Abstract
Analysis of histones, especially histone H1, is severely limited by immunological reagent availability. This paper describes the application of cellular fractionation with LC-MS for profiling histones in the cytosol and upon chromatin. First, we show that linker histones enriched by cellular fractionation gives less nuclear contamination and higher histone content than when prepared by nuclei isolation. Second, we profiled the soluble linker histones throughout the cell cycle revealing phosphorylation increases as cells reach mitosis. Finally, we monitored histone H1.2–H1.5 translocation to the cytosol in response to the CDK inhibitor flavopiridol in primary CLL cells treated ex vivo. Data shows all H1 variants translocate in response to drug treatment with no specific order to their cytosolic appearance. The results illustrate the utility of cellular fractionation in conjunction with LC-MS for the analysis of histone H1 throughout the cell.
Keywords: Histone H1, LC-MS, Cellular Compartmentalization
INTRODUCTION
Histones are evolutionarily conserved proteins responsible for the condensation and organization of the DNA in the nucleus. The basic repeating unit of chromatin, the nucleosome core particle, is an octamer made up of two copies of each core histone (H2A, H2B, H3 and H4) and 146 base pairs of superhelical DNA [1-10]. Higher order chromatin structure is achieved by binding of the fifth histone, histone H1, to linker DNA stabilizing the chromatin fiber [11-14]. The histone H1 family has four conserved somatic variants (H1.2, H1.3, H1.4 and H1.5) with seven additional variants having more sporadic expression (H1.0, H1.1, H1.X, H1t, H1T2, H1ILS1, and H1.oo) [15-27]. Although histone H1 has been shown to stabilize the chromatin fibers into higher order chromatin structure, the overall role of specific histone H1 isoforms are still under much debate.
In addition to the function of histone H1 on chromatin structure, the histone H1.2 isoform has been shown to have extra-chromatin function [28-30]. Confocal imaging of mitotic fibroblasts have shown histone H1.2 localized to the cytosol demonstrating compartmental mobility in vitro [30]. Furthermore, several groups have illustrated histone H1.2 is released to the cytoplasm following X-ray induced double strand breaks prompting a Bak dependent release of cytochrome C from the mitochondria and activation of caspase-3, caspase-7 [29,31,32]. Giné et al. reported translocation of histone H1.2 to the cytoplasm in response to therapeutic treatment in chronic lymphocytic leukemia primary cells (CLL) [28]. Although cytoplasmic function of histone H1.2 has been established, the molecular signaling, transport chaperones and cofactors of this process are still to be determined.
Nuclei and hydroxyapatite isolation followed by acid extraction have been the most widely used methods of enrichment for histones [33-37]. While these methods allow for the analysis of the histones bound to the chromatin through high salt or acid extraction, potentially important cytosolic histones are lost [33,35,38,39]. In addition to limited extraction procedures for cytosolic histones, commercial antibody selection and selectivity for the histones, especially histone H1, are inadequate due to high sequence homology and the high degree of post-translational modifications leading to a loss in antibody specificity.
Mass spectral analysis of histones bypasses the limitations set forth by antibody dependent methods and selection. As a result, mass spectrometry has become widely used for protein and post-translational modification sequence identification through global proteomics and enriched protein populations. In this work, we describe a method for examination of histone in the various compartments of the cell. Using mass spectrometry, we analyzed and compared linker histones enriched from the cytosol using both cellular fractionation against those enriched, from the cytosol, using nuclei isolation. The results demonstrate the utility of cellular fractionation combined with LC-MS for the analysis of histone H1 throughout the cell. Specific examples are presented that monitor soluble H1 isoform profiles across cellular compartments in response to drug treatment. Overall our data establishes an alternative method for histone enrichment from soluble cellular compartments for monitoring the extra-chromatin movement of the linker histone H1.
EXPERIMENTAL
Preparation of Cell Lines
Ramos cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured as described by Liu et al. [40]. Briefly, cells were incubated at 37 °C with 5 % CO2 in RPMI-1640 (Life Technologies, Grand Island, NY) supplemented with 10 % fetal bovine serum (FBS) (Sigma Aldrich, St. Louis, MO), 50 U/ml penicillin-G, 50 μg/ml streptomycin, and 2.05 mM L-Glutamine (Life Technologies, Grand Island, NY). Asynchronous cells were harvested in quadruplicate for LC-MS analysis.
Cell Synchronization
Ramos cells were synchronized by double thymidine block as described by Bostock et al. [41]. In short, thymidine (Sigma Aldrich, St. Louis, MO) prepared in sterile PBS (Life Technologies, Grand Island, NY) was added to cell cultures to 2 mM. Cultures were incubated for 18 h, washed twice with PBS, and resuspended in warmed fresh culture media and grown for 9 h. Thymidine block was repeated for 16 h. Cells were released from G1/S arrest by washing twice with PBS and resuspended in fresh warm culture media. Cell aliquots were harvested at 0 h, 3.5 h and 7 h post-release for flow cytometry and histone extraction.
For analysis of histones at various stages in cell cycle, the soluble histones were isolated from the cytoslic component of the cellular fractionation. However, during mitosis, dissolution of the nuclear membrane limits the inability to distinguish the cytosolic and nuclear fractions. Therefore, we refer histones isolated from these enrichments as the soluble histones, which are distinct from those bound to the chromatin.
Acquisition and Preparation of Primary Cells
Written consent was obtained from The Ohio State University Institutional Review Board and patients with previously diagnosed, as described by the NCI criteria, Chronic Lymphocytic Leukemia (CLL) were identified [42]. Peripheral blood mononuclear cells (PBMC) were isolated using density gradient centrifugation (Ficoll-Paque Pus, Pharmacia Biotech, Piscataway, NJ) as described previously [43,44]. B-cells were negatively selected from the PBMC using the RosetteSep® Human B Cell Enrichment Cocktail (STEMCELL Technologies Inc., BC, Canada). Enriched cells were immediately cultured at 1×107 cells/ml in RPMI-1640 media as described above. For drug treatments, primary CLL cells were spun down washed once with PBS and resuspended in RPMI-1640 media supplemented with 10% heat inactivated human AB serum (Valley Biomedical Inc, Winchester, VA). Cells were treated with 1 μM flavopiridol (National Cancer Institute, Bethesda, MD) or left untreated for 0 h, 2 h or 4 h. Aliquots were taken for flow cytometry, western blot and LC-MS analysis.
Histone Extraction by Nuclei Isolation
Histones were extracted as described by Ren et al. from approximately 1×108 cells [45,46]. Briefly, cells were spun down, washed 1x with ice-cold PBS and placed in 1 ml of NP-40 extraction buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5 % NP-40, 0.15 mM Spermine, 0.5 mM Spermidine (aq)) supplemented with enzyme inhibitors (Sigma Protease Inhibitor Cocktail, 100 mM phenylmethyl sulfonyl fluoride, Serine/Threonine Phosphatase Inhibitor Cocktail, Tyrosine Phosphatase Inhibitor Cocktail, Sigma Aldrich, St. Louis, MO). Nuclei were pelleted at 200 × g, washed 1x with ice-cold PBS and allowed to stand in 400 μl of 0.4 N H2SO4 for 30 minutes on ice. Histones were precipitated from the supernatant overnight at -20 °C in ice-cold acetone. Histone pellets were washed once with acetone and resuspended in HPLC-grade water.
Cellular Fractionation Histone Extraction
Approximately 1×108 cells were spun down, washed with ice-cold PBS, and resuspended in CERII buffer (NE-PER, Pierce, Rockford, IL). Suspensions were allowed to stand on ice for 10 min. CERII buffer supplemented with enzyme inhibitors was added to the suspension and allowed to stand for 1 min on ice. Solutions were spun at 21,000 × g for 5 min and the supernatant was transferred to a fresh tube. The pellet was washed with ice-cold PBS and resuspended in NER buffer supplemented with enzyme inhibitors. Nuclei were vortexed at max for 10 sec every 10 min for 40 min total. Following, the suspensions were spun at 21,000 × g for 10 min, supernatants were transferred to fresh tubes and the pellet was washed with ice-cold PBS. Histones were extracted from the remaining DNA through the addition of 400 μl of 0.4 N H2SO4 for 30 min on ice. Additionally, histones were acid extracted from the cytosolic fractions by adding 0.8 N H2SO4 at equal volumes to the protein solution. While it has been debated that acid liable modifications, such as phosphorylation, can be lost during acid extraction, we have shown that acid extraction and hydroxyapatite enrichment yield nearly identical profiles [47,48]. Extracts were spun at 15,000 × g for 10 min, supernatants were transferred to a fresh tube and diluted solution to 2 ml with ice-cold acetone. Samples were precipitated overnight at -20 °C, washed with ice-cold acetone and diluted with HPLC-grade water.
Flow Cytometry
Ramos cells were subjected to cell cycle analysis by DNA content through propidium iodide (PI) uptake following release from double thymidine block as described by Whitfield et al. [49]. Briefly, cells were washed 1x with PBS and fixed with 80 % ethanol (aq) for 10 min. Samples were washed 1x with PBS and stained for 30 min with PI staining solution (50 μg/ml propidium iodide, 10 mM Tris pH 7.5, 5 mM MgCl2, 10 μg/ml DNase free RNase). All samples were stored in the dark for up to 24 h at 4 °C until analysis.
Primary CLL cells were analyzed for Annexin V and propidium iodide negativity. Following treatment, cells were washed 1x with PBS and Annexin V and PI staining solution in binding buffer (Beckman Coulter, Brea, CA) was added. Binding occurred for 15 min in the dark at room temperature. Samples were immediately analyzed following staining. All flow cytometry analyses were completed on a Beckman Coulter FC500 and computationally analyzed using FlowJo cytometry software (Beckman Coulter, Brea, CA, FlowJo, Tree Star Inc., Ashland, OR).
Western Blotting
Protein concentration of cytosolic extracts were determined by Bradford assay (Bio-Rad, Richmond, CA) [50]. Equivalent amounts of the cytosolic fractions were run in 10 % SDS-PAGE gels, transferred to nitrocellulose, and blotted for histone H1.2, Lamin B1 (Abcam, Cambridge, MA), Actin and BRG1 (Santa Cruz Biotechnology, Santa Cruz, CA).
Liquid Chromatography Mass Spectrometry (LC-MS)
Histones extracted from primary CLL cells and cell lines were subjected to LC-MS analysis. Characterization was performed by HPLC separation (Dionex, Waltham, MA) in line with a MicroMass Q-TOF (MicroMass, Milford, MA) mass spectrometer. Approximately 20 μg of histones were separated on a 1.0 × 150mm C18 column (Discovery Bio wide pore C18 column, 5 μm, 300 Å, Supelco, USA) with a modified gradient described by Wang et al. [46]. Mobile phase A contained 0.05 % TFA (Pierce, Rockford, IL) in HPLC water (J.T. Baker, Center Valley, PA). Mobile phase B contained 0.05 % TFA in acetonitrile (EMD Millipore, Billerica, MA). Starting at 20 % B, the gradient increased linearly to 30 % at 2 minutes, 35 % at 10 minutes, 50 % B at 30 minutes, 60 % at 35 minutes and 95 % at 36 minutes. The column was held at 95 % B for 4 minutes. Column equilibration was conducted for 15 minutes at 20 % B. Peaks corresponding to the histone H1’s (H1.5 RT: 24.5-25.2, H1.2-H1.4 RT: 26.9-27.6) were deconvoluted by the MaxEnt algorithm and analyzed using the MassLynx software 4.0 (Waters Corp., Milford, MA). Statistical analysis was performed using Prism GraphPad software (GraphPad Software Inc., La Jolla, CA).
RESULTS AND DISCUSSION
Comparison of Histone Extraction Methods
Sample preparation is an important but often overlooked aspect of histone characterization. Many of the current extraction and isolation methods for histones have been in use since the 1970’s [33-36,38,39]. The most common methods extract histones from isolated nuclei using acid or high ionic strength buffers followed by organic solvent precipitation. Nuclei are enriched through gentle lysis of the cell membrane, leaving the nuclear membrane intact. Acid or high ionic buffers added to the nuclei disrupt the histone-DNA interactions solubilizing the proteins. While nuclei isolation allows for the analysis of chromatin bound histones, there is increasing evidence that extra-chromatin histones have functional roles [28,29,33-36]. Konishi et al. demonstrated that histones could be isolated from cellular compartments and subsequently characterized by LC-MS [29]. They isolated and characterized linker histones from cytoplasmic and nucleosolic fractions from mouse thymus tissue through homogenization. While homogenization is shown to be effective for characterization of histones by LC-MS in the cellular compartments, our work utilizes a commercially available nuclear and cytosolic isolation kit for a more robust extraction of proteins from these compartments. Additionally, we improve upon the established approach by allowing for simultaneous LC-MS profiling of linker and core histones and by including the characterization of chromatin bound histone fraction.
In the following sections, the power of cellular fractionation and analysis of linker histones is illustrated. The first example provides a comparison between cytosolic histones profiles obtained by the nuclear isolation approach and the cellular fractionation approach. The second example examines changes in the soluble histone, those not bound to the chromatin, across the cell cycle using Ramos Burkitt’s lymphoma cells. The final example monitors linker histone translocation across cellular compartments in primary Chronic Lymphocytic Leukemia (CLL) cells treated with kinase inhibitors. These data demonstrate the utility of cellular fractionation as a robust method to characterize soluble histones.
It is necessary to first establish cross-contamination of histones between the nucleus and the cytosol for each approach. The cytosolic fraction contains the least amount of histones and thus is more prone to contamination by soluble nuclear histones and chromatin. To compare enrichment of the cytosolic fraction obtained by nuclei isolation and cellular fractionation, we performed an immunoblot on the cytosolic fraction obtained by each method probing for a protein only found in the nucleus, Lamin B1 (Figure 1A). The western blots show less Lamin B1 in the cytosol of those samples isolated through cellular fractionation when compared to nuclei isolation. While it is not possible to completely eliminate nuclear cross-contamination in the cytosol of the fractionation method, our data shows that the nuclear contamination is significantly reduced as it is observed only in overexposed gel images (Supplemental Data 1). Additionally, the blot in Figure 1A shows higher histone H1.2 content in the cytosol from the cellular fractionation method. Cytosolic histones obtained by both approaches were further characterized by LC-MS. Representative spectra obtained by each isolation method are presented in Figure 1B & 1C (additional replicate samples are provided in Supplemental Data 2). These data show the fractionation method yields cytosolic histones in higher abundance than the traditional nuclei isolation method, supporting the western blot data. Only one nuclei isolation replicate yielded enough histones for LC-MS characterization, Figure 1B. These results demonstrate cellular fractionation yields cytosolic extracts with less nuclear contamination and greater histone content than the cytosolic extracts prepared by traditional nuclei isolation.
Figure 1.
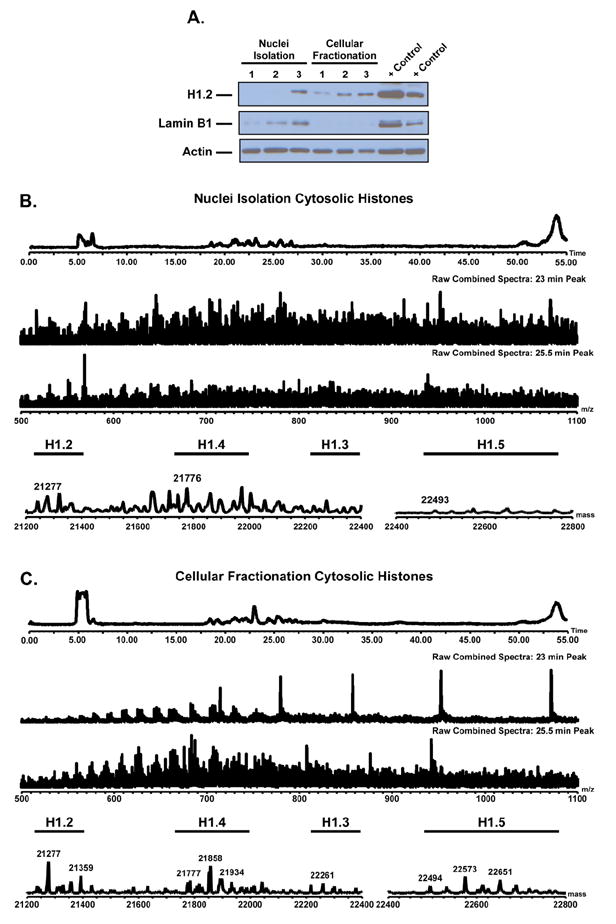
Western blot and representative LC-MS spectra for cytosolic histones prepared by nuclei isolation and cellular fractionation. A) Western blot of cytosolic lysate from each isolation method (n = 3). Blots shown for histone H1.2, Actin (Loading Control), Lamin B1 (Nuclear Control). Blot shows cellular fractionation contains less nuclear specific protein and more histone H1.2 than the nuclei isolation method. LC-MS of histones extracted from the cytosol of each isolation approach B) Nuclei isolation C) Cellular fractionation (n = 4). Spectra show histone H1 is more prevalent in the cytosol of the cellular fractionation enrichments. Results demonstrate the cellular fractionation provides preparations with less nuclear contamination and greater histone content.
To ensure the method of cellular fractionation does not bias the histone profiles, we compared the chromatin bound histone fraction to histones obtained by traditional nuclei isolation. Supplemental Data 3 shows the LC-MS spectra of the highly phosphorylated somatic histone H1 isoforms (H1.2, H1.3, H1.4 and H1.5) enriched using non-ionic NP-40 nuclei isolation vs. the commercial cellular fractionation kit. The spectra for each isolation method show similar peak abundances and extent of phosphorylations (identified by a shift in mass of 80 Da) for both extraction methods. Additionally, both spectra show evidence of an allelic variant (A142T) of histone H1.2 denoted by a change in 30 Da as reported previously for HeLa cells by Zheng et al. [51]. These data qualitatively suggest the cellular fractionation method does not bias the histone relative abundance profiles of those histones isolated from the chromatin when compared to traditional nuclei isolation. Collectively, the data shows the cellular fractionation method is capable of isolating more soluble histone H1 with less nuclear cross-contamination than the nuclei isolation method while preserving the relative abundance profile of chromatin bound histone H1 isoforms.
Distribution of Histone H1 as a Function of the Cell Cycle
Historically, it has been established that the core histones were primarily synthesized in the cytosol during S phase of the cell cycle in parallel with the DNA synthesis [52,53]. These newly produced histones exhibit regular patterns of post-translation modifications [54]. For example, newly synthesized histone H4 are acetylated in the cytosol at lysines 5 and 12 of its N-terminal tail [54]. While core histone post-translational modifications have been noted in the cytosol of replication dependent synthesis, no such data exists for the linker histone H1 variants [55].
Similarly to the core histones, histone H1 variants have been shown to have primarily DNA synthesis dependent in addition to a smaller proportion independent of DNA synthesis [56-59]. Linker histone isoforms have been shown to be synthesized in variable amounts throughout S phase of the cell cycle [60,61]. Additionally, H1 variants can be produced differently depending on the cell state, i.e. proliferating or quiescent cells [59,62-65]. Because histone H1 variants are synthesized in the cytosol primarily during DNA replication across S phase of the cell cycle, we utilized the samples from double thymidine cell synchronization with LC-MS analysis to characterize the soluble and chromatin bound histone H1 profiles [56]. However, cell cycle analysis is limited by the inability to distinguish the cytosolic and nuclear fractions during mitosis due to the dissolution of the nuclear membrane. Therefore, we refer to these histones as the soluble histones, which are distinct from those bound to the chromatin.
Figure 2 Left Column shows the cell cycle analysis by propidium iodide uptake (DNA content) for Ramos cells synchronized by double thymidine block (A. Unsynchronized, B. G1/S phase, C. S phase, D. G2/M phase). As the cells progress from G1/S through S phase into G2/M, the content of DNA doubles (from 2N to 4N) which can be measured through an increase in propidium iodide fluorescent intensity, as shown by a shift along the x-axis in Figures 2B-2D [41]. Figure 2D also shows a small peak in the 2N region of the graph. This is attributed to cells that complete replication and cycle back to the G1/S phase and/or cells that never resumed the cell cycle following release of the thymidine block. Figure 2 Right Column shows the LC-MS spectra for the soluble histone H1 isoforms isolated from the cytosolic enrichment of the synchronized cells (Supplemental Data 4). As expected, only a small amount of histone H1 was enriched from this fraction [66]. Although low abundant, the histone H1 isoforms in the soluble fraction were enriched in each phase of the cell cycle. Variants H1.5 and H1.4 were found in across the cell cycle whereas H1.2 and H1.2(A142T) were only found in the G2/M fraction [51]. The abundance of the singly phosphorylated histone H1.4 isoform was variable across the different phases with a decrease in S phase. Histone H1.5 was doubly phosphorylated in G1/S and S phase and triply phosphorylated in G1/M phase. It is well established that phosphorylation of chromatin bound histone H1 increases as cells progress through the cell cycle with maximum phosphorylation occurring during the mitotic phase [34,67-72]. Gréen et al., through the use of confocal microscopy and a pan phospho-H1 antibody, have shown phosphorylation of histone H1 in the cytosolic space during the cell cycle [30]. Our data demonstrates for the first time that the number of phosphorylation on specific cytosolic histone H1 variants (H1.5) also increases in G2/M.
Figure 2.
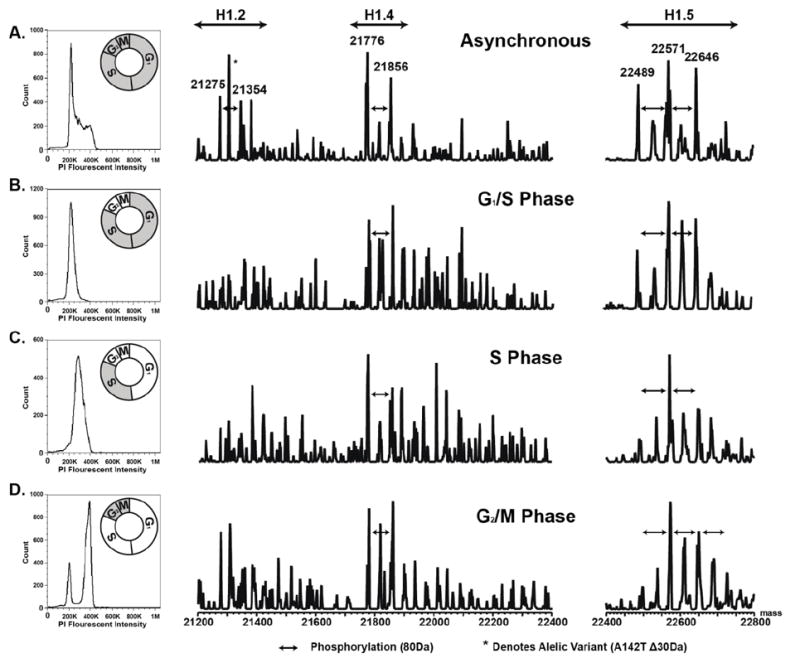
Flow cytometry of propidium iodide uptake from Ramos cells (Left). LC-MS spectra of histone H1 isolated from the soluble nuclei free fraction of Ramos cells (Right). A) Unsynchronized. B) 0h time point (G1/S). C) 3.5h time point (S). D) 7h time point (G2/M). Results demonstrate the enrichment and LC-MS analysis of nuclei free histone H1 from the cytosol of asynchronous and synchronized cells.
The LC-MS spectra for the chromatin bound histone H1 isoforms is shown in Figure 3 (Supplemental Data 4). As expected, these data support the existing body of data that show phosphorylation of chromatin bound histone H1 increases as cells traverse the cell cycle [34,67-72]. The most evident increase in phosphorylation was that of the histone variant H1.5. While not as profound, histone variants H1.2 and H1.4 also exhibited increased phosphorylation abundance in G2/M. These data clearly show the increase in histone H1’s phosphorylation on chromatin in G2/M of the cell cycle, which is consistent with established hypotheses [34,67-72]. Collectively, these experiments illustrate the utility of the fractionation method for LC-MS analysis of those histones on and off of the chromatin. These data establish the soluble histone H1 is variably phosphorylated across the cell cycle.
Figure 3.
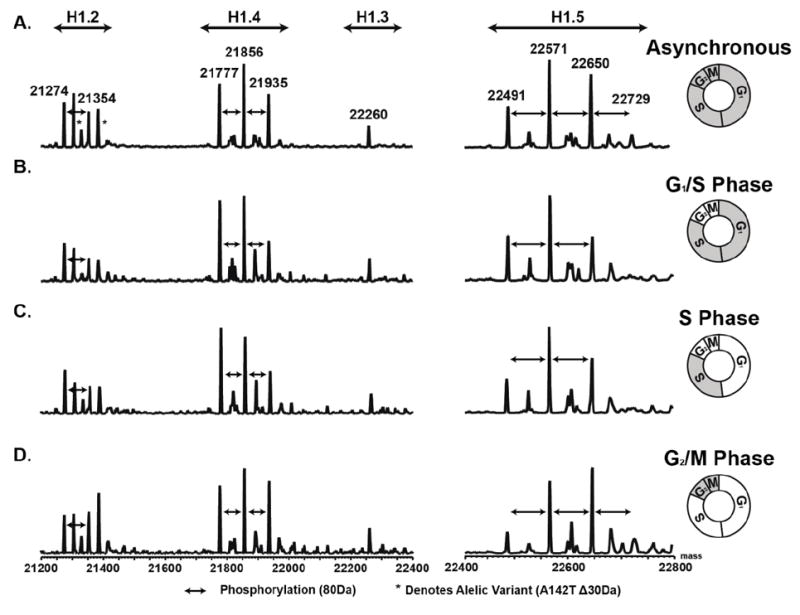
LC-MS spectra of histone H1 isolated from the chromatin of unsynchronized and synchronized Ramos cells. A) Unsynchronized. B) 0h time point (G1/S). C) 3.5h time point (S). D) 7h time point (G2/M). Data demonstrates the isolation LC-MS analysis of histone H1 from the chromatin of asynchronous and synchronized cells.
Characterization of Histone H1 Translocation
Research into extra-chromatin histone H1 has established a significant importance in the radiation induced double strand break apoptotic pathway [29,31,32]. Additionally, histone H1.2 has been reported to translocate from the nucleus to the cytoplasm in response to drug treatments in chronic lymphocytic leukemia (CLL) primary cells [28]. These established roles of histone H1.2 in the cytoplasm led us to evaluate the role of the linker histone H1 in primary CLL patient ex vivo cell response to the therapeutically relevant cyclin dependent kinase inhibitor flavopiridol.
To assess the role of histone H1 in the cytosol, we treated primary CLL B-cells with 1 μM flavopiridol in vitro. Following treatment, soluble and chromatin bound histones were isolated for LC-MS and an aliquot of cytosolic lysate was analyzed by western blot. Figure 4A-C shows representative mass spectra of soluble histones extracted from the cytosol of flavopiridol (1 μM) ex vivo treated primary CLL cells at 0 h, 2 h and 4 h time points. These data, and Supplemental Figures 5A-C & 6A-C, show a time dependent increase in the soluble histone H1 isoforms in the cytosol with flavopiridol treatment. While there is no specific order to when each specific isoform becomes present in the cytosol, nearly all of the isoforms are present at the 4 h time point, which is in line with a decrease in viability (Annexin V/propidium iodide negativity, Figure 4D). Western blotting of the cytosolic lysate with an antibody against histone H1.2 revealed a time dependent increase in H1.2 in this compartment of the cell (Figure 4D). These data suggest histone H1 becomes present in the cytosol as the CLL cell viability decreases in response to 1 μM flavopiridol treatment. Similarly, the media control sample showed results comparable to the flavopiridol treated sample in both western blot and LC-MS analyses of the cytosolic fraction (Supplemental Figure 5A-D).
Figure 4.
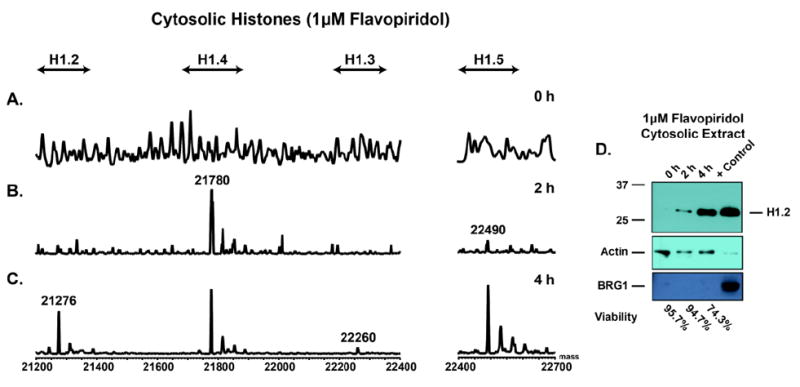
LC-MS spectra of histone H1 isolated from the cytosol of flavopiridol treated CLL primary cells (patient 1 of 3). A) 0 h time point. B) 2 h time point. C) 4 h time point. D) Representative western blot of cytosolic lysate from flavopiridol treated cells. Blots for histone H1.2, Actin (Loading Control), and BRG1 (Nuclear Control). Data shows the LC-MS is consistent with the western blot of the corresponding cytosolic extracts. Additionally, the data demonstrates an increase in histone H1 in the cytosol with flavopiridol treatment as viability decreases.
The LC-MS profiles for the chromatin bound histone H1’s were similar at each flavopiridol time point (Figure 5, Supplemental Figure 5E-G). The core histones were only observed in the chromatin bound fraction. Replicates for two additional independent CLL patient samples show similar results (Supplemental Figures 6-9) for both the cytosolic and chromatin bound histones. These data show the fractionation method allows for the analysis of soluble histones in the cytosol, by not only western blotting, but also by LC-MS. Additionally, the LC-MS and western blots show an increase in histone H1 in the cytosol following flavopiridol and media treatment inline with decreasing CLL cell viability.
Figure 5.
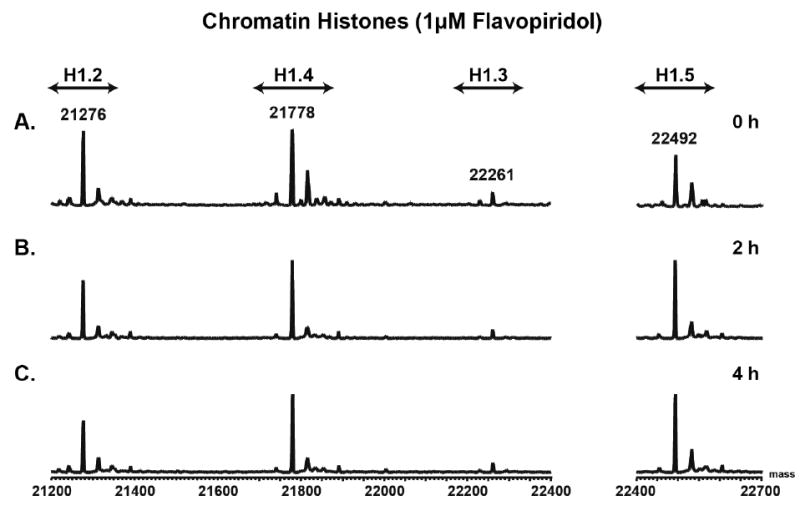
Representative LC-MS spectra of histone H1 isolated from the chromatin of flavopiridol treated CLL primary cells (patient 1 of 3). A) 0 h time point. B) 2 h time point. C) 4 h time point. Data shows the LC-MS spectra show no difference in the chromatin bound histone H1’s following flavopiridol treatment.
LC-MS spectra, for both the cytosolic histones and the chromatin bound histones, show no significant phosphorylation for peripheral CLL cells. This observation can be explained as most circulating CLL cells are in a quiescent state [73]. It is established histone H1 becomes phosphorylated through S phase with the maximum phosphorylation in M phase [34,67-72]. As a result, these cells do not have appreciable H1 phosphorylation.
As noted by Gréen et al., phosphorylated histone H1 in addition to unmodified H1.2 and H1.5 have been identified in the cytoplasm by confocal microscopy at specific times during the cell cycle in normal human fibroblasts [30]. Here, we similarly identify soluble histone H1.2 in the cytosol during G2/M phase. Konishi et al. and Giné et al. have also reported that cytosolic histone H1.2 induces apoptosis in response to X-ray induced double stranded breaks, as well as genotoxic and non-genotoxic therapies [28,29]. Collectively, these studies suggest there is a level of regulation beyond mere H1.2 localization. This type of regulation could be an H1.2 post-translational modification(s), such as phosphorylation, or a chaperone sequestering cytoplasmic H1.2 from inducing apoptosis. Much of this work has relied on the availability of antibodies that recognize histone variants and their isoforms. The combined cellular fractionation and LC-MS provides a broader more unbiased assessment of histone profiles in the compartments.
With increasing evidence for the functionality of extra-chromatin histone H1, the need for methods to bypass the limitations in H1 antibody selection and specificity are paramount. LC-MS and LC-MS/MS are the most commonly used approaches to circumvent limited immuno-reagent availability. The method presented here illustrates cellular fractionation combined with LC-MS for histone profiling. We have established that cellular fractionation protocol that is capable of enriching soluble cytosolic linker histones for anlaysis by LC-MS. Thus, we were able to monitor the profile changes in these histones during the cell cycle in synchronized Ramos cells and in primary CLL cells treated with a clinically relevant CDK inhibitor.
Supplementary Material
SIGNIFICANCE.
This paper demonstrates the first time application of cellular fractionation to characterize cytosolic histone H1 by liquid chromatography mass spectrometry (LC-MS). Using the Ramos Burkett’s lymphoma cell line, cellular fractionation was shown to give less nuclear contamination and higher histone content than preparations by nuclei isolation. Further application of the cellular fractionation approach was shown by using primary chronic lymphocytic leukemia (CLL) cells to monitor the movement of histone H1 across cellular compartments in response to the cyclin dependent kinase inhibitor flavopiridol. Collectively, these data establish a mass spectrometric method for exploration into the function of cytosolic histone H1.
Highlights.
-
➢
efficient method for isolation of histone H1 from cellular compartments.
-
➢
demonstrates compatibility with LC-MS analysis.
-
➢
shows similar LC-MS of histone H1 with the nuclei isolation method.
-
➢
monitored histone H1 compartmental movement in primary CLL cells.
Acknowledgments
This study was funded by the Leukemia and Lymphoma Society’s Specialized Center of Research (SCOR) grant #7004-11.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–2. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA. 1975;72:2626–30. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- 5.Whitlock JP, Simpson RT. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976;15:3307–14. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- 6.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 7.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–31. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 8.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–52. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 10.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker Histones Stabilize the Intrinsic Salt-Dependent Folding of Nucleosomal Arrays: Mechanistic Ramifications for Higher-Order Chromatin Folding. Biochemistry. 1998;37:14776–87. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 12.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Holde KE. Chromatin. New York: Springer-Verlag; 1989. [Google Scholar]
- 14.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–27. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carozzi N, Marashi F, Plumb M, Zimmerman S, Zimmerman A, Coles LS, et al. Clustering of human H1 and core histone genes. Science. 1984;224:1115–7. doi: 10.1126/science.6719136. [DOI] [PubMed] [Google Scholar]
- 16.Eick S, Nicolai M, Mumberg D, Doenecke D. Human H1 histones: conserved and varied sequence elements in two H1 subtype genes. Eur J Cell Biol. 1989;49:110–5. [PubMed] [Google Scholar]
- 17.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101:284–94. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 18.Drabent B, Franke K, Bode C, Kosciessa U, Bouterfa H, Hameister H, et al. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm Genome. 1995;6:505–11. doi: 10.1007/BF00356166. [DOI] [PubMed] [Google Scholar]
- 19.Drabent B, Kardalinou E, Doenecke D. Structure and expression of the human gene encoding testicular H1 histone (H1t) Gene. 1991;103:263–8. doi: 10.1016/0378-1119(91)90284-i. [DOI] [PubMed] [Google Scholar]
- 20.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, et al. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci USA. 2005;102:2808–13. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka H, Matsuoka Y, Onishi M, Kitamura K, Miyagawa Y, Nishimura H, et al. Expression profiles and single-nucleotide polymorphism analysis of human HANP1/H1T2 encoding a histone H1-like protein. Int J Androl. 2006;29:353–9. doi: 10.1111/j.1365-2605.2005.00600.x. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–64. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- 23.Yan W. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proceedings of the National Academy of Sciences. 2003;100:10546–51. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto T, Horikoshi M. Cloning of the cDNA encoding a novel subtype of histone H1. Gene. 1996;173:281–5. doi: 10.1016/0378-1119(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 25.Happel N, Schulze E, Doenecke D. Characterisation of human histone H1x. Biol Chem. 2005;386:541–51. doi: 10.1515/BC.2005.064. [DOI] [PubMed] [Google Scholar]
- 26.Doenecke D, Tönjes R. Differential distribution of lysine and arginine residues in the closely related histones H1 ° and H5. J Mol Biol. 1986;187:461–4. doi: 10.1016/0022-2836(86)90446-8. [DOI] [PubMed] [Google Scholar]
- 27.Albig W, Kardalinou E, Drabent B, Zimmer A, Doenecke D. Isolation and characterization of two human H1 histone genes within clusters of core histone genes. Genomics. 1991;10:940–8. doi: 10.1016/0888-7543(91)90183-f. [DOI] [PubMed] [Google Scholar]
- 28.Giné E, Crespo M, Muntañola A, Calpe E, Baptista MJ, Villamor N, et al. Induction of histone H1.2 cytosolic release in chronic lymphocytic leukemia cells after genotoxic and non-genotoxic treatment. Haematologica. 2008;93:75–82. doi: 10.3324/haematol.11546. [DOI] [PubMed] [Google Scholar]
- 29.Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y, et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–88. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 30.Gréen A, Lönn A, Peterson KH, Ollinger K, Rundquist I. Translocation of histone H1 subtypes between chromatin and cytoplasm during mitosis in normal human fibroblasts. Cytometry A. 2010;77:478–84. doi: 10.1002/cyto.a.20851. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Vela A, Korsmeyer SJ. Proapoptotic histone H1.2 induces CASP-3 and -7 activation by forming a protein complex with CYT c, APAF-1 and CASP-9. FEBS Lett. 2007;581:3422–8. doi: 10.1016/j.febslet.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 32.Okamura H, Yoshida K, Amorim BR, Haneji T. Histone H1.2 is translocated to mitochondria and associates with Bak in bleomycin-induced apoptotic cells. J Cell Biochem. 2008;103:1488–96. doi: 10.1002/jcb.21537. [DOI] [PubMed] [Google Scholar]
- 33.Jamaluddin M, Mohananphilip, Chandra HS. A rapid and gentle method for the salt extraction of chromatin core histones H2A, H2B, H3 and H4 from rat liver nuclei. J Biosci. 1979;1:49–58. [Google Scholar]
- 34.D’Anna JA, Gurley LR, Deaven LL. Dephosphorylation of histones H1 and H3 during the isolation of metaphase chromosomes. Nucleic Acids Res. 1978;5:3195–208. doi: 10.1093/nar/5.9.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weintraub H, Palter K, Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975;6:85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- 36.Shaw BR, Herman TM, Kovacic RT, Beaudreau GS, Van Holde KE. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci USA. 1976;73:505–9. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su X, Jacob NK, Amunugama R, Hsu P-H, Fishel R, Freitas MA. Enrichment and characterization of histones by two-dimensional hydroxyapatite/reversed-phase liquid chromatography-mass spectrometry. Anal Biochem. 2009;388:47–55. doi: 10.1016/j.ab.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MURRAY K. The acid extraction of histones from calf thymus deoxyribonucleoprotein. J Mol Biol. 1966;15:409–19. doi: 10.1016/s0022-2836(66)80116-x. [DOI] [PubMed] [Google Scholar]
- 39.DAVISON PF, JAMES DW, Shooter KV, BUTLER JA. The histones of calf thymus deoxyribonucleoprotein. II Electrophoretic and sedimentation behaviour and a partial fractionation. Biochim Biophys Acta. 1954;15:415–24. doi: 10.1016/0006-3002(54)90045-1. [DOI] [PubMed] [Google Scholar]
- 40.Liu Q, Zhao X, Frissora F, Ma Y, Santhanam R, Jarjoura D, et al. FTY720 demonstrates promising preclinical activity for chronic lymphocytic leukemia and lymphoblastic leukemia/lymphoma. Blood. 2008;111:275–84. doi: 10.1182/blood-2006-10-053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bostock CJ, Prescott DM, Kirkpatrick JB. An evaluation of the double thymidine block for synchronizing mammalian cells at the G1-S border. Exp Cell Res. 1971;68:163–8. doi: 10.1016/0014-4827(71)90599-4. [DOI] [PubMed] [Google Scholar]
- 42.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O’Brien S, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–7. [PubMed] [Google Scholar]
- 43.Byrd JC, Shinn C, Ravi R, Willis CR, Waselenko JK, Flinn IW, et al. Depsipeptide (FR901228): a novel therapeutic agent with selective, in vitro activity against human B-cell chronic lymphocytic leukemia cells. Blood. 1999;94:1401–8. [PubMed] [Google Scholar]
- 44.Byrd JC, Shinn C, Waselenko JK, Fuchs EJ, Lehman TA, Nguyen PL, et al. Flavopiridol induces apoptosis in chronic lymphocytic leukemia cells via activation of caspase-3 without evidence of bcl-2 modulation or dependence on functional p53. Blood. 1998;92:3804–16. [PubMed] [Google Scholar]
- 45.Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. Peptide mass mapping of acetylated isoforms of histone H4 from mouse lymphosarcoma cells treated with histone deacetylase (HDACs) inhibitors. J Am Soc Mass Spectrom. 2005;16:1641–53. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Harshman SW, Liu S, Ren C, Xu H, Sallans L, et al. Assaying pharmacodynamic endpoints with targeted therapy: flavopiridol and 17AAG induced dephosphorylation of histone H1.5 in acute myeloid leukemia. Proteomics. 2010;10:4281–92. doi: 10.1002/pmic.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujitaki JM, Fung G, Oh EY, Smith RA. Characterization of chemical and enzymatic acid-labile phosphorylation of histone H4 using phosphorus-31 nuclear magnetic resonance. Biochemistry. 1981;20:3658–64. doi: 10.1021/bi00515a055. [DOI] [PubMed] [Google Scholar]
- 48.Su X, Jacob NK, Amunugama R, Lucas DM, Knapp AR, Ren C, et al. Liquid chromatography mass spectrometry profiling of histones. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;850:440–54. doi: 10.1016/j.jchromb.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, et al. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol. 2010;189:407–15. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borun TW, Gabrielli F, Ajiro K, Zweidler A, Baglioni C. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975;4:59–67. doi: 10.1016/0092-8674(75)90134-8. [DOI] [PubMed] [Google Scholar]
- 53.Robbins E, Borun TW. The cytoplasmic synthesis of histones in hela cells and its temporal relationship to DNA replication. Proc Natl Acad Sci USA. 1967;57:409–16. doi: 10.1073/pnas.57.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–41. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–95. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plumb M, Marashi F, Green L, Zimmerman A, Zimmerman S, Stein J, et al. Cell cycle regulation of human histone H1 mRNA. Proc Natl Acad Sci USA. 1984;81:434–8. doi: 10.1073/pnas.81.2.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Appels R, Ringertz NR. Metabolism of F1 histone in G1 and G0 cells. Cell Differ. 1974;3:1–8. doi: 10.1016/0045-6039(74)90035-9. [DOI] [PubMed] [Google Scholar]
- 58.Gurley LR, Walters RA, Tobey RA. The metabolism of histone fractions. IV Synthesis of histones during the G1-phase of the mammalian life cycle. Arch Biochem Biophys. 1972;148:633–41. doi: 10.1016/0003-9861(72)90182-8. [DOI] [PubMed] [Google Scholar]
- 59.Tarnowka MA, Baglioni C, Basilico C. Synthesis of H1 histones by BHK cells in G1. Cell. 1978;15:163–71. doi: 10.1016/0092-8674(78)90092-2. [DOI] [PubMed] [Google Scholar]
- 60.Hohmann P, Cole RD. Hormonal effects on amino acid incorporation into lysine-rich histones in the mouse mammary gland. J Mol Biol. 1971;58:533–40. doi: 10.1016/0022-2836(71)90369-x. [DOI] [PubMed] [Google Scholar]
- 61.Sizemore SR, Cole RD. Asynchronous appearance of newly synthesized histone H1 subfractions in HeLa chromatin. J Cell Biol. 1981;90:415–7. doi: 10.1083/jcb.90.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurley LR, Walters RA, Tobey RA. The metabolism of histone fractions. Arch Biochem Biophys. 1974;164:469–77. doi: 10.1016/0003-9861(74)90057-5. [DOI] [PubMed] [Google Scholar]
- 63.Zlatanova J. Lack of coupling between DNA and histone synthesis in growth-arrested Friend erythroleukemia cells. Mol Cell Biochem. 1981;35:49–53. doi: 10.1007/BF02358187. [DOI] [PubMed] [Google Scholar]
- 64.Pehrson JR, Cole RD. Histone H1 subfractions and H10 turnover at different rates in nondividing cells. Biochemistry. 1982;21:456–60. doi: 10.1021/bi00532a006. [DOI] [PubMed] [Google Scholar]
- 65.D’Anna JA, Gurley LR, Tobey RA. Syntheses and modulations in the chromatin contents of histones H1 degrees and H1 during G1 and S phases in Chinese hamster cells. Biochemistry. 1982;21:3991–4001. doi: 10.1021/bi00260a014. [DOI] [PubMed] [Google Scholar]
- 66.Zlatanova JS, Srebreva LN, Banchev TB, Tasheva BT, Tsanev RG. Cytoplasmic pool of histone H1 in mammalian cells. J Cell Sci. 1990;96(Pt 3):461–8. doi: 10.1242/jcs.96.3.461. [DOI] [PubMed] [Google Scholar]
- 67.Gurley LR, Walters RA, Tobey RA. Sequential phsophorylation of histone subfractions in the Chinese hamster cell cycle. J Biol Chem. 1975;250:3936–44. [PubMed] [Google Scholar]
- 68.Gurley LR, D’Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 69.Matsumoto Y, Yasuda H, Mita S, Marunouchi T, Yamada M. Evidence for the involvement of H1 histone phosphorylation in chromosome condensation. Nature. 1980;284:181–3. doi: 10.1038/284181a0. [DOI] [PubMed] [Google Scholar]
- 70.Ajiro K, Borun TW, Cohen LH. Phosphorylation states of different histone 1 subtypes and their relationship to chromatin functions during the HeLa S-3 cell cycle. Biochemistry. 1981;20:1445–54. doi: 10.1021/bi00509a007. [DOI] [PubMed] [Google Scholar]
- 71.Hohmann P, Tobey RA, Gurley LR. Phosphorylation of distinct regions of f1 histone. Relationship to the cell cycle. J Biol Chem. 1976;251:3685–92. [PubMed] [Google Scholar]
- 72.Bradbury EM, Inglis RJ, Matthews HR, Sarner N. Phosphorylation of very-lysine-rich histone in Physarum polycephalum. Correlation with chromosome condensation. Eur J Biochem. 1973;33:131–9. doi: 10.1111/j.1432-1033.1973.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 73.Reed JC. Molecular biology of chronic lymphocytic leukemia. Semin Oncol. 1998;25:11–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


