Abstract
Objectives and design
A vaccine capable of providing cross-clade, sterilizing protection has been the holy grail of HIV-1 prevention and control since the beginning of the pandemic. A major component of this effort has been the identification and characterization of broadly neutralizing antibodies (bNAbs). Recent advances in bNAb isolation, structure-based engineering, and vector-mediated gene transfer have led to increased interest in bypassing the immune system by expressing neutralizing antibodies directly in muscle. To assess the neutralization potency and coverage of a panel of second-generation bNAbs, we cloned and phenotypically characterized 227 primary HIV-1 envelopes from 23 mother-to-child transmission (MTCT) pairs.
Methods
Viral envelopes were tested for in-vitro neutralization sensitivity using a standard pseudotype assay system. A 50% inhibitory concentration (IC50) at least 10 μg/ml was used to define neutralization resistance.
Results
The combination of antibodies PG16 and NIH45–46G54W had the broadest activity with the highest neutralization potency, achieving full coverage of 87% of transmission pairs (at a median sampling depth of 10 envelopes per pair) and 96% of recently infected infants in a very conservative analysis.
Conclusions
Our data strongly support the inclusion of NIH45–46G54W, or a more extensively modified variant, in future proof-of-principle immunoprophylaxis or gene therapy-based trials. Furthermore, until robust sequence-based resistance detection becomes available, it will be necessary to conduct deeper phenotypic screening of primary isolates in order to determine the prevalence of minor resistant variants to help in selecting the best reagents for clinical trials.
Keywords: AIDS, antibodies, HIV, neutralization, paediatrics, prevention of mother-to-child transmission, vaccine
Introduction
Despite three decades of effort, we still do not have a universal HIV-1 vaccine, and it will be difficult to fully control the pandemic without one. Most chronically HIV-infected individuals, and most vaccine immunogens tested to date, produce a somewhat narrow, strain-specific response that does not afford significant protection against heterologous challenge [1–3]. Fortunately, some chronically HIV-infected individuals do eventually produce an antibody response with the cross-clade activity desirable in a vaccine [2,4,5]. Thus, a major component of the vaccine development effort is the isolation and characterization of broadly neutralizing antibodies (bNAbs) in order to decipher the molecular characteristics, and eventually the ontogeny, of such a response [6–9].
Some bNAbs have been shown to confer sterilizing protection via passive immunoprophylaxis in animal challenge models [10–14], and delay virologic rebound during treatment interruption in humans [15]. Furthermore, proof-of-concept studies in mice and macaques have demonstrated protection when bNAbs are expressed using vector-mediated gene transfer, raising the possibility that difficulties in immunogen design might be bypassed by the induction of durable immunity through gene therapy approaches [16–19]. Given the substantial costs, risks, and regulatory issues involved in deploying this technology in human clinical trials, let alone eventually treating the hundreds of millions of people at risk for HIV acquisition, selection of the most broadly active and potent antibody or combination of antibodies is a vital prerequisite.
To quantify the degree of ‘broadness’ of the available bNAbs, studies to date have generally sampled a single or a very small number of viral isolates from a large number of patients [20–22]. However, it is important to note that the patients from whom these bNAbs were originally isolated do not fully control their own virus, and circulating quasispecies variants from one patient studied longitudinally show evidence of ongoing escape from the contemporaneous antibody repertoire [23]. Thus, it is important to determine the frequency of pre-existing minor variants resistant to the relevant bNAbs before they are deployed within a population, as such variants could result in treatment failure. Unfortunately, due to the complexity of the targeted epitopes and our incomplete knowledge of the envelope trimer structure, robust sequence-based resistance screening is currently impossible. Consequently, direct phenotypic testing of multiple primary isolates from both chronically and newly infected patients is required to characterize the patterns of baseline resistance present in the target population.
For much of the past two decades, there have been only four bNAbs in our armamentarium, all isolated from subtype B-infected patients: 4E10, 2F5, b12, and 2G12 [24–27]. These antibodies have limited activity in non-B subtypes, with the exception of 4E10, which neutralizes most strains with low to moderate potency [28]. These limitations greatly diminish their usefulness in the epicenter of the pandemic (sub-Saharan Africa) where subtype C dominates. However, since 2009, there has been a rapid increase in the pace of identification of anti-HIV bNAbs [20,22,29–31], resulting in a much needed bolus of potent new antibodies and essentially reversing the problem; now they are being developed faster and in greater numbers than can be feasibly put into clinical trials.
Many of these antibodies target two highly conserved sites on gp120: either a quaternary epitope including V1–V3 (PG series) or the CD4-binding site (VRC, NIH45, 3BNC, and 8ANC series), and have good activity against nonsubtype B viruses [29–31]. Given that antibodies against these two sites do not interfere with each other [21], but antibodies to each individual site compete amongst themselves [30,31], we sought to determine what combination of available bNAbs had the highest potency and broadest coverage against a large panel of primary isolates from subtype C-infected women and their infants. Due to the relatively large number of molecular envelope clones available from each transmission pair (median of 10 unique envelopes per pair), this cohort allowed us to look at coverage and neutralization sensitivity at greater depth than has been previously reported.
Our study found the combination of PG16 and the engineered variant NIH45–46G54W was the broadest and most potent, neutralizing all tested variants from the greatest number of transmission pairs, and a striking 96% of recently infected infants. Since vector-mediated gene transfer obviates the need to elicit a ‘natural’ antibody response, our data strongly support the use of engineered variants like NIH45–46G54W, or even more extensively modified antibodies, as the basis for future human clinical trials.
Materials and methods
Patient population
All women were participants in the Zambia Exclusive Breastfeeding Study (ZEBS); a clinical trial for the prevention of mother-to-child transmission (MTCT) that has been previously described [32]. Briefly, antiretroviral drug-naive, pregnant, HIV-infected women were prospectively enrolled and maternal blood was collected at study entry; and maternal blood and breast milk were collected at specified intervals following delivery. Infant blood was collected at 3 months after delivery. Antiretroviral drug exposure during the study was limited to single-dose peripartum nevirapine, given in accordance with Zambian government guidelines at the time. No patients had been treated with bNAbs or HIV Immune Globulin (HIVIG). Twenty-three mother/infant transmission pairs were included in the analyses.
Envelope cloning
Molecular cloning of full-length HIV gp160 genes has been described previously [33]. Envelopes were obtained from breast milk cells, plasma RNA, and PBMC DNA; previous work has shown a lack of genetic compartmentalization for these sample types [33]. All envelope sequences were screened for re-sampling, recombination, inter-patient, and reference strain contamination as described in the preceding reference. Phylogenetic trees were constructed using DIVEIN [34] under the GTR+I+G maximum likelihood model and visualized/edited in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Cells and antibodies
293T/17 retroviral packaging cells were purchased from the American Type Culture Collection (ATCC). TZM-bl indicator cells, a HeLa clone expressing high levels of CD4, CXCR4, and CCR5, as well as luciferase and beta-galactosidase reporters under the control of the HIV promoter, were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: (cat# 8129) courtesy of Dr John C Kappes, Dr Xiaoun Wu, and Tranzyme Inc. Cells were maintained in Dulbecco’s Modified Eagles Media (DMEM; Mediatech, Corning, NY) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, California, USA), 100 U/ml penicillin-streptomycin (Mediatech, Corning, new York, USA), and 2 mmol/l L-glutamine (Mediatech) at 37°C + 5% CO2. Antibodies PG9 and PG16 were provided by Dr Dennis Burton (The Scripps Research Institute), and the IAVI Protocol G team; VRC01 and VRC03 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (cat#s 12033 and 12032, respectively) from Dr John Mascola; NIH45–46, 8ANC195, and 3BNC117 were provided by Dr Michel Nussenz-weig (The Rockefeller University); and NIH45–46G54W was provided by Dr Pamela Bjorkman (California Institute of Technology).
Virus neutralization assay
Neutralization experiments were performed and IC50 values calculated as previously described [35]. In summary, pseudotype virus (2000 infectious units) produced in 293T/17 cells by co-transfection of an env-deficient backbone and env-expression plasmid, were preincubated 1 h with serial 5×dilutions of antibody, then added to TZM-bl cells and incubated for 48 h before luciferase activity was measured [35].
Data analysis
Every IC50 analyzed is the mean of at least two independent experiments with 3× or less variation. Fold-change in potency by IC50 ratio was calculated in Excel (Microsoft, Seattle, Washington, USA), with all IC50 values at least 10 μg/ml censored at 10. Resistance was defined as an IC50 at least 10 μg/ml prior to data collection. For per-patient and per-pair coverage we chose the conservative analytical approach of assigning to each patient (or pair) the highest IC50 value of any clone from that patient (or pair) for each antibody. Every data point from a patient was treated independently; thus if two unique clones from a single patient were resistant to two different antibodies, that patient was scored as resistant to both. Coverage was defined as the percentage of patient-pairs for which every clone was sensitive (IC50 <10 μg/ml) to a given antibody.
To compare potency of different antibodies, IC50 ratios were calculated for all 227 clones individually for each combination of interest. To estimate the effect of reduced sampling depth on coverage, we set the detected frequency of resistant variants within a given patient-pair as the true frequency, and then calculated the cumulative probability of sampling only sensitive clones from that pair at a given depth. These probabilities were then summed across all 23 pairs and used to construct coverage curves for each antibody.
Results
Clinical and molecular characteristics of study cohort
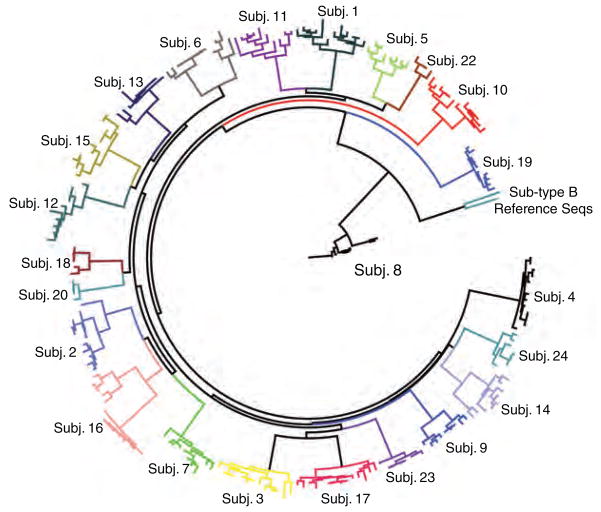
Twenty-three mother–infant pairs were included in this study; 22 were infected with subtype C HIV-1 and one (Subj. 8) was infected with subtype G. Median maternal plasma HIV RNA was 171 931 copies/ml and median maternal CD4 cell count was 146 cells/μl. Infant viral RNA load and CD4 percentages were not determined. A total of 227 full-length gp160 s were tested for neutralization sensitivity, with a median of seven maternal and three infant clones per pair (Table 1). Envelope sequences from each chronically infected woman and her infant formed a distinct monophyletic cluster (Fig. 1).
Table 1.
Patient characteristics.
| Patient ID | Maternal HIV RNA (copies/ml of plasma) | Maternal CD4+ (cells/μl) | Number of maternal clones phenotyped | Number of infant clones phenotyped |
|---|---|---|---|---|
| Subj. 1 | 300 000 | 76 | 11 | 1 |
| Subj. 2 | 200 656 | 154 | 8 | 3 |
| Subj. 3 | 206 763 | 94 | 10 | 3 |
| Subj. 4 | 50 291 | 332 | 10 | 3 |
| Subj. 5 | 298 310 | 110 | 7 | 1 |
| Subj. 6 | 143 217 | 276 | 6 | 6 |
| Subj. 7 | 270 691 | 317 | 8 | 2 |
| Subj. 8 | 117 007 | 103 | 7 | 3 |
| Subj. 9 | 104 209 | 198 | 6 | 3 |
| Subj. 10 | 509 607 | 300 | 10 | 3 |
| Subj. 11 | 83 060 | 137 | 8 | 2 |
| Subj. 12 | 45 907 | 52 | 7 | 3 |
| Subj. 13 | 39 801 | 138 | 8 | 2 |
| Subj. 14 | 375 319 | 94 | 6 | 3 |
| Subj. 15 | 18 220 | 246 | 6 | 5 |
| Subj. 16 | 211 792 | 118 | 11 | 3 |
| Subj. 17 | 47 602 | 291 | 10 | 4 |
| Subj. 18 | 750 001 | 91 | 2 | 3 |
| Subj. 19 | 358 315 | 219 | 7 | 3 |
| Subj. 20 | 6855 | 299 | 1 | 3 |
| Subj. 22 | 43 301 | 88 | 2 | 2 |
| Subj. 23 | 537 736 | 318 | 4 | 4 |
| Subj. 24 | 9925 | 263 | 4 | 3 |
| Median | 171 937 | 146 | 7 | 3 |
Fig. 1. Phylogenetic tree of all envelope sequences included in the study.
A phylogenetic tree including all 227 gp160 sequences and several reference sequences was constructed in DIVEIN [34] and annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Loop-directed antibodies PG9 and PG16 have similar breadth but PG16 is more potent
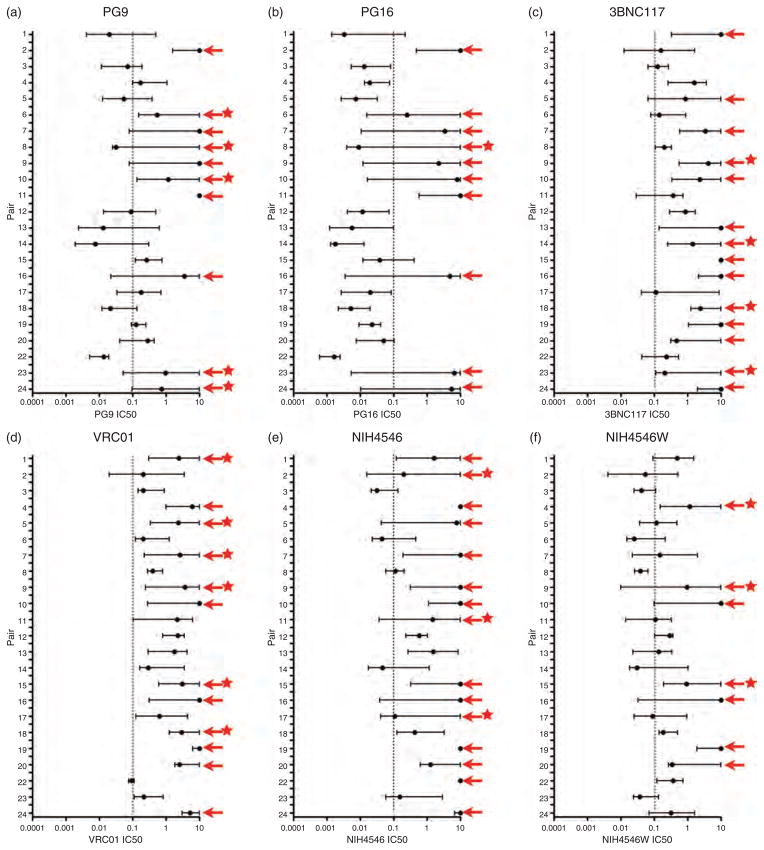
PG9 and PG16, the first of the ‘second-generation’ monoclonal antibodies described, are somatic variants that target distinct but partially overlapping quaternary epitopes on gp120 that include the V1, V2, and V3 loops [30,36,37]. As expected, we observed a high degree of concordance between these two bNAbs, with only 2.2% (5/227) of isolates resistant to one but sensitive to the other. When mothers and their infected infants were analyzed separately, we found PG9 neutralized all viruses from 72% (33/46) of patients (i.e. 72% coverage), whereas PG16 had 67% (31/46) coverage of our cohort (Table 2). When results were pooled by transmission-pair (mother and infant considered together) we observed 100% concordance with 43% (10/23) of pairs harboring resistance to both antibodies and the remaining 57% (13/23) of pairs harboring virus fully sensitive to both (Figs 2a, b and 3a, b). PG16 was approximately 4.4× more potent than PG9 by IC50 ratio (data not shown).
Table 2.
Maximum per-patient IC50, in μg/ml, by antibody.
| Patient ID | PG9 | PG16 | 3BNC117 | VRC01 | NIH4546 | NIH4546W |
|---|---|---|---|---|---|---|
| Subj. 7 (Infant) | 0.1080 | 0.0175 | 4.3186 | 0.5333 | 0.9952 | 0.0976 |
| Subj. 7 (Maternal) | >10 | >10 | >10 | >10 | >10 | 2.0101 |
| Subj. 24 (Infant) | >10 | >10 | >10 | 6.3204 | >10 | 1.6403 |
| Subj. 24 (Maternal) | 2.9000 | >10 | >10 | >10 | >10 | 1.0398 |
| Subj. 11 (Infant) | >10 | >10 | 0.0599 | 0.1825 | 0.0737 | 0.0223 |
| Subj. 11 (Maternal) | >10 | >10 | 0.7230 | 6.2516 | >10 | 0.3285 |
| Subj. 2 (Infant) | 3.5078 | 0.5571 | 0.1586 | 0.2110 | 2.9741 | 0.0539 |
| Subj. 2 (Maternal) | >10 | >10 | 1.6674 | 3.5054 | >10 | 0.5135 |
| Subj. 23 (Infant) | >10 | >10 | >10 | 0.8126 | 2.9215 | 0.1339 |
| Subj. 23 (Maternal) | 0.5880 | 5.1743 | 0.2460 | 0.2352 | 0.2020 | 0.0638 |
| Subj. 6 (Infant) | 3.4062 | >10 | 0.2253 | 0.3139 | 0.1687 | 0.0554 |
| Subj. 6 (Maternal) | >10 | >10 | 0.8988 | 1.2734 | 0.4653 | 0.2132 |
| Subj. 8 (Infant) | 0.0293 | 0.0069 | 0.1411 | 0.3156 | 0.0951 | 0.0389 |
| Subj. 8 (Maternal) | >10 | >10 | 0.3323 | 0.7832 | 0.2050 | 0.0663 |
| Subj. 3 (Infant) | 0.1946 | 0.0839 | 0.2012 | 0.2309 | 0.0628 | 0.1011 |
| Subj. 3 (Maternal) | 0.1803 | 0.0299 | 0.2689 | 0.9005 | 0.1383 | 0.1116 |
| Subj. 12 (Infant) | 0.0257 | 0.0060 | 0.9163 | 2.9528 | 1.0225 | 0.3755 |
| Subj. 12 (Maternal) | 0.5102 | 0.0747 | 1.7322 | 3.5183 | 0.8930 | 0.3636 |
| Subj. 22 (Infant) | 0.0193 | 0.0025 | 0.5399 | 0.1136 | >10 | 0.7267 |
| Subj. 22 (Maternal) | 0.0182 | 0.0022 | 0.1379 | 0.0795 | >10 | 0.1580 |
| Subj. 17 (Infant) | 0.0539 | 0.0061 | 0.1309 | 0.8770 | 0.1803 | 0.1563 |
| Subj. 17 (Maternal) | 0.7016 | 0.0873 | 8.8465 | 4.4965 | >10 | 0.9527 |
| Subj. 13 (Infant) | 0.0123 | 0.0099 | >10 | 1.4622 | 8.7000 | 0.3475 |
| Subj. 13 (Maternal) | 0.6361 | 0.0987 | >10 | 4.2253 | 5.3264 | 0.3403 |
| Subj. 14 (Infant) | 0.0075 | 0.0017 | >10 | 0.2942 | 0.0227 | 0.0301 |
| Subj. 14 (Maternal) | 0.3048 | 0.0131 | 1.5427 | 3.5120 | 1.1442 | 1.0242 |
| Subj. 18 (Infant) | 0.0592 | 0.0096 | 2.4144 | 2.9543 | 0.4284 | 0.1850 |
| Subj. 18 (Maternal) | 0.1352 | 0.0200 | >10 | >10 | 3.2767 | 0.5097 |
| Subj. 1 (Infant) | 0.0161 | 0.0018 | 2.8495 | 1.0703 | 0.1225 | 0.1056 |
| Subj. 1 (Maternal) | 0.4971 | 0.2215 | >10 | >10 | >10 | 1.5289 |
| Subj. 5 (Infant) | 0.2599 | 0.0106 | 0.0667 | 0.3387 | 0.0435 | 0.0445 |
| Subj. 5 (Maternal) | 0.3838 | 0.0329 | >10 | >10 | >10 | 0.4886 |
| Subj. 4 (Infant) | 0.1701 | 0.0226 | 2.2765 | >10 | >10 | >10 |
| Subj. 4 (Maternal) | 1.0833 | 0.0766 | 3.6266 | >10 | >10 | 3.0279 |
| Subj. 20 (Infant) | 0.4471 | 0.1027 | 0.5591 | 3.0342 | 1.5592 | 0.3680 |
| Subj. 20 (Maternal) | 0.0415 | 0.0076 | >10 | >10 | >10 | >10 |
| Subj. 15 (Infant) | 0.3206 | 0.1802 | >10 | >10 | >10 | >10 |
| Subj. 15 (Maternal) | 0.7601 | 0.4157 | >10 | 4.6893 | >10 | 0.9210 |
| Subj. 19 (Infant) | 0.2090 | 0.0415 | >10 | >10 | >10 | >10 |
| Subj. 19 (Maternal) | 0.2503 | 0.0417 | >10 | >10 | >10 | >10 |
| Subj. 16 (Infant) | >10 | >10 | >10 | >10 | >10 | >10 |
| Subj. 16 (Maternal) | >10 | >10 | >10 | >10 | >10 | >10 |
| Subj. 9 (Infant) | >10 | >10 | 5.2797 | 4.8655 | >10 | 1.7975 |
| Subj. 9 (Maternal) | >10 | >10 | >10 | >10 | >10 | >10 |
| Subj. 10 (Infant) | 0.3102 | 0.0878 | >10 | >10 | >10 | >10 |
| Subj. 10 (Maternal) | >10 | >10 | >10 | >10 | >10 | >10 |
| % Coverage | 72% | 67% | 57% | 65% | 50% | 78% |
Fig. 2. Median and range (IC50 in μg/ml) by patient-pair for each antibody.
Black circle indicates per-patient median IC50, while bars extend from the lowest to the highest IC50 of the isolates from each pair. Red arrows indicate which patient-pairs harbored at least one isolate fully resistant (IC50 ≥ 10 μg/ml) to that antibody. Red stars indicate patient-pairs from which resistant envelopes were considered a minority (≤20% frequency). A dotted grey line has been placed at 0.1 μg/ml, which has been suggested as the threshold for good in vivo neutralization activity [20]. (a) PG9. (b) PG16. (c) 3BNC117. (d) VRC01. (e) NIH45–46. (f) NIH45–46G54W.
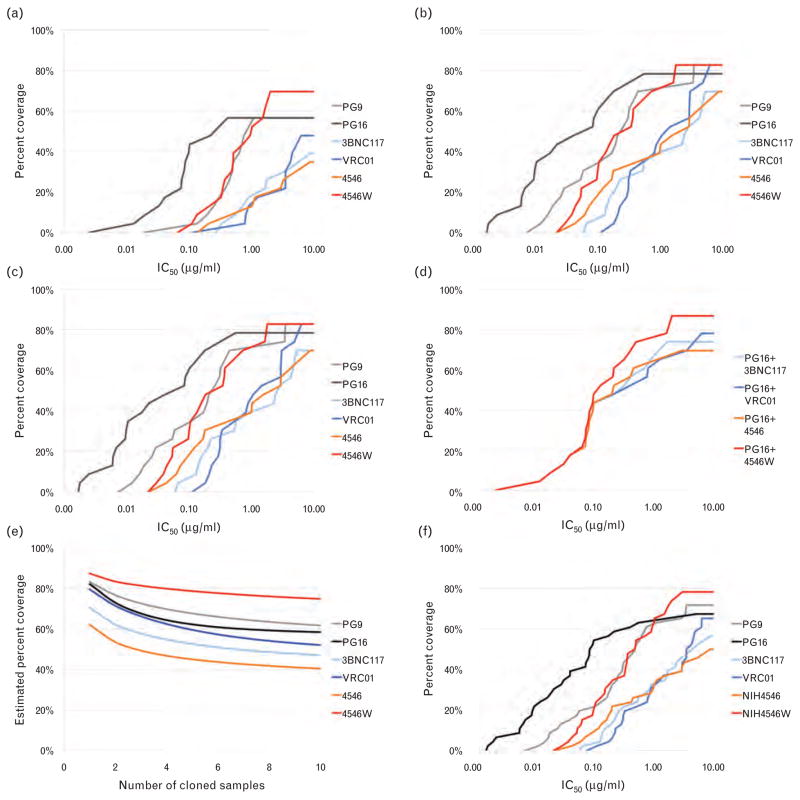
Fig. 3. Coverage of patient isolates at different antibody concentrations and sampling depths.
Coverage maps were constructed based on the IC50 value of the most resistant isolate from each patient (or pair), with full resistance defined as at least 10 μg/ml. Curves indicate the percentage of patients from whom every tested isolate was neutralized at the indicated antibody concentration. (a) Percentage coverage of the cohort on a per-patient basis (mothers and infants treated separately, n =46 patients). (b) Percentage coverage of the cohort on a per-pair basis (infant and maternal data pooled, n =23 pairs). (c) Percentage coverage of the infant isolates only (n =23 infants). (d) Percentage coverage of the cohort on a per-pair basis (n =23 pairs) by the combination of PG16 and each of the CD4bs-targeted antibodies. (e) Percentage coverage of the infant isolates only (n =23 infants) by the combination of PG16 and each of the CD4bs-targeted antibodies. (f) Effect on coverage estimates if sampling depth were reduced from a median of n =10 clones per pair down to n =1 clone per transmission pair.
Potent CD4 binding-site antibodies VRC01, NIH45–46, and 3BNC117 display similar breadth and potency
VRC01, VRC03, and NIH45–46 (4546) were isolated from the same patient using different sets of PCR primers, and target the CD4 binding site (CD4bs) on gp120 [29,31]. 8ANC195 and 3BNC117, also targeting the CD4bs, were isolated from two additional patients [29]. VRC03 and 8ANC195, both of which have been described as having limited breadth and potency relative to the other CD4bs bNAbs [29,31], also had minimal activity in our cohort and were excluded from further analysis (data not shown).
In the per-patient analysis, VRC01 had complete coverage (neutralizing all envelopes from a given patient) of 65% (30/46) of patients, 4546 had 50% (23/46) coverage, and 3BNC117 had 57% (26/46) coverage (Table 2). VRC01, 4546, and 3BNC117 also had similar breadth in the more conservative per-pair analysis, with overall coverage of 48% (VRC01), 35% (4546), and 39% (3BNC117) (Figs 2c–e and 3a, b). There was also significant overlap, with most transmission-pairs that harbored virus resistant to one CD4bs bNAb also harboring variants resistant to the other two, though not in every case. On a per-clone basis, all three bNAbs were similarly potent, with median IC50 ratios of 0.85–1.00 (data not shown).
Engineered bNAb NIH45–46G54W has significantly improved breadth and potency relative to other CD4bs antibodies
NIH45–46G54W (4546W) is the first example of structure-based engineering of a neutralizing antibody. It is a variant of NIH45–46 designed to improve breadth and potency by inserting a tryptophan into the ‘Phe43 pocket’, a critical site involved in gp120 binding to CD4. The design and binding properties of 4546W have been described [22]. The glycine-to-tryptophan substitution increased coverage of this antibody relative to the parental variant, from 50% (23/46) to 78% (36/46) in the per-patient analysis (Table 2), and doubled it from 35% (8/23) to 70% (16/23) in the more conservative per-pair analysis (Figs 2f and 3a, b). In our cohort, 4546W was approximately 2.7-fold more potent (by IC50 ratio) than 4546, and approximately 3.7 and 6.1× more potent than 3BNC117 and VRC01, respectively. Importantly, no isolates previously sensitive to 4546 were found to be resistant to 4546W.
A combination of NIH45–46G54W and PG16 achieves the highest coverage overall and against early-transmitted isolates
We next sought to determine which combination of a loop-directed and CD4bs-directed bNAb would give the best overall coverage with the highest neutralization potency. PG9 and PG16 had equivalent coverage, but PG16 neutralized with substantially greater potency. Of the CD4bs bNAbs tested, 4546W had clear advantages in both breadth and potency in our conservative per-pair analysis. The combination of PG16 and 4546W had effective per-pair coverage of 87%, the highest of any two-antibody combination available. PG16 and VRC01 was the next most effective combination, with 78% overall coverage in the per-pair analysis (Fig. 3d).
Since we had data on a large number of early transmitted isolates, we examined the baseline coverage of these antibodies both alone and in combination to simulate a prophylactic intervention. Against all 68 early transmitted isolates (from 23 transmission pairs) we found antibody coverage was within a similar range (65–78%) for all bNAbs tested (Fig. 3c). The combination of PG16 with either VRC01 or 4546Woffered the best coverage, with all variants from 96% of infants (22/23) susceptible to at least one bNAb. Both 4546 and 3BNC117 achieved 87% coverage in conjunction with PG16 (Fig. 3e). Considering the median IC50 ratios of just the early-transmitted isolates, 4546W was approximately 2.1× more potent than parental 4546 and approximately 5× more potent than VRC01.
Minor variants resistant to NIH45–46G54W are uncommon and associated with resistance to other CD4bs bNAbs
The ability to detect pre-existing minor variants that harbor resistance is a critical factor in the success or failure of treatment with existing antiretroviral drugs including entry inhibitors such as maraviroc [38–40]. Thus, whereas broad sampling at shallow depth gives a good estimate of the prevalence of dominant resistance, it cannot determine the frequency of minor variants that could, under selection, rapidly outgrow, and compromise the effectiveness of an inhibitor. Whereas we do not consider a median sampling depth of 10 clones per pair to be exhaustive, this cohort does give us the opportunity to probe for minority variants within quasispecies resistant to these new antibodies in a way that has not been previously described.
When we set a ‘low frequency’ cut-off of 20% or less, we identified only three pairs harboring minor variants resistant to 4546, 4546W, four with minor variants resistant to 3BNC117, and six pairs harboring a minority population resistant to VRC01. Importantly, the presence of a minor population resistant to 4546W was associated with low and/or high frequency resistance to at least two of the other CD4bs antibodies. In contrast, there were several cases where minor variants resistant to 4546, 3BNC117, or VRC01 were detected in the absence of resistance to multiple other CD4bs antibodies (Fig. 2c–f).
High intra-pair and intra-patient variation and its effect on the relationship between sampling depth and coverage
We observed a surprising degree of intra-pair variation in susceptibility with all tested antibodies, with a median fold difference of 14.7–38.1× between the most resistant and most sensitive clone within a pair (data not shown). In one example (Subj. 13) sampling of 10 unique clones identified a 265-fold difference in IC50 for PG9, without detecting outright resistance (range 0.0024–0.6361 μg/ml) (Fig. 2a).
Given the high degree of intra-patient variation, we next estimated the probability of detecting at least one resistant variant for each transmission pair over a range of sampling depths and constructed a plot showing estimated coverage with a sampling depth ranging from 1 to 10 clones per pair (Fig. 3f). Coverage estimates produced by sampling only a single clone per pair ranged from 62–87%, with PG9, PG16, and 4546W scoring above 80% and VRC01 scoring 79%. These coverage estimates are at least 20% higher than our actual results at a median of 10 clones per pair, which are still likely to be overestimates.
Discussion
We found that loop-targeted antibodies PG9 and PG16 neutralized envelopes from our cohort of MTCT pairs with 57% coverage in our conservative per-patient analysis. Resistance to one PG antibody was highly correlated with resistance to the other, which is expected given that they are somatic variants targeting distinct but overlapping epitopes. PG16 neutralized viruses from this cohort more potently than PG9 by almost half a log10, suggesting it would be the better choice for initial clinical studies, at least for prevention of MTCT (pMTCT) in a population predominantly infected with subtype C.
Qualitatively, the battery of CD4-binding site antibodies fell into three categories: VRC03 and 8ANC195 performed poorly; VRC01, 3BNC117, and 4546 performed moderately well and neutralized with similar potencies; and 4546W had the broadest coverage and highest neutralization potency of any CD4bs antibody in our study. Additionally, minor variants resistant to 4546W were uncommon, and patients harboring 4546W-resistant envelopes typically harbored variants resistant to the other CD4bs antibodies as well. A caveat is that our median sampling depth of 10 clones per transmission pair, whereas more extensive than other studies, is not definitive, and likely underestimates the true prevalence of pre-existing resistance to these reagents. Our data argue in favor of using 4546W (or a further modified variant) for future clinical studies, since resistance to 4546W appears to be rare and associated with resistance to the CD4bs antibody class in general. Studies of additional CD4 binding site antibodies would be required to confirm this characteristic of 4546W. This observation is compatible with a more conserved mechanism of neutralization for 4546W (via the ‘Phe43 pocket’) and a higher genetic barrier to resistance, but our study did not directly address these issues.
Our data indicate that the combination of 4546W and PG16 is likely to be the most effective pair of antibodies for pMTCT, and perhaps preventive studies in general in subtype C-infected populations. This combination has the greatest breadth in both the conservative per-pair analysis and the restricted subset of early transmitted isolates (neutralizing all tested variants from 22/23 infected infants). Moreover, these antibodies had the highest neutralization potency (measured by IC50) of all the antibodies we tested from their respective classes. Several other CD4bs antibodies (VRC01, 3BNC117, and 4546) also had relatively broad coverage of the early transmitted isolates, but at 2–5× reduced potency compared to 4546W.
Women in our study were HIV-positive at the time of enrollment and seroconversion dates were not available. Their low CD4 cell counts suggest late-stage chronic infection, which is generally associated with greater genetic diversity, likely contributing to the frequent detection of pre-existing bNAb-resistant variants. Envelopes from the 23 epidemiologically linked infants exhibited the restricted genetic diversity typical of recent infection, and were better covered by the tested bNAbs. The generalizability of our findings to other forms of transmission is unclear and warrants further investigation.
The study represents, to our knowledge, the deepest sampling of HIV-infected persons in the context of second-generation bNAbs. Our data are qualitatively similar to a recent study [21] that sampled approximately 200 patients from multiple clades at a median depth of 1 clone per patient and found the combination of a loop-targeted (PG series) and CD4bs antibody (VRC01, VRC-PG04) had very good coverage of a diverse panel of global strains, though that study did not include the 4546W variant for comparison. It is important to emphasize here the heterogeneity of our data and the effect of sampling depth on coverage estimates that we observed. Antibody susceptibility within some patient-pairs differed by several orders of magnitude, which indicates substantial phenotypic variation exists even within patients described by our analyses as fully sensitive. At a sampling depth of 1 clone per pair, our results would be very similar to that study [21], with VRC01, PG9 and PG16 all achieving coverage in the approximately 75–85% range at an IC50 less than 10 μg/ml, whereas our actual median sampling depth of 10 clones identified resistant variants in an additional 25–30% of patients. Given that even minor resistant variants in the 2% prevalence range have been implicated in virologic failure with other therapeutic agents, including the entry inhibitor maraviroc [39,40], our data encourage tempering of expectations. HIV’s propensity for rapid escape, due to the exceptional plasticity of its envelope, suggests modified reagents that raise the genetic barrier to resistance may have a disproportionate advantage against diverse quasispecies. Our current inability to screen for bNAb resistance using high-throughput genetic methods requires isolates to be phenotypically characterized. Thus, it will be important to pair studies like this one with broader surveys such as the one referenced above [21].
Considering the difficulty the field has experienced in immunogen design for the induction of broadly neutralizing antibodies, and the consequent interest in using a gene therapy-based approach to bypass the immune system, we see no reason to constrain future proof-of-principle immunotherapy or immunoprophylaxis studies to ‘naturally’ occurring antibodies only. In this context, our data indicate that engineered antibody 4546W is the most broad and potent CD4 binding site antibody currently available in the setting of HIV-1 subtype C pMTCT, and it, or an even more potent variant, should be considered for inclusion in any future clinical trial.
Acknowledgments
We thank the participants and staff of the Zambia Exclusive Breastfeeding Study, without whose generosity none of this work would have been possible. We also thank the NIH AIDS Research and Reference Reagent Program and Drs M. Sharp and D. Finzi for their support of this project.
Funding: This work was supported by the National Institute of Child Health and Development (R01 HD 39611, R01 HD 40777, R01 HD 57617) and the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) U01 AI 68632, and the University of Washington Center for AIDS Research Computational biology Core (P30 AI 27757). G.M.A. is an Elizabeth Glaser Pediatric AIDS Foundation Scientist.
Overall support for IMPAACT was provided by the National Institute of Allergy and Infectious Diseases (NIAID) (U01 AI 68632), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institute of Mental Health (NIMH) (AI 68632).
Additional funding provided by the University of Washington Center for AIDS Research (CFAR), an NIH-funded program (P30 AI 27757) which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: Designed experiments: K.J.N., G.M.A. Performed experiments: K.J.N., C.C. Analyzed data: K.J.N., C.C., E.R.S., L.H., L.K. Contributed reagents/materials/tools: L.H., M.S., C.K., D.M.T., J.I.M., L.K. Wrote manuscript: K.J.N., G.M.A., L.K.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 2.Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 2007;3:e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 6.Montefiori DC. Neutralizing antibodies take a swipe at HIV in vivo. Nat Med. 2005;11:593–594. doi: 10.1038/nm0605-593. [DOI] [PubMed] [Google Scholar]
- 7.Douek DC, Kwong PD, Nabel GJ. The rational design of an AIDS vaccine. Cell. 2006;124:677–681. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavine CL, Lao S, Montefiori DC, Haynes BF, Sodroski JG, Yang X. High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection. J Virol. 2012;86:2153–2164. doi: 10.1128/JVI.06201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 11.Ferrantelli F, Hofmann-Lehmann R, Rasmussen RA, Wang T, Xu W, Li PL, et al. Postexposure prophylaxis with human monoclonal antibodies prevented SHIV89. 6P infection or disease in neonatal macaques. AIDS. 2003;17:301–309. doi: 10.1097/00002030-200302140-00003. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Smith BA, Baba TW, Liska V, et al. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J Virol. 2001;75:7470–7480. doi: 10.1128/JVI.75.16.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89. 6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 15.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 16.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson PR, Schnepp BC, Zhang J, Connell MJ, Greene SM, Yuste E, et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph A, Zheng JH, Chen K, Dutta M, Chen C, Stiegler G, et al. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti-HIV antibody. J Virol. 2010;84:6645–6653. doi: 10.1128/JVI.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis AD, Chen R, Montefiori DC, Johnson PR, Clark KR. Generation of neutralizing activity against human immunodeficiency virus type 1 in serum by antibody gene transfer. J Virol. 2002;76:8769–8775. doi: 10.1128/JVI.76.17.8769-8775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doria-Rose NA, Louder MK, Yang Z, O’Dell S, Nason M, Schmidt SD, et al. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol. 2012;86:3393–3397. doi: 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Wang C, O’Dell S, Li Y, Keele BF, Yang Z, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 25.Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 26.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 27.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray ES, Meyers T, Gray G, Montefiori DC, Morris L. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 2006;3:e255. doi: 10.1371/journal.pmed.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn L, Aldrovandi GM, Sinkala M, Kankasa C, Semrau K, Mwiya M, et al. Effects of early, abrupt weaning on HIV-free survival of children in Zambia. N Engl J Med. 2008;359:130–141. doi: 10.1056/NEJMoa073788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath L, Conway S, Jones L, Semrau K, Nakamura K, Walter J, et al. Restriction of HIV-1 genotypes in breast milk does not account for the population transmission genetic bottleneck that occurs following transmission. PLoS One. 2010;5:e10213. doi: 10.1371/journal.pone.0010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, et al. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques. 2010;48:405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura KJ, Gach JS, Jones L, Semrau K, Walter J, Bibollet-Ruche F, et al. 4E10-resistant HIV-1 isolated from four subjects with rare membrane-proximal external region polymorphisms. PLoS One. 2010;5:e9786. doi: 10.1371/journal.pone.0009786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JA, Li JF, Wei X, Lipscomb J, Irlbeck D, Craig C, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzner KJ, Giulieri SG, Knoepfel SA, Rauch P, Burgisser P, Yerly S, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–247. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 40.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Jensen MA, et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis. 2011;203:237–245. doi: 10.1093/infdis/jiq030. [DOI] [PMC free article] [PubMed] [Google Scholar]