figure 2C). In addition, the sni1 mutant had elevated expression of DDR-related genes such as BRCA1, RAD51, PARP1, RAD17 and RPA (Culligan et al., 2006) (Figure 2D). These results indicate that in the sni1 mutant, DDR is constitutively activated.
Figure 2. Mutation in SNI1 or SA Treatment Induces DNA Damage.
(A and B) The sni1 mutant shows more DNA damage than WT in a comet assay. (A) Representative pictures of the comet assay. The scale bar is 100 μm. (B) Quantification of the percentage of DNA in the comet tails. The data are presented as mean ± SEM (n > 200).
(C) Spontaneous cell death in sni1 seedlings indicated by trypan blue staining.
(D) Relative expression of DDR-related genes in sni1 compared to WT. The results are shown as mean ± SD (n = 3).
(E–H) SA treatment induces DNA damage in both WT (E and F) and the npr1 mutant (G and H). Comet assay was performed 4 h after plants were treated with water or 1 mM SA. The scale bar is 100 μm. The data are presented as mean ± SEM (n > 200). ***, P < 0.001 (Student’s t-test, two-tailed). Two-week-old plants were used in all the experiments in this figure.
As the sni1 mutant was originally identified based on its defense phenotypes, the DDR phenotypes in the sni1 mutant suggest a connection between DDR and immune responses. To test whether DDR is an intrinsic part of the plant immune responses, we performed comet assays on WT Arabidopsis seedlings treated with the hormone SA to trigger the immune response. As shown in Figures 2E and 2F, we found that in SA-treated seedlings, DNA damage was significantly increased compared to the mock-treated ones. Since NPR1 is a major regulator of SA-mediated responses (Cao et al., 1997), we also examined DNA breakage in the npr1 mutant after SA treatment. As shown in Figures 2G and 2H, SA treatment could still induce DNA damage in the npr1 mutant, suggesting that this response is NPR1-independent and represents a new SA signaling pathway.
The SMC5/6 Complex Negatively Regulates the DNA Damage Sensors RAD17 and ATR
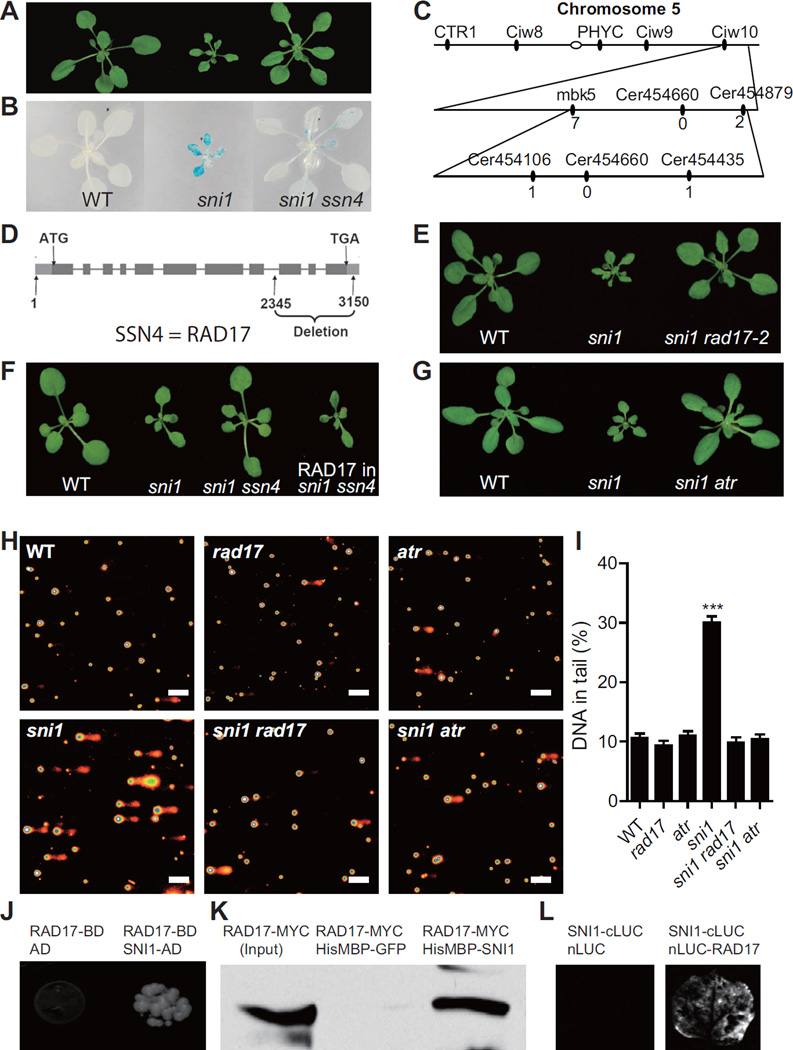
Our biochemical data suggest that DDR is involved in the Arabidopsis immune response. This is consistent with our previous data obtained through a genetic screen for suppressors of sni1 (ssn). We found that mutations in the DDR genes RAD51D (SSN1), SWS1 (SSN2), BRCA2A (SSN3) and RAD51 could suppress the sni1 mutant phenotypes and also compromised disease resistance (Durrant et al., 2007; Song et al., 2011; Wang et al., 2010). However, all of these SSN proteins function in the late steps of HR (Martin et al., 2006). It is possible that their role in defense gene expression is independent of HR pathway. To address this question, we sought for upstream DDR components that may function with or be regulated by the SMC5/6 complex. We focused on ssn4, which is another ssn mutant that could suppress both the stunted growth and basal PR1 and PR2 expression in sni1 (Figures 3A, 3B, S2A and S2B). The ssn4 mutation was mapped between markers Cer454106 and Cer454435 (Figure 3C). Sequencing analysis of the candidate genes in this region revealed that there is an 805 bp deletion in RAD17, a known DNA damage sensor gene (Zhou and Elledge, 2000), in the ssn4 mutant (Figure 3D). To confirm that RAD17 is the SSN4 gene, we crossed sni1 with a T-DNA insertion mutant rad17-2 and found the same effect on plant morphology as the ssn4 deletion mutant (Figure 3E). In addition, the sni1 ssn4 double mutant phenotype could be complemented by expressing the WT RAD17 gene (Figure 3F). Consistently, the phenotype of the asap1 mutant could also be suppressed by rad17 as well as the other ssn mutants (Figure S2C).
Figure 3. The SMC5/6 Complex Negatively Regulates RAD17 and ATR.
(A and B) The ssn4 mutant suppresses the sni1 morphology (A) and basal defense gene expression (B). In (B), the blue color indicates expression of the defense gene reporter PR2:GUS.
(C) Map-based cloning of SSN4. The number of recombinants between the markers and SSN4 are shown below each marker.
(D) The gene structure of RAD17. The boxes represent exons and the lines represent introns. Start codon (ATG) and stop codon (TGA) are shown. The deletion mutation in ssn4 is from nucleotide 2345 to nucleotide 3150.
(E) The T-DNA insertion mutant rad17-2 suppresses the sni1 morphology.
(F) Transforming the RAD17 gene into the sni1 ssn4 double mutant can suppress the ssn4 phenotype and restore the sni1 morphology.
(G) The mutation in ATR suppresses the sni1 morphology.
(H and I) Comet assays. (H) Representative pictures of the comet assay. The scale bar is 100 μm. (I) Quantification of the percentage of DNA in the comet tails. The data are presented as mean ± SEM (n > 200). ***, P < 0.001 (one-way ANOVA followed by Dunnett's multiple comparison test).
(J) RAD17 interacts with SNI1 in the Y2H assay. The yeast growth on media lacking Trp, Leu, Ade and His indicates interaction. AD, activation domain; BD, DNA-binding domain.
(K) RAD17-MYC can be pulled down by HisMBP-SNI1. HisMBP-SNI1 and HisMBP-GFP were expressed in E. coli and purified. RAD17-MYC was in vitro translated. The blots were detected with an anti-MYC antibody.
(L) RAD17 interacts with SNI1 in the split luciferase assay. See also Figure S2.
In mammals, RAD17 is regulated by ATM (ataxia telangiectasia, mutated) and/or ATR (ATM and RAD3-related), the central regulators of DDR (Bao et al., 2001). Through genetic crosses, we found that the sni1 phenotypes could also be suppressed by the atr mutation, but not by atm (Figures 3G and S2D). This is consistent with the finding that ATR and ATM play distinct roles in DDR in Arabidopsis (Culligan et al., 2006). Given that other SSN genes are involved in HR, it is likely that RAD17 and ATR are the upstream regulators of the HR pathway.
Since RAD17 and ATR are known to suppress genomic instability, the observation that rad17 and atr mutants are sni1 suppressors was surprising given the elevated DNA damage observed in sni1. To further elucidate the relationship between the SMC5/6 complex and RAD17 and ATR in HR, we performed comet assays on these mutants. As shown in Figures 3H and 3I, there was no significant increase in DNA damage in the rad17 and atr mutants compared to WT, indicating that even though the HR pathway is blocked in these mutants, other DDR pathways, such as NHEJ, are sufficient to repair DNA damage. Strikingly, the elevated level of DNA damage in the sni1 mutant was also suppressed by rad17 and atr, suggesting that the increased DNA damage observed in sni1 mutant is caused by RAD17/ATR activation.
Based on these genetic epistasis data, we hypothesized that the SMC5/6 complex is a negative regulator of RAD17 and ATR. To test the possibility that this regulation is through a physical interaction between SNI1 and RAD17, we performed yeast two-hybrid (Y2H) analysis. We found that yeast expressing the RAD17 bait and the SNI1 prey could grow in the selective medium (Figure 3J), indicating that they can interact with each other. We also carried out pull-down assays using purified recombinant HisMBP (maltose-binding protein)-SNI1 protein and in vitro translated RAD17-MYC protein and observed that RAD17 could be pulled down by HisMBP-SNI1, but not the HisMBP-GFP control (Figure 3K). To validate this finding in planta, we performed the split luciferase assay. The activity of the reconstituted luciferase (Figure 3L) supports our hypothesis that SNI1 and RAD17 physically interact. However, how the binding of SNI1 to RAD17 suppresses its function has yet to be discovered.
DDR Potentiate Plant Immunity
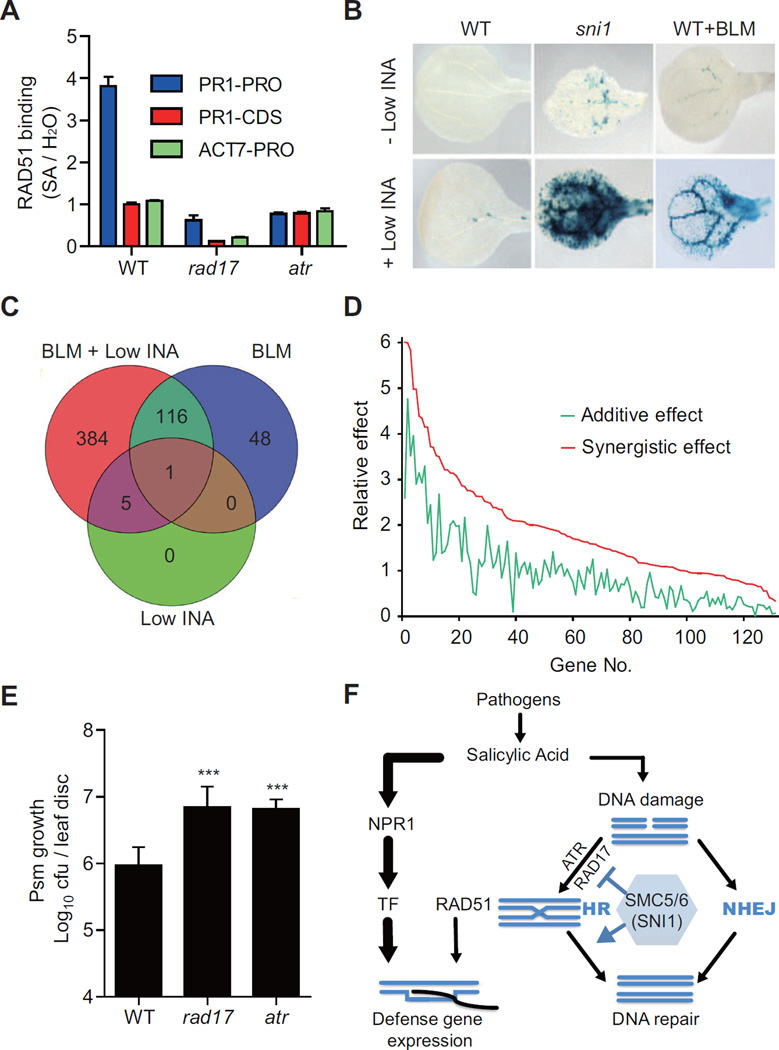
Our previous studies suggest that SNI1 (i.e., the SMC5/6 complex) may negatively affect the recruitment of HR proteins to DNA (Song et al., 2011; Wang et al., 2010). In chromatin immunoprecipitation (ChIP) experiments, RAD51 could bind to defense gene promoters after SA treatment (Song et al., 2011; Wang et al., 2010). To test whether this association is a consequence of SA-induced DDR, we performed ChIP on RAD51 in the DNA damage sensor mutants, rad17 and atr. As shown in Figure 4A, in WT plants, RAD51 could specifically bind to the PR1 promoter upon SA induction. This association appeared to be gene-specific and promoter-specific because it was neither observed at the ACTIN7 (ACT7) gene promoter nor in the coding region of PR1. Interestingly, this SA-induced association was significantly compromised in the rad17 and atr mutants, indicating that RAD17 and ATR are two upstream regulators required for not only the activation of HR repair pathway, but also control the recruitment of HR proteins to defense gene promoters.
Figure 4. DNA Damage Responses Potentiate Plant Immunity.
(A) The binding of RAD51 to the PR1 promoter depends on RAD17 and ATR. ChIP assays were performed in WT, rad17 and atr treated with water or 1 mM SA for 16 h. The ChIP samples were subjected for qPCR analysis for the promoter or the coding region of PR1 (PR1-PRO or PR1-CDS) and the promoter region of ACTIN7 (ACT7-PRO). The fold enrichment between SA-treated and H2O-treated samples is shown. The error bars represent SEM (n = 3).
(B–D) DNA-damaging agent bleomycin (BLM) and low dose of the immune inducer INA synergistically induce defense gene expression. Plants were grown on medium with 4 μg/ml BLM and/or low INA (10 μM) for 9 days. (B) PR2:GUS expression. WT+BLM, WT plants treated with BLM. (C and D) Whole-genome microarray analysis. (C) Venn diagram analysis of induced genes (Fold change > 2, P < 0.05). (D) Simulation-based analysis of the 131 synergistically induced genes. The green line represents the sum of the BLM effect and the INA effect (additive), and the red line represents the effect of BLM+INA co-treatment (synergistic). The numbers on the X-axis represent genes listed in Table S4.
(E) The rad17 and atr mutants are more susceptible to the bacterial pathogen Psm ES4326 than WT. cfu, colony forming unit. Error bars represent 95% confidence intervals (n = 7). ***, P < 0.001 (Student’s t-test, two-tailed).
(F) A working model that shows how DDR facilitates defense gene expression independent of NPR1. TF, transcription factor. HR, homologous recombination. NHEJ, non-homologous end joining. See also Figure S3, Tables S4 and S5.
The association of HR proteins with defense gene promoters may indicate a role in the transcription of these genes. To test this possibility, we examined the defense gene expression in plants treated with a suboptimal dose of the immune inducer INA and the DNA-damaging agent bleomycin (BLM). As shown in Figure 4B, INA or BLM treatment alone only slightly induced the expression of the defense gene reporter PR2:GUS in WT. However, when BLM and INA were applied together, marked induction of PR2:GUS expression was observed. The requirement of BLM was alleviated in the sni1 mutant, in which the low dose of INA was sufficient to induce the reporter expression. Similar results were obtained for another defense marker gene PR1 by qRT-PCR (Figure S3A). This effect was not specific to BLM; plants treated with another genotoxic agent, hydroxyurea, also significantly enhanced INA-mediated defense gene expression (Figure S3B). These results indicate that there is a synergistic interaction between DDR and SA signaling.
To evaluate the synergism at the whole-genome level, we performed an expression profiling experiment using the Affymetrix ATH1 genome array. Interestingly, while the low dose of INA or BLM alone induced only 6 and 165 genes, respectively, INA and BLM co-treatment induced 506 genes (Fold change > 2, P < 0.05). Among these genes, 384 were only induced in co-treated samples (Figure 4C). We performed a simulation-based analysis (see Supplemental Experimental Procedures for detail) to distinguish between additive and synergistic effects. In total, we found that 131 genes, including PR1 and PR2, were synergistically induced (Figure 4D and Table S4, available online). Gene ontology analysis revealed that the most enriched category of these genes was defense response (P < 1.82×10−13, Table S5, available online).
The synergistic effect of BLM and INA on defense gene expression further indicates that DDR is an intrinsic mechanism of immune responses. In strong support of this idea, mutant of either RAD17 or ATR, two DNA damage sensor genes, was found to be significantly more susceptible to the bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326 than WT (Figure 4E). This is consistent with the previous data showing that the mutants of downstream HR components, rad51d, sws1, brca2a and rad51, are also more susceptible to Psm infection (Durrant et al., 2007; Song et al., 2011; Wang et al., 2010).
DISCUSSION
In this study, we identified SNI1 and ASAP1 as the two missing subunits of the SMC5/6 complex in Arabidopsis. Interestingly, in the sni1 mutant, activation of the HR pathway (Figure 2D) and an increase in DNA damage were both observed (Figures 2A and 2B). These conflicting phenotypes may be explained by a dual role for the SMC5/6 complex in DDR. In this study, we found that this complex negatively regulates the RAD17/ATR-mediated HR repair pathway through a physical interaction. The SMC5/6 complex is also known to facilitate resolution of DNA intermediates, such as Holliday junctions (Chavez et al., 2010). Therefore, in the sni1 mutant, even though HR is hyperactive, the damaged DNA is “stuck” at the end step of HR, due to unresolved DNA intermediates. In the sni1 ssn mutants, however, the HR repair pathway is blocked and the damaged DNA can be repaired through other DNA repair pathways, such as NHEJ.
The involvement of DDR in plant immune response was discovered unexpectedly in the sni1 mutant, which appears to simulate the physiological immune-induced state. Treatment of WT plants with the immune signal SA can also induce DNA damage as well as activate the HR pathway. Based on our findings, we propose a working model to explain the role of DDR in plant immunity (Figure 4F). Pathogen infection triggers the production of the immune signal SA, which induces DNA damage such as DSBs. It is known that DSBs can be repaired through either HR or NHEJ (Chapman et al., 2012). As DNA damage sensor proteins, RAD17 (SSN4) and ATR activate the HR pathway involving RAD51D (SSN1), SWS1 (SSN2), BRCA2A (SSN3) and RAD51, which can be recruited either to the site of DNA damage for repair or to defense gene promoters to facilitate gene expression. Although SA functions mainly through NPR1, the DDR-mediated defense pathway represents an NPR1-independent pathway. These two pathways function together to ensure maximum defense gene expression. The SMC5/6 complex, containing SNI1, has dual functions in HR. It not only represses HR through its interaction with SSNs, but also promotes HR by facilitating the resolution of Holliday junctions.
DDR is an evolutionarily ancient stress response, upon which other defense mechanisms could have been built. It is well-known that both biotic and abiotic stresses can trigger the release of reactive oxygen species, which inevitably leads to DNA damage. During evolution, plants might have adapted to use the DDR machinery as a part of the defense mechanism to facilitate gene expression as well as to maintain genome stability.
In animals, it has also been shown that DNA-damaging treatment can activate the immune systems (Gasser et al., 2005; Menendez et al., 2011). These findings indicate that the crosstalk between DDR and the immune responses is a common feature in both plants and animals. Recently, large-scale clinical trials have shown that long-term use of aspirin (a SA derivative) has surprising effects in preventing and reducing death by various types of cancer. However, the mechanism remains largely unknown (Thun et al., 2012). Our study shows that SA treatment can lead to activation of DDR, the basis of many cancer therapies, raising the possibility that the anti-cancer effects of aspirin involve DDR.
EXPERIMENTAL PROCEDURES
SNI1 Complex Purification and Protein Identification
SNI1 complex purification was performed as described (Rubio et al., 2005) using IgG Sepharose 6 Fast Flow beads (GE Healthcare) and Ni-NTA resin (Qiagen). The purified protein complex was submitted for LC-MS/MS analysis at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
3D Protein Structure Prediction
The sequences of Arabidopsis SNI1, ASAP1 and fission yeast NSE5, NSE6 were submitted to the I-TASSER server (Zhang, 2008) to predict their 3D structures. Pairwise structure alignments were performed using TM-align (Zhang and Skolnick, 2005). The multiple structural alignments were performed using MISTRAL (Micheletti and Orland, 2009).
Comet Assay
Comet assay was performed using CometAssay Kit (Trevigen). The comets were visualized by staining with SYBR Green I, captured with Zeiss LSM 510 upright confocal microscope at Light Microscopy Core Facility, Duke University and analyzed with CometScore (Tritek).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (Gendrel et al., 2002). Immunoprecipitation was performed using a monoclonal RAD51 antibody (Gentex) and Dynabeads® Protein G (Invitrogen). The purified ChIP samples were subject to qPCR analysis. Fold of enrichment was calculated using the comparative Ct method (Schmittgen and Livak, 2008) using the input samples as normalizers.
Microarray Analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen), labeled with MessageAmp Premier RNA Amplification Kit (Ambion) and hybridized with GeneChip Arabidopsis ATH1 Genome Array (Affymetrix) at the Duke Microarray Facility. The microarray data were normalized using Gene-Spring GX Software (RMA algorithm; Agilent).
Pathogen Infection
Pseudomonas syringae ES4326 infection was performed as previously described (Durrant et al., 2007). Three-week-old plants were infiltrated with Psm ES4326 suspension (OD600 nm = 0.0002). The leaves were harvested 3 days after infection.
Supplementary Material
HIGHLIGHTS.
SNI1 is a subunit of the SMC5/6 complex
The SMC5/6 complex negatively regulates RAD17 and ATR
Mutation in SNI1 or salicylic acid treatment induces DNA damage
DNA damage and salicylic acid synergistically induce defense gene expression
ACKNOWLEDGMENTS
We thank Dr. Kevin M. Culligan for sharing the atr and atm mutants; Dr. Qinhong Wang for critiquing the manuscript. This work was funded by grants from NSF (IOS-0744602), NIH (GM069594-05), and Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (through Grant GBMF3032) to X.D., and a fellowship to W.E.D. from the International Human Frontier Science Program Organization.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE44793).
SUPPLEMENTAL INFORMATION
Supplemental Information includes 3 figures, 5 tables, Supplemental Experimental Procedures and Supplemental References and can be found with this article online.
REFERENCES
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong XN. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MRG, Boulton SJ. Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chavez A, George V, Agrawal V, Johnson FB. Sumoylation and the Structural Maintenance of Chromosomes (Smc) 5/6 Complex Slow Senescence through Recombination Intermediate Resolution. J. Biol. Chem. 2010;285:11922–11930. doi: 10.1074/jbc.M109.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong XN. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA. 2007;104:4223–4227. doi: 10.1073/pnas.0609357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of Salicylic-Acid for the Induction of Systemic Acquired-Resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Graham TA, Ferkey DM, Mao F, Kimelman D, Xu WQ. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat. Struct. Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang YL, Clarke JD, Li Y, Dong XN. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks. PloS Genet. 2011;7 doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheletti C, Orland H. MISTRAL: a tool for energy-based multiple structural alignment of proteins. Bioinformatics. 2009;25:2663–2669. doi: 10.1093/bioinformatics/btp506. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song JQ, Dong XN. A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Wohschlegel J, McDonald WH, Yates JR, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair. 2009;8:499–506. doi: 10.1016/j.dnarep.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen YP, Saijo Y, Liu YL, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C–T method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Song JQ, Durrant WE, Wang S, Yan SP, Tan EH, Dong XN. DNA Repair Proteins Are Directly Involved in Regulation of Gene Expression during Plant Immune Response. Cell Host & Microbe. 2011;9:115–124. doi: 10.1016/j.chom.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong XN. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nat. Rev. Clin. Oncol. 2012;9:259–267. doi: 10.1038/nrclinonc.2011.199. [DOI] [PubMed] [Google Scholar]
- Wang S, Durrant WE, Song JQ, Spivey NW, Dong XN. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. USA. 2010;107:22716–22721. doi: 10.1073/pnas.1005978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 Complex Promotes Sister Chromatid Alignment and Homologous Recombination after DNA Damage in Arabidopsis thaliana. Plant Cell. 2009;21:2688–2699. doi: 10.1105/tpc.108.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BBS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.