Abstract
Diabetic nephropathy is a common complex disease with a clear genetic predisposition. Human gene association studies are beginning to bear fruit by identifying gene loci that increase diabetic nephropathy risk. Chua et al. report a similar study in diabetic mice that reveals a major nephropathy locus on chromosome 8. Could this be a human nephropathy gene? Time will tell, but such findings will at least improve the use of mouse models of human kidney disease.
Mouse models have become a major focus of diabetic nephropathy research during the past decade (1, 2). A major reason for this emphasis has been the ease of genetic manipulation in the mouse to either enhance or interrupt the abnormal signaling and pathologic lesions of diabetic nephropathy. This increasing reliance on the mouse has occurred even in the face of the nearly impenetrable resistance to nephropathy of the C57BL/6 mouse (1, 2), the workhorse of genetic murine research. While most studies of nephropathy in mice have focused on candidate pathways and single gene manipulation to test the effects of those pathways on diabetic changes, recently several studies have used less direct and more open-ended approaches to gain insight into diabetic kidney disease. These include more systemic approaches in which candidate pathways are not interrogated but in which all associations with the ultimate nephropathy phenotype are sought. One such example is illuminated in the report by Chua, et al. (this issue).
This elegantly comprehensive study used well established gene mapping methods with an overlay of genome-wide expression (GWE) analysis. These tools have been critical for analysis of non-Mendelian diseases in humans for which there are complex genetic susceptibilities, such as type 1 and type 2 diabetes, diabetic nephropathy itself (3, 4) and other forms of chronic kidney disease. Such studies, including genome-wide association studies (GWAS) and other similarly broad genetic approaches, have identified genes and gene loci associated with nephropathy in humans that lie outside the usual pantheon of pathways known to be disrupted in chronic kidney disease. Indeed, there has been a bit of an explosion of reports in the last 2 years that have identified new nephropathy genes in humans using these techniques. Perhaps the most dramatic of these are the identification of uromodulin (5), the gene encoding Tamm-Horsfall protein, and of MYH9 (4, 6), a non-muscle myosin gene, as major susceptibility loci for progression of chronic kidney disease. Indeed, variants of MYH9 may explain up to 70% of nondiabetic forms of end-stage renal disease in African Americans (4, 6, 7). Ironically, one of the reports identifying this locus of non-diabetic kidney disease was from the Family Investigation of Nephropathy and Diabetes (FIND) Research Group (4). The FIND group has yet to find such a powerful locus for diabetic nephropathy susceptibility as MYH9 is for non-diabetic disease, although a recent report identified several suggestive diabetic nephropathy loci (8). Nonetheless, it is certain that these approaches have directed our attention to genes and gene products that had hitherto been ignored as possible major participants in progression of kidney disease. Thus, these reports have already charted new fields of study and are likely to lead rapidly to a diagnostic reclassification and perhaps novel approaches to therapy for many patients with chronic kidney disease.
As Chua and associates correctly argue, the pursuit of similar studies in mice can complement these pathfinding analyses in humans by taking advantage of the genetic and environmental uniformity of mice and the easy availability of relevant tissues from these models. Another advantage, perhaps too obvious to be noted by the authors, is that genome-wide susceptibility mapping including genome-wide expression analysis (which has not yet been completed by the investigators) is orders of magnitude less expensive and time-consuming than similar studies in humans. Chua’s findings were completed by a single research group whereas the human GWAS data have involved well-funded multi-center consortia at work for the better part of decade. Of course, the mouse studies, even though relatively parsimonious might be regarded as profligate were they not to identify genes that are relevant to human progressive nephropathy.
So how did Chua, et al. do in that regard? While the jury is still out, it seems relatively likely that they have hit on an important locus for nephropathy. Their strategy was well reasoned. Avoiding the trap of the nephropathy resistant C57BL/6 mouse by turning it into a perfect negative control, the investigators used the db/db type 2 diabetic nephropathy model on a relatively nephropathic genetic background, the FVB mouse (more on that later). They analyzed linkage of the clinical nephropathy phenotype to genetic loci identified by single nucleotide polymorphisms in the diabetic progeny of FVB and C57BL/6 mice. In contrast to the expected identification of a number of relatively small genetic contributors to nephropathy, the investigators found a single FVB locus on chromosome 8 which remarkably accounted for a major amount of the histopathology differences between mice. Importantly, there was no association between this locus and non-nephropathy phenotypes such as the degree of hyperglycemia. The authors are currently investigating genes in the region to identify the nephropathy gene. Then they and other investigators will be able to ascertain whether the paralog of this gene is also involved in human nephropathy.
While promising, there are some reasons to be concerned that the chromosome 8 gene may not be directly relevant to human diabetic nephropathy. Part of this concern has to do with the peculiar issues of mouse nephropathy. As already noted, the C57BL/6 mouse is particularly resistant to all types of kidney disease (at some point those protective loci need to be identified). The FVB diabetic mouse develops more significant nephropathy than the C57BL/6 mouse, but nephropathy has been variable in this strain (1) and therefore appears subject to environmental effects that could make any genetic susceptibility locus less applicable to human disease. Moreover, this strain and virtually all mouse strains develop only early glomerulopathy and do not develop substantial tubulointerstitial fibrosis or a progressive decline in glomerular filtration rate so the chromosome 8 gene may be important only for such early changes (1, 2). In addition, the diabetic FVB mice in this study developed a sudden increase in albuminuria between 21 and 24 weeks of age, the acuity of which is atypical of human diabetic nephropathy. None of this is to deny that the authors have identified an important murine nephropathy locus which will likely teach us quite a bit about the development of albuminuria and glomerulosclerosis, it just may not instruct us as much about human diabetic nephropathy, per se.
Indeed, there is a two-way street between such mouse and human studies. Traffic in both directions can aid in elucidating the fundamental processes of diabetic nephropathy and other chronic kidney diseases. Besides determining whether nephropathy loci in mice are present in humans with disease, nephropathy genes and gene products identified in human studies can be placed in susceptible murine strains, like db/db FVB or diabetic DBA/2 mice (1), and the effects on nephropathy determined. In addition interactions between mouse and human nephropathy genes can be interrogated in murine studies. Finally, putatively critical gene expression changes in human kidney diseases can be validated in mouse models. This interspecies exchange freeway should help us move closer to truly understanding, treating and preventing diabetic nephropathy.
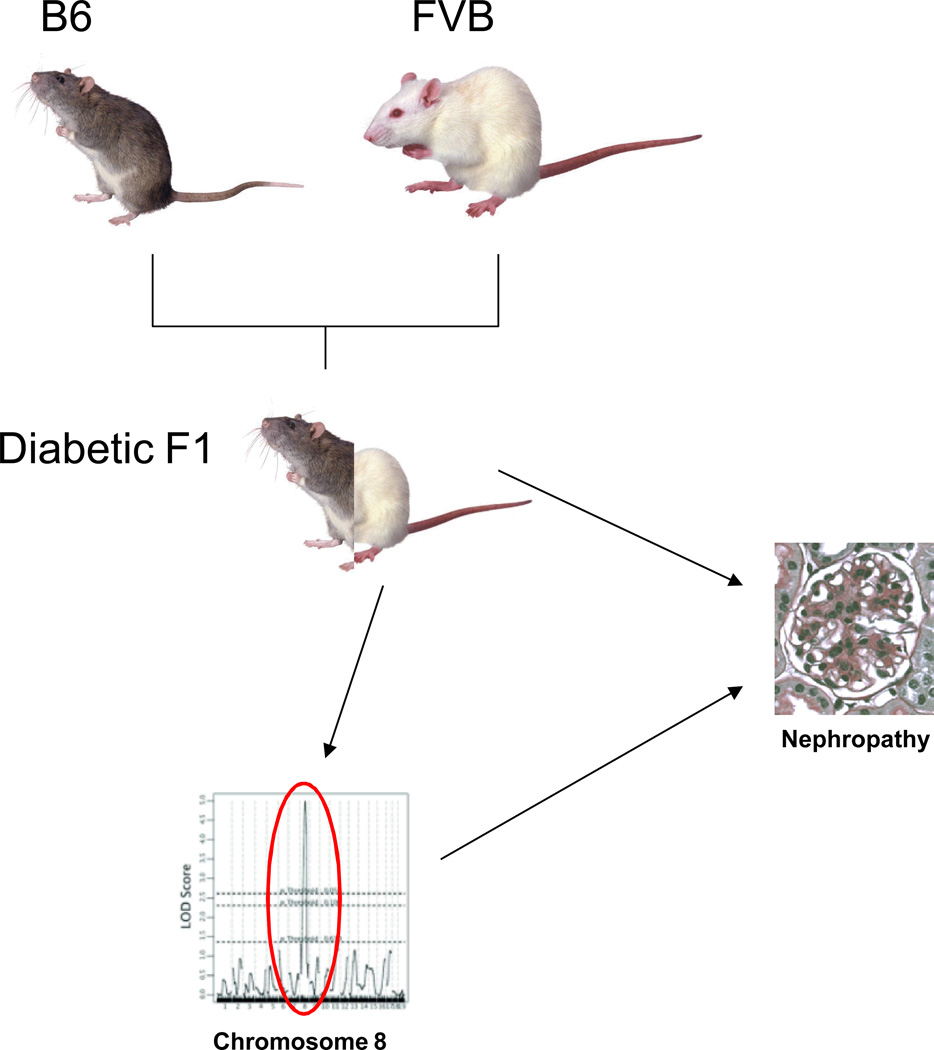
Figure 1.
Resistant C57Bl/6 (B6) mice were backcrossed with nephropathic FVB/N (FVB) mice to produce diabetic progeny (diabetic F1) which were analyzed for histopathologic features of nephropathy and genotyped for over 100 single nucleotide polymorphisms. A single locus on chromosome 8 showed a major linkage to the combined nephropathy histology score suggesting a major contribution of this single gene to the development of nephropathy in FVB diabetic mice.
References
- 1.Brosius FC, 3rd, Alpers CE, Bottinger EP, Breyer MD, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20:2503–2512. doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breyer MD, Bottinger E, Brosius FC, 3rd, Coffman TM, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2005;16:27–45. doi: 10.1681/ASN.2004080648. [DOI] [PubMed] [Google Scholar]
- 3.Malhotra A, Igo RP, Jr, Thameem F, Kao WH, et al. Genome-wide linkage scans for type 2 diabetes mellitus in four ethnically diverse populations-significant evidence for linkage on chromosome 4q in African Americans: the Family Investigation of Nephropathy and Diabetes Research Group. Diabetes Metab Res Rev. 2009;25:740–747. doi: 10.1002/dmrr.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao WH, Klag MJ, Meoni LA, Reich D, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nature genetics. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kottgen A, Glazer NL, Dehghan A, Hwang SJ, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nature genetics. 2009 doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp JB, Smith MW, Nelson GW, Johnson RC, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nature genetics. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostrom MA, Freedman BI. The Spectrum of MYH9-Associated Nephropathy. Clin J Am Soc Nephrol. doi: 10.2215/CJN.08721209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schelling JR, Abboud HE, Nicholas SB, Pahl MV, et al. Genome-wide scan for estimated glomerular filtration rate in multi-ethnic diabetic populations: the Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2008;57:235–243. doi: 10.2337/db07-0313. [DOI] [PubMed] [Google Scholar]