Abstract
Three-dimensional (3D) gradients of biochemical and physical signals in macroscale, degradable hydrogels have been engineered that can regulate photoencapsulated human mesenchymal stem cell (hMSC) behavior. This simple, cytocompatible and versatile gradient system may be a valuable tool for researchers in biomaterials science to control stem cell fate in 3D and guide tissue regeneration.
Keywords: biomaterials, gradients, alginate, hydrogels, stem cells, tissue engineering
Cells continuously receive biochemical and biophysical stimuli from their microenvironment. These environmental stimuli drive cellular behavior and function during development and tissue regeneration.[1,2] Biochemical signals such as growth factors have been shown to guide many developmental processes[3] and have also been used to control cell behavior for regenerative medicine applications.[4] Cell interactions with the extracellular matrix (ECM)[5] and physical signals[2,6,7] such as matrix rigidity and mechanical stimuli can also have strong effects on cellular phenotype and tissue formation. While biochemical and physical signals play an important role in controlling cell behavior, the presentation of signals that control human development and regeneration are complex, often existing in gradients.[8] Gradients of signals can also regulate cell function when presented by engineered biomaterial systems.[7,9–11] The ability to precisely control the spatial location of biochemical and physical signals may better facilitate the capacity to regulate cell fate and even to recapitulate gradients present during normal physiologic processes to permit regional control of cell function for the design of cell based therapies for use in regenerative medicine.
Tissue engineering is a promising field for the development of biologic replacement tissues for failing or damaged native tissues.[12,13] One prominent approach in tissue engineering is the seeding of cells into or onto a three-dimensional biomaterial such as a hydrogel that partially recapitulates native cellular environments, can be injected or implanted into a defect site in a patient and guides cell behavior until new tissue is formed to ultimately restore lost functionality.[12,14] Hydrogels, which are 3D, crosslinked, insoluble hydrophilic polymer networks formed via crosslinking of water soluble polymers, have found widespread application in tissue engineering as their aqueous, structured environment can partially mimic a cell’s natural ECM.[15,16] They are attractive scaffolds for controlling cell function, as a variety of technologies and chemistries have been developed for tailoring their biochemical and physical properties.[7,11,17,18–20] However, the methods used to modulate hydrogel properties typically result in isotropic presentation of the cell signals because they are created from homogeneously mixed solutions. Since the complex processes of development and healing are usually at least partially predicated on the spatially controlled presentation of these cellular signals, the ability to incorporate signal gradients into hydrogel scaffolds may open the door to answering important questions about the role of these gradients on cell behavior and the development of morphologically complex tissues, as well as facilitating the engineering of more biomimetically functional tissues. Recently, a variety of strategies have been developed to engineer spatial gradients of biological signals in biomaterials.[10,21,22–24] However, these systems often employ long and sophisticated fabrication procedures that necessitate expertise and expensive equipment, cannot be used to fabricate larger, clinically relevant macro-scale 3D gradient biomaterials, involve fabrication procedures, high voltage electric fields or cytotoxic precursor solutions which induce cell death during 3D encapsulation, and/or cannot form linear gradients. In this report, we present an easy, versatile approach to form a variety of physical and biochemical 3D linear gradients in macroscale hydrogels with encapsulated cells for regulating their behavior, which be of great value in regenerative medicine and for basic biology research.
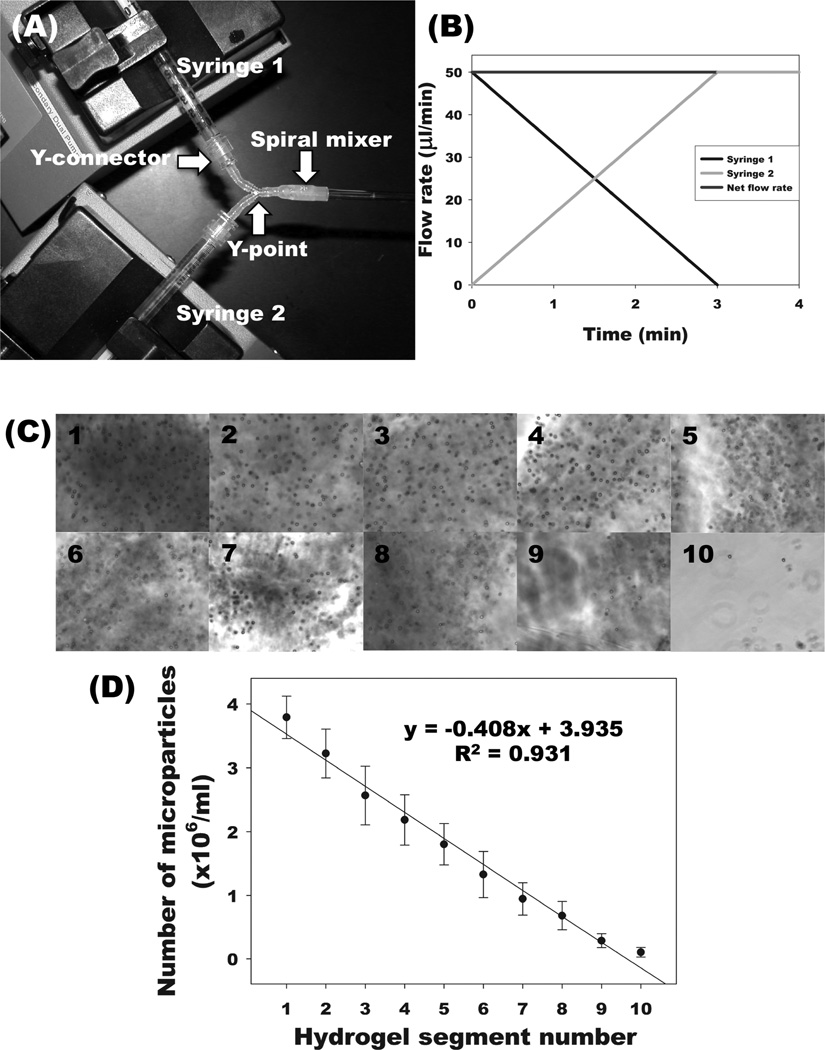
Recently, we reported on a photocrosslinked, biodegradable, alginate hydrogel system with no inherent biological activity.[20] The photopolymerization process employed to crosslink the macromers permits easy encapsulation of cells with high viability. We have shown the capacity to control the mechanical properties and degradation rate of this material,[20,25] the ability to incorporate cellular adhesivity independently of physical properties[19,26] and the sustained delivery of heparin binding growth factors.[18] In the present study, we have engineered a simple and rapid approach to create physical and biochemical signaling gradients within these hydrogels using two programmable syringe pumps, as shown in Figure 1A. Since it is difficult to visualize the physical or biochemical gradients in hydrogels quickly or directly, gradients are often characterized by radiolabeling or by fluorescently tagging a component in the hydrogels.[27] Here, we utilize a simpler method for visualizing and confirming linear gradient formation within centimeter scale biodegradable, photocrosslinked alginate hydrogels using a microparticle-based system.[28] By inversely varying the flow rates (Figure 1B), a linear gradient of microparticles was formed, from being comprised predominantly of hydrogel with microparticles from syringe 1 at one end, to mostly hydrogel without microparticles from syringe 2 at the other end. This permits identification of the composition of the gel being pumped out at any time, and allows for verification that the gel formed contains a linear gradient along the entire length. Optical microscopic images obtained with sequential, evenly spaced hydrogel segments visually exhibit the formation of a microparticle gradient in a photocrosslinked alginate hydrogel (Figure 1C). Quantification of the number of microparticles in each segment demonstrated that a fairly linear gradient of microparticles in the hydrogels was produced (Figure 1D).
Figure 1.
Fabrication of microparticle-based gradient alginate hydrogels. (A) Photograph of gradient making system. (B) Flow rates of two syringes to pump a linear gradient for a 5-cm length × 2-mm diameter alginate hydrogel. After linear gradient pumping for 3 min, an additional 50 µL of alginate solution, which is the volume from the Y point to the beginning of quartz tube, was further pumped into a spiral mixer for 1 min. (C) Photomicrographs of microparticles in cross-sections of gradient alginate hydrogel segments. Segments 1–10 represent sequential segments of the gel. (D) Quantification of microparticles in each segment of gradient alginate hydrogels.
A dual pump gradient generating system has been reported previously[24] in which two types of poly(lactide-co-glycolide) (PLGA) microparticles are infused into a mold at varying rates, thus generating a continuous spatial gradient. However, this fabrication process requires the use of ethanol to physically attach adjacent microspheres and form a solid scaffold, thus preventing the inclusion of cells within the microparticle suspensions. To improve the cytocompatibility of the microsphere fusing process, sub-critical CO2 sintering has been used in the presence of cells.[29] The system described in our report utilizes a non-cytotoxic photocrosslinking mechanism that allows cells to be safely suspended in the alginate solutions and also encapsulated throughout the entirety of macro-scale hydrogel constructs with independently tunable physical and biochemical properties.
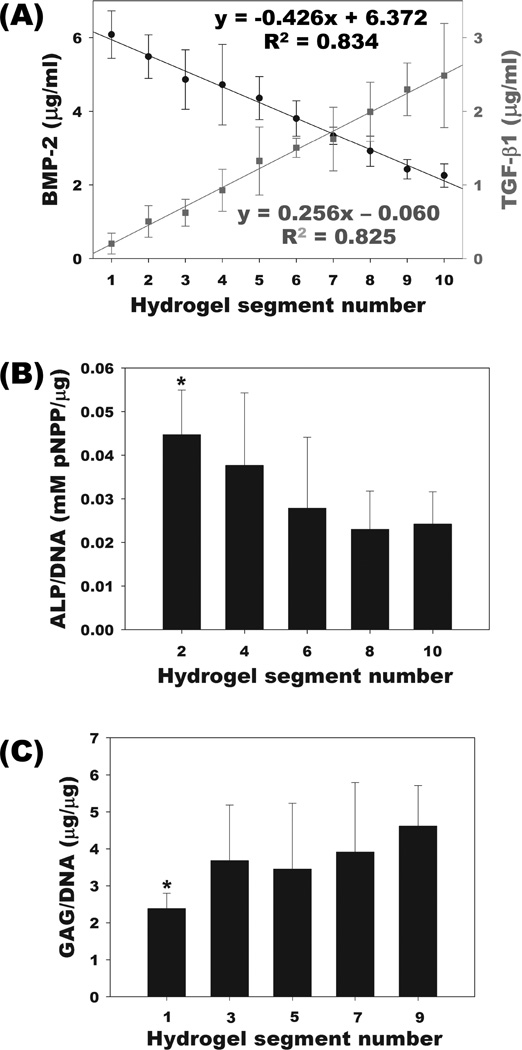
Spatial and temporal control over the presentation of bioactive factors within a 3D scaffold are critical design criteria for providing complex signals to regulate the fate of encapsulated cells during tissue forming processes such as angiogenesis, neurogenesis, wound healing, chondrogenesis and osteogenesis. To partially mimic the tissue complexity found in vivo, there has been a great deal of interest in spatial patterning of bioactive signaling molecules within 3D scaffolds.[22,28,30,31] One potentially important application of spatial and temporal gradients of bioactive growth factors is in osteochondral tissue engineering to regenerate the bone and cartilage interface.[32] In this study, gradients of multiple growth factors were generated to examine their capacity to control the differentiation of hMSCs down the osteogenic and chondrogenic lineages. An affinity-based growth factor gradient system based on photocrosslinked heparin-alginate (HP-ALG) hydrogels was utilized to allow for prolonged retention and presentation of heparin binding growth factors to incorporated hMSCs.[18] The same gradient making system presented in Figure 1 was used to prepare HP-ALG hydrogels with incorporated gradients of heparin binding growth factors BMP-2, a potent osteogenic growth factor,[33] and TGF-β1, a chondrogenic growth factor,[34] in opposite directions. When the concentration of growth factors in segments of gradient HP-ALG hydrogels was quantified, linear gradient distributions of BMP-2 and TGF-β1 in opposite directions were observed (Figure 2A). In response to the incorporated growth factors, hMSCs underwent osteogenic and chondrogenic differentiation during a 2-week culture period and the levels of alkaline phosphatase (ALP) activity and glycosaminoglycan (GAG) production were quantified along the gradient HP-ALG hydrogels as measures of osteogenic and chondrogenic differentiation, respectively. As the BMP-2 concentration increased, ALP expression significantly increased along the BMP-2 gradient (Figure 2B). In contrast, GAG production of encapsulated hMSCs significantly increased as the TGF-β1 concentration increased (Figure 2C). While the changes in these hMSC differentiation markers were not linear, they did follow the expected trend with highest ALP activity and GAG production in the regions of hydrogels with highest BMP-2 and TGF-β1 content, respectively.
Figure 2.
Characterization of BMP-2 and TGF-β1 gradient alginate hydrogels and response of photoencapsulated hMSCs in growth factor gradient hydrogels. (A) Quantification of BMP-2 (closed circle) and TGF-β1 (closed square) content in each segment of the growth factor gradient alginate hydrogels. (B) ALP activity of photoencapsulated hMSCs in even-numbered segments and (C) GAG content of photoencapsulated hMSCs in odd-numbered segments of the growth factor gradient alginate hydrogels. *p<0.05 compared with segment 9 for GAG/DNA and segment 10 for ALP/DNA, respectively.
There are few previous reports of scaffolds having a growth factor gradient presented within a 3D scaffold for bone and cartilage tissue engineering. Wang et al. reported microsphere-mediated growth factor gradients in polymer scaffolds. BMP-2 and insulin-like growth factor-1 (IGF-1) were encapsulated in PLGA and silk microspheres, which were further incorporated in hydrogels with human bone marrow derived mesenchymal stem cells (hMSC) using a two chamber gradient maker, to induce hMSC differentiation.[28] This system exhibited greater chondrogenic and osteogenic differentiation along the gradient of increasing BMP-2 and decreasing IGF-1.[28] However, it did not achieve osteogenic and chondrogenic differentiation in opposite directions. Suciati et al. engineered layered PLGA microsphere-based scaffolds, using poly(ethylene glycol) as a plasticizer, to create stepped regions of growth factors within 3D scaffolds.[35] They demonstrated zonal release of BMP-2 from these layered PLGA microsphere-based scaffolds. However, for this system it is technically challenging to fabricate the growth factor-encapsulated PLGA microspheres and scaffolds, and a sintering step is required for their preparation, preventing cell encapsulation during fabrication. Opposing gradients of BMP-2 and TGF-β1 in PLGA scaffolds have been engineered to try to regenerate osteochondral defects in an animal model.[23] The gradient induced enhanced osteochondral differentiation of hMSCs compared to blank scaffolds in the rabbit femoral medial condyle. Although these gradients induced osteogenic and/or chondrogenic differentiation of hMSCs, these systems required preprocessing to encapsulate growth factor in microparticles to achieve the growth factor gradients, which may induce growth factor inactivation during encapsulation.[36] In contrast, here we present inverse gradients of BMP-2 and TGF-β1 in an affinity-based drug delivery system using photocrosslinked heparin-functionalized alginate hydrogels that allow for controlled and prolonged presentation of bioactive growth factors to incorporated cells by simple mixing into macromer solution prior to gelation. This approach permitted controlled osteogenic and chondrogenic differentiation in opposing directions. Additionally, this system could be applied to make gradients with tunable release profiles of other growth factors that have affinities for heparin, such as fibroblast growth factor-2 and vascular endothelial growth factor,[18] which when presented together in gradient combinations could enhance regeneration for other tissue types.
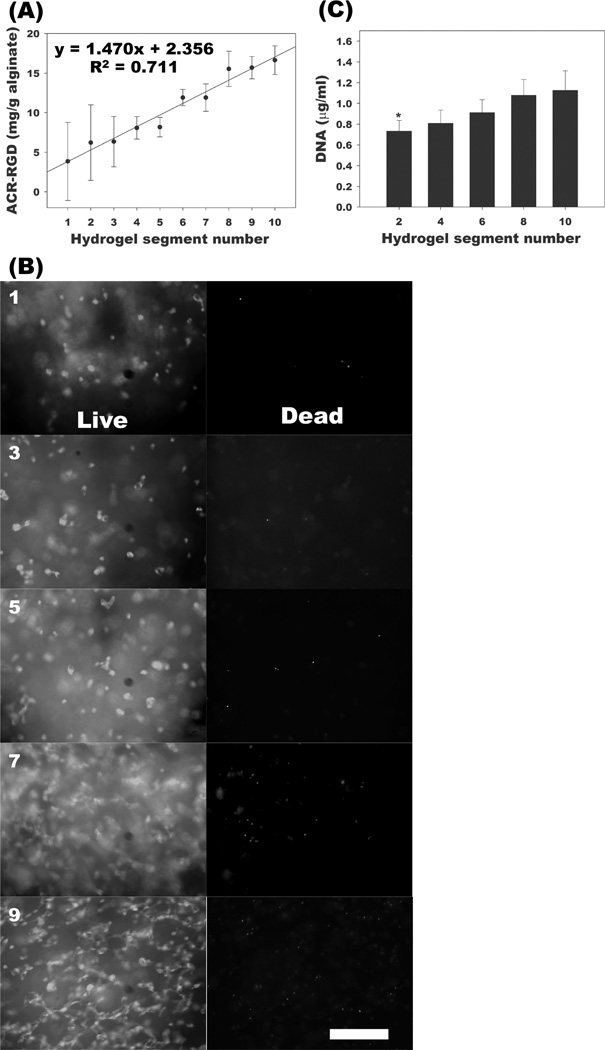
Next, the dual-pump system was used to generate a gradient of peptides containing the cell adhesion ligand, RGD, covalently coupled to the alginate hydrogel backbone. This system could then be used to examine whether having such spatial control over cell-biomaterial interactions could result in gradient changes in cell responses like cell number, alignment and migration, which are known to be affected by these interactions.[9,10,37] The establishment of the RGD peptide gradient was confirmed by ninhydrin assay on each segment of the gel (Figure 3A). When hMSCs were cultured in the adhesion ligand gradient hydrogels, a higher cell number was seen in regions of higher RGD concentration compared to regions of lower RGD concentration as demonstrated qualitatively by live/dead staining (Figure 3B) and quantitatively by PicoGreen DNA assay (Figure 3C) after 2 weeks of culture. Since photocrosslinkable peptides can easily be incorporated into this system, various adhesion ligand types, concentrations and combinations could also potentially be used to form gradients to investigate their influence on cell behaviors within 3D hydrogel systems. While a number of other studies have investigated the effects of gradients of adhesion ligands without the encapsulation of cells,[9,10] the present work involves 3D cellular encapsulation. Importantly, here, 3D hydrogels are used to demonstrate an increase in hMSC number up a 3D RGD gradient, whereas previous work had demonstrated similar results in 2D.[38] Not only do hydrogels created by a dual-pump system allow for the formation of 3D gradients of adhesion ligands, but the fabrication technique also permits the encapsulation of cells within the hydrogels.
Figure 3.
Characterization of RGD gradient alginate hydrogels and response of photoencapsulated hMSCs in RGD gradient hydrogels. (A) Quantification of RGD via ninhydrin assay in each segment of the RGD gradient hydrogels. (B) Representative live (FDA, green) / dead (EB, red) photomicrographs of photoencapsulated hMSCs in odd-numbered segments and (C) quantification of DNA content in even-numbered segments of RGD gradient alginate hydrogels with photoencapsulated hMSCs after 2 weeks culture in vitro. The scale bar indicates 200 µm. *p<0.05 compared with segment 8 and 10.
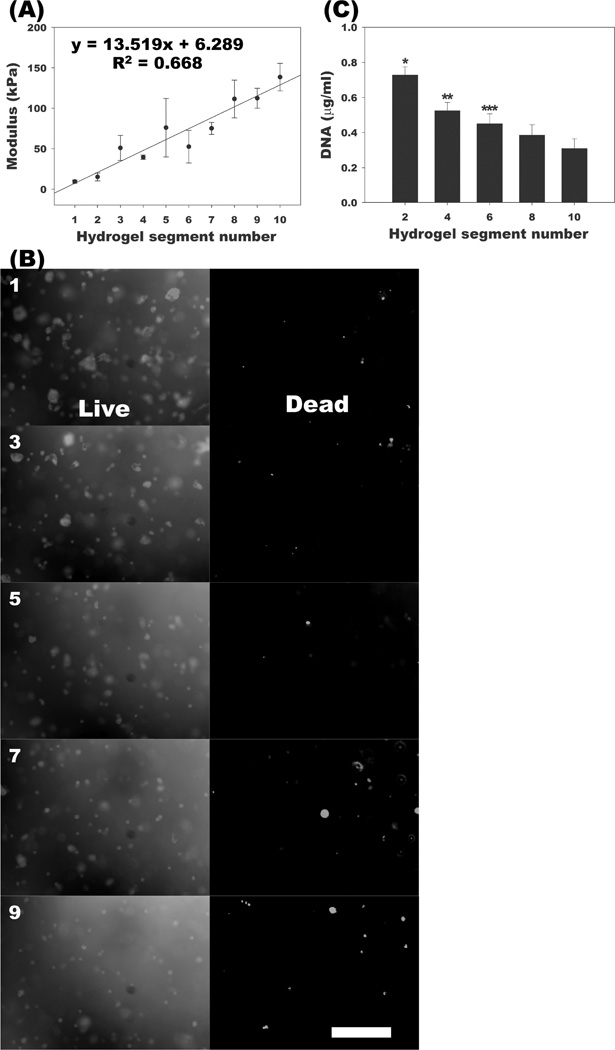
Lastly, stiffness gradients were established in the biodegradable, photocrosslinked alginate hydrogels to evaluate the effect of linear spatial changes in this physical property on the number of encapsulated hMSCs. Increasing the degree of alginate methacrylation increases resultant hydrogel crosslinking density and has previously been shown to increase hydrogel compressive modulus.[20] Using the dual-syringe pump system, hydrogels were made with a spatial gradient of crosslinking density, and the compressive modulus increased linearly over a 15-fold range from ~10 kPa to ~150 kPa with increasing alginate methacrylation (Figure 4A). This gradient hydrogel was then used as a scaffold for 3D screening of cellular growth response to changes in hydrogel stiffness. To examine cell viability during the photocrosslinking process and culture, photoencapsulated hMSCs in the hydrogels were evaluated using a live/dead assay after 2-week culture (Figure 4B). High cell viability was observed throughout all segments. To evaluate hMSC changes in cell number in the gradient hydrogels, the amount of DNA present was quantified in even-numbered segments after 2-week culture. The DNA content of gradient hydrogel segments was significantly higher in softer regions compared to the stiffer regions, with DNA decreasing in progressively stiffer regions (Figure 4C).
Figure 4.
Characterization of stiffness gradient alginate hydrogels and response of photoencapsulated hMSCs in stiffness gradient hydrogels. (A) Modulus of each segment of stiffness gradient hydrogels. (B) Representative live/dead photomicrographs of photoencapsulated hMSCs in odd-numbered segments and (C) quantification of DNA content in even-numbered segments of stiffness gradient alginate hydrogels with photoencapsulated hMSCs after 2 weeks culture in vitro. The scale bar indicates 200 µm. *p<0.05 compared with the other segments. **p<0.05 compared with segments 8 and 10. ***p<0.05 compared with segment 10.
The stiffness, crosslinking density, and degradation rate of hydrogels are all important in dictating cell growth.[15] Cell proliferation is enhanced in a matrix microenvironment that permits the diffusion of nutrients and waste.[15] These processes are mainly affected by the permeability and porosity of the ECM.[15] The permeability and porosity of hydrogels can be controlled by varying their crosslinking density;[31] as the crosslinking density increases, the permeability and porosity of hydrogels decrease.[39] Therefore, a high crosslinking density in hydrogels reduces the supply of nutrients to the encapsulated cells, prevents the removal of deleterious wastes, and consequently may affect cell growth. The degradation rate of the hydrogels impacts changes in the crosslinking density, and therefore nutrient and waste transport, over time as well. The softer regions of the gels were crosslinked to a lesser extent and degrade more rapidly; both of these characteristics would improve transport and could ultimately enhance cell growth. Early work on the stiffness of biomaterials demonstrated that fibroblast migration on the surface of polyacrylamide gels is influenced by gradients of matrix stiffness.[11] The current system may allow for the study of 3D cell migration in a gradient of matrix stiffness. More recent work using polyacrylamide gels has shown that the differentiation of hMSCs seeded on the surface of these gels can be controlled by matrix stiffness in the range of 1–100kPa.[7] Investigating the role of substrate mechanics in 3D, hydrogels of homogeneous stiffness in the range 2.5–110kPa have been used to demonstrate the role of the matrix stiffness in controlling encapsulated stem cell fate.[40] The present work allows for cell encapsulation in a cytocompatible 3D hydrogel system with a gradient of stiffnesses in a similar range (10–150kPa), which will permit the effects of such a gradient on cell differentiation to be investigated in future studies.
In summary, we have demonstrated a simple yet versatile method to form and quantify physical and biochemical 3D linear gradients within centimeter scale, biodegradable, photocrosslinked alginate hydrogels. Three-dimensional gradients of growth factor concentration, adhesion ligand density and stiffness in alginate hydrogels have been formed that can regulate encapsulated hMSC number and/or osteochondral differentiation. This flexible, cell compatible gradient making approach, which is amenable to a variety of hydrogel systems, may be a valuable tool for researchers in biomaterials science to control stem cell fate in 3D and guide tissue regeneration.
Experimental
Fabrication and characterization of hydrogels with microparticle gradients
To fabricate continuous linear microparticle-based gradient hydrogels, 25% actual methacrylated alginate[20] solutions (2% w/v) in DMEM containing Irgacure 2959 photoinitiator (0.05% w/v) with or without microparticles (5×106 particles/ml) were loaded separately into two syringes. The syringes were then installed in a custom gradient fabrication apparatus comprised of 2 programmable syringe pumps (NE-1000X, New Era Pump System Inc., Farmingdale, NY, (Figure 1A). The methacrylated alginate solutions were pumped with inversely varying flow rates (Figure 1B) through a spiral mixer (TAH Industries Inc., Kensington, MD) to a cylindrical quartz tube (2 mm internal diameter and 100 mm length, National Scientific, Rockwood, TN), and photocrosslinked with 365nm UV light (Model EN-280L; Spectroline, Westbury, NY) at ~1 mW/cm2 for 10 min to form the gradient hydrogels. The photocrosslinked microparticle-based gradient hydrogels were cut into 10 equal segments using a razor blade and individually placed in 5 N NaOH for 4 hrs to digest them. After 4 hrs digestion, the number of microparticles in each digested solution was counted using a hemocytometer to characterize the gradient (N=5).
Fabrication and characterization of growth factor gradient hydrogels
To fabricate growth factor gradient alginate hydrogels, BMP-2 and TGF-β1 were incorporated into HP-ALG hydrogels. Methacrylated heparin[18] (0.18% w/v) and methacrylated alginate (1.82% w/v, 10 % actual methacrylation) were dissolved in DMEM with 0.05% w/v photoinitiator. BMP-2 (10 µg/ml) and TGF-β1 (5 µg/ml) were dissolved in 2 separate HP-ALG macromer solutions. The macromer solutions were pumped through a spiral mixer into a quartz tube, and photocrosslinked as described above. Photocrosslinked growth factor gradient HP-ALG hydrogels were cut into 10 equal segments using a razor blade. To extract the BMP-2 and TGF-β1, each segment was dissolved in 250 µl of 1 N NaOH under stirring at 4 °C until clear solution was obtained. The amount of BMP-2 and TGF-β1 in the solution was quantified using enzyme-linked immunosorption assay (ELISA) kits (Human BMP-2 and TGF-β1 Duoset; R&D Systems, Minneapolis, MN) (N=5).
Fabrication and characterization gradient hydrogels with covalently coupled RGD-containing peptides
To fabricate alginate hydrogels containing a gradient of peptides containing the Arg-Gly-Asp (RGD) sequence, acrylated Gly-Arg-Gly-Asp-Ser-Pro (GRGDSP) were incorporated into photocrosslinked alginate hydrogels.[26] Methacrylated alginate (10 % actual methacrylation) solutions (2% w/v) in DMEM containing photoinitiator (0.05% w/v) with or without acrylated RGD (20 mg/g alginate) were loaded separately into two syringes. The macromer solutions were then pumped through a spiral mixer into a cylindrical quartz tube and photocrosslinked as described above. The photocrosslinked RGD gradient hydrogels were cut into 10 equal segments using a razor blade. To characterize the RGD gradients, a ninhydrin assay was used. After each segment was dissolved in 5 ml of 1 M sodium acetate buffer (pH 5), the amount of free amino groups in the solution was determined according to a previously reported method.[19] Unmodified methacrylated alginate and glycine (Fisher) were used as the control and the standard, respectively (N=5).
Fabrication and characterization of stiffness gradient hydrogels
To fabricate continuous stiffness gradient alginate hydrogels, two syringes loaded with macromer solutions (2% w/v in DMEM with 0.05% w/v photoinitiator) of two different degrees of methacrylation (25% and 10% actual) were positioned in the syringe pumps. The macromer solutions were pumped through a spiral mixer into a cylindrical quartz tube, and photocrosslinked as described above. Photocrosslinked stiffness gradient hydrogels were cut into 10 equal segments using a razor blade. The elastic moduli of each hydrogel segment were determined by performing uniaxial, unconfined constant strain rate compression tests at room temperature using a constant crosshead speed of 5%/sec on a Rheometrics Solid Analyzer (RSAII; Rheometrics, Piscataway, NJ) equipped with a 10 N load cell. Elastic moduli were calculated from the slope of stress versus strain plots, limited to the linear first 5% of strain of plots (N=3).
hMSC isolation and preparation
A human bone marrow aspirate was harvested from the posterior iliac crest of a donor after informed consent under a protocol approved by the University Hospitals of Cleveland Institutional Review Board. The hMSCs were isolated from the bone marrow aspirate and cultured in the Skeletal Research Center Mesenchymal Stem Cell Facility as previously described.[41]
Encapsulation of hMSC
hMSCs (passage number 2) were photoencapsulated in growth factor, RGD adhesion ligand or stiffness gradient alginate hydrogels by suspending them in macromer solutions. To fabricate growth factor gradient alginate/hMSCs hydrogel constructs, BMP-2 (5 µg/ml) and TGF-β1 (500 ng/ml) were separately dissolved in HP-ALG macromer solutions [0.18% w/v methacrylated heparin and 1.82% w/v methacrylated alginate (10 % actual methacrylation) in DMEM with 0.05% w/v photoinitiator and 5×106 cells/mL hMSCs] and the two solutions were loaded into syringes on the two separate pumps. To fabricate RGD gradient alginate/hMSCs constructs, methacrylated alginate (10 % actual methacrylation) solutions (2% w/v in DMEM with 0.05% w/v photoinitiator and 5×106 cells/mL hMSCs) with or without acrylated-RGD (20 mg/g alginate) were loaded separately into two syringes. To fabricate stiffness gradient alginate/hMSCs hydrogel constructs, two syringes were loaded with RGD-modified methacrylated alginate macromer solutions[19] (2% w/v in DMEM with 0.05% w/v photoinitiator and 5×106 cells/mL hMSCs) at two different alginate methacrylation levels (25% and 10%). The solutions were pumped through a spiral mixer into a quartz tube, and photocrosslinked as described above. The resulting gradient alginate/hMSC hydrogel constructs were removed from the quartz tube, placed in tissue culture flasks (T25, Corning, Tewksbury, MA) with 10 mL of DMEM containing 10% FBS, and cultured in a humidified incubator at 37°C with 5% CO2 for 2 weeks. After culture, the gradient alginate/hMSCs constructs were cut into 10 segments using a razor blade. The odd-numbered RGD and stiffness gradient hydrogel segments were used for a live/dead assay, and the even-numbered segments were used for a DNA assay. The odd-numbered growth factor gradient hydrogel segments were used for a GAG assay, and the even-numbered segments for an ALP activity assay. The DNA content in each segment of growth factor gradient alginate/hMSCs constructs was also measured (N=3).
Assays of hMSCs encapsulated in gradient alginate hydrogels
The ALP activity of hMSCs photoencapsulated in even-numbered segments of growth factor gradient alginate hydrogels was measured using SensoLyte p-nitrophenylphosphate (pNPP) ALP Assay kit (AnaSpec Inc., Fremont, CA) according to manufacturer’s instructions. The even-numbered segments of growth factor gradient hydrogel/hMSCs constructs were homogenized at 35000 rpm for 30 sec using a TH homogenizer (Omni International, Marietta, GA) and lysed by sample freezing and thawing repeated three times, and the lysates were cleared by centrifugation at 16200×g for 10 min using a microcentrifuge (accuSpin Micro 17R, Fisher). 25 µl of supernatant was incubated with 150 µl of ALP substrate containing pNPP at 37°C for 30 min. The reaction was stopped by adding 25 µl of 3 N NaOH to the substrate reaction solution. The absorbance of the samples was read at 405 nm on a plate reader. Hoechst 33258 dye (0.1 µg/mL in nuclease-free water; Acros Organics, Morris Plains, NJ) was used for the DNA assay as previously described.[19] Calf thymus DNA standards (Rockland Immunochemicals, Gilbertsville, PA) were prepared with 0–4 µg/mL DNA in nuclease-free water. After the centrifugation of papain-digested samples, 100 µL of supernatant was mixed with 100 µL of the prepared dye solution. Fluorescence intensity of the dye-conjugated DNA solution was measured in 96-well plates on a plate reader (358nm excitation and 452nm emission; Safire, Tecan, Austria), and the DNA content was calculated from a standard curve generated with the calf thymus DNA. Each segment’s ALP activity measurement was normalized to the DNA content (N=3).
The GAG contents of hMSCs photoencapsulated in odd-numbered segments of growth factor gradient alginate hydrogels was measured using the dimethylmethylene blue (Sigma) assay in 96-well plates.[42] The odd-numbered segments of growth factor gradient hydrogel/hMSCs constructs were washed with PBS, homogenized, and digested in 1 mL papain buffer solution (25µg/mL papain [Sigma], 2mM cysteine [Sigma], 50mM sodium phosphate [Fisher], and 2mM ethylenediaminetetraacetic acid [Fisher], pH 6.5, in nuclease-free water) at 65°C overnight. After centrifuging at 16200×g for 10 min using a microcentrifuge, 50 µL of supernatant was mixed with 250 µL of dye containing 16 mg/L dimethylmethylene blue and 3.04 g/L glycine (pH 1.5) in each well. The absorbance was read at 595 nm using the plate reader. Chondroitin-6-sulfate (Sigma) from shark cartilage was used to construct the standard curve. The DNA content was measured as described above. Each segment’s GAG content measurement was normalized to the DNA content (N=3).
The viability and morphology of photoencapsulated hMSCs in odd-numbered segments of the RGD and stiffness gradient alginate hydrogels were investigated using a live/dead assay comprised of fluorescein diacetate (FDA; Sigma) and ethidium bromide (EB; Sigma). FDA stains the cytoplasm of viable cells green, whereas EB stains the nuclei of nonviable cells orange-red. The staining solution was freshly prepared by mixing 1 mL of FDA solution (1.5 mg/mL of FDA in dimethyl sulfoxide; Research Organics, Cleveland, OH) and 0.5 mL of EB solution (1 mg/mL of EB in PBS) with 0.3mL of PBS (pH 8). The cut segments were transferred into 24-well tissue culture plates with 1 ml DMEM containing 10% FBS, and 20µL of staining solution was added into each well and incubated for 3–5min at room temperature. Stained hydrogel-cell constructs were imaged using a fluorescence microscope (ECLIPSE TE 300; Nikon, Tokyo, Japan) equipped with a digital camera (Retiga-SRV; Qimaging, Burnaby, BC, Canada) from three different fields in the center of cell/hydrogel constructs (N=3). The even-numbered segments of the RGD and stiffness gradient alginate hydrogel/hMSCs constructs were homogenized and lysed by repeated sample freezing and thawing three times, and the lysates were cleared by centrifugation at 16200×g for 10 min using a microcentrifuge. After centrifuging, 100 µl of supernatant was mixed with 100 µl of 1×Tris-EDTA buffer (Invitrogen, Carlsbad, CA) containing fluorescent PicoGreen® reagent (Invitrogen) and incubated at room temperature for 30 min. Fluorescence intensity of the dye-conjugated DNA solution was measured in 96-well plates on a plate reader (480 nm excitation and 520 nm emission), and the DNA content was calculated from a standard curve generated with known amounts of calf thymus DNA (Invitrogen) (N=3).
Statistical analysis
All quantitative data is expressed as mean ± standard deviation. Statistical analysis was performed with one-way analysis of variance (ANOVA) with Tukey honestly significant difference post hoc test using Origin software (OriginLab Co., Northampton, MA). A value of p < 0.05 was considered statistically significant.
Acknowledgements
The authors gratefully acknowledge funding from the National Institute of Arthritis And Musculoskeletal And Skin Diseases of the National Institutes of Health under Award Number R01AR061265 and the National Institute of Dental & Craniofacial Research of the National Institutes of Health under Award Number R56DE022376. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Oju Jeon, Department of Biomedical Engineering, Case Western Reserve University Cleveland, OH 44106 (USA).
Daniel S. Alt, Department of Biomedical Engineering, Case Western Reserve University Cleveland, OH 44106 (USA)
Stephen W. Linderman, School of Medicine, Washington University St. Louis, MO 63130 (USA)
Eben Alsberg, Email: eben.alsberg@case.edu, Department of Biomedical Engineering, Case Western Reserve University Cleveland, OH 44106 (USA); Department of Orthopaedic Surgery, Case Western Reserve University Cleveland, OH 44106 (USA).
References
- 1.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Patwari P, Lee RT. Circ. Res. 2008;103:234. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanz-Ezquerro JJ, Tickle C. Differentiation. 2001;69:91. doi: 10.1046/j.1432-0436.2001.690203.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, Im GI. Tissue Eng. Part A. 2009;15:1543. doi: 10.1089/ten.tea.2008.0368. [DOI] [PubMed] [Google Scholar]
- 5.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]; Kang SW, Cha BH, Park H, Park KS, Lee KY, Lee SH. Macromol. Biosci. 2011;11:673. doi: 10.1002/mabi.201000479. [DOI] [PubMed] [Google Scholar]; Lei Y, Gojgini S, Lam J, Segura T. Biomaterials. 2011;32:39. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paletta JR, Bockelmann S, Walz A, Theisen C, Wendorff JH, Greiner A, Fuchs-Winkelmann S, Schofer MD. J. Mater. Sci. Mater. Med. 2010;21:1363. doi: 10.1007/s10856-009-3947-2. [DOI] [PubMed] [Google Scholar]
- 6.Mauck RL, Nicoll SB, Seyhan SL, Ateshian GA, Hung CT. Tissue Eng. 2003;9:597. doi: 10.1089/107632703768247304. [DOI] [PubMed] [Google Scholar]; Mauck RL, Seyhan SL, Ateshian GA, Hung CT. Ann. Biomed. Eng. 2002;30:1046. doi: 10.1114/1.1512676. [DOI] [PubMed] [Google Scholar]; Baker BM, Shah RP, Huang AH, Mauck RL. Tissue Eng. Part A. 2011;17:1445. doi: 10.1089/ten.tea.2010.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen X, Roberts CJ, Stevens MM. Eur. Cell Mater. 2009;18:1. doi: 10.22203/ecm.v018a01. [DOI] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Sant S, Hancock MJ, Donnelly JP, Iyer D, Khademhosseini A. Can. J. Chem. Eng. 2010;88:899. doi: 10.1002/cjce.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zeng X, Goetz JA, Suber LM, Scott WJ, Schreiner CM, Robbins DJ. Nature. 2001;411:716. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 9.Kang CE, Gemeinhart EJ, Gemeinhart RA. J. Biomed. Mater. Res. A. 2004;71:403. doi: 10.1002/jbm.a.30137. [DOI] [PubMed] [Google Scholar]
- 10.Sundararaghavan HG, Burdick JA. Biomacromolecules. 2011;12:2344. doi: 10.1021/bm200415g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo CM, Wang HB, Dembo M, Wang YL. Biophys. J. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 13.Beck J, Angus R, Madsen B, Britt D, Vernon B, Nguyen KT. Tissue Eng. 2007;13:589. doi: 10.1089/ten.2006.0183. [DOI] [PubMed] [Google Scholar]; Nunes SS, Song H, Chiang CK, Radisic M. J. Cardiovasc. Transl. Res. 2011 doi: 10.1007/s12265-011-9307-x. [DOI] [PubMed] [Google Scholar]; Kuo CK, Li WJ, Mauck RL, Tuan RS. Curr. Opin. Rheumatol. 2006;18:64. doi: 10.1097/01.bor.0000198005.88568.df. [DOI] [PubMed] [Google Scholar]
- 14.Carletti E, Motta A, Migliaresi C. Methods Mol. Biol. 2011;695:17. doi: 10.1007/978-1-60761-984-0_2. [DOI] [PubMed] [Google Scholar]
- 15.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv. Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geckil H, Xu F, Zhang X, Moon S, Demirci U. Nanomedicine (Lond) 2010;5:469. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen FM, An Y, Zhang R, Zhang M. J. Control. Release. 2011;149:92. doi: 10.1016/j.jconrel.2010.10.021. [DOI] [PubMed] [Google Scholar]; Leslie-Barbick JE, Moon JJ, West JL. J. Biomater. Sci. Polym. Ed. 2009;20:1763. doi: 10.1163/156856208X386381. [DOI] [PubMed] [Google Scholar]; Saik JE, Gould DJ, Watkins EM, Dickinson ME, West JL. Acta Biomater. 2011;7:133. doi: 10.1016/j.actbio.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hern DL, Hubbell JA. J. Biomed. Mater. Res. 1998;39:266. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]; Ho MH, Wang DM, Hsieh HJ, Liu HC, Hsien TY, Lai JY, Hou LT. Biomaterials. 2005;26:3197. doi: 10.1016/j.biomaterials.2004.08.032. [DOI] [PubMed] [Google Scholar]; Levesque SG, Shoichet MS. Biomaterials. 2006;27:5277. doi: 10.1016/j.biomaterials.2006.06.004. [DOI] [PubMed] [Google Scholar]; Wan LQ, Jiang J, Arnold DE, Guo XE, Lu HH, Mow VC. Cell. Mol. Bioeng. 2008;1:93. doi: 10.1007/s12195-008-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Biomacromolecules. 2005;6:386. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon O, Powell C, Solorio LD, Krebs MD, Alsberg E. J. Control. Release. 2011;154:258. doi: 10.1016/j.jconrel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon O, Powell C, Ahmed SM, Alsberg E. Tissue Eng. Part A. 2010;16:2915. doi: 10.1089/ten.TEA.2010.0096. [DOI] [PubMed] [Google Scholar]
- 20.Jeon O, Bouhadir KH, Mansour JM, Alsberg E. Biomaterials. 2009;30:2724. doi: 10.1016/j.biomaterials.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Hahn MS, Miller JS, West JL. Adv. Mater. 2006;18:2679. [Google Scholar]; Nguyen EH, Schwartz MP, Murphy WL. Macromol. Biosci. 2011;11:483. doi: 10.1002/mabi.201000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du YN, Hancock MJ, He JK, Villa-Uribe JL, Wang B, Cropek DM, Khademhosseini A. Biomaterials. 2010;31:2686. doi: 10.1016/j.biomaterials.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dormer NH, Singh M, Zhao L, Mohan N, Berkland CJ, Detamore MS. J. Biomed. Mater. Res. A. 2012;100:162. doi: 10.1002/jbm.a.33225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh M, Morris CP, Ellis RJ, Detamore MS, Berkland C. Tissue. Eng. Part C Methods. 2008;14:299. doi: 10.1089/ten.tec.2008.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon O, Alt DS, Ahmed SM, Alsberg E. Biomaterials. 2012;33:3503. doi: 10.1016/j.biomaterials.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon O, Alsberg E. Tissue Eng. Part A. 2013;19:1424. doi: 10.1089/ten.tea.2012.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burdick JA, Khademhosseini A, Langer R. Langmuir. 2004;20:5153. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]; Vozzi G, Lenzi T, Montemurro F, Pardini C, Vaglini F, Ahluwalia A. Mol. Biotechnol. 2012;50:99. doi: 10.1007/s12033-011-9411-9. [DOI] [PubMed] [Google Scholar]; Miller ED, Li K, Kanade T, Weiss LE, Walker LM, Campbell PG. Biomaterials. 2011;32:2775. doi: 10.1016/j.biomaterials.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piraino F, Camci-Unal G, Hancock MJ, Rasponi M, Khademhosseini A. Lab. Chip. 2012;12:659. doi: 10.1039/c2lc20515g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Wenk E, Zhang X, Meinel L, Vunjak-Novakovic G, Kaplan DL. J. Control. Release. 2009;134:81. doi: 10.1016/j.jconrel.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh M, Sandhu B, Scurto A, Berkland C, Detamore MS. Acta Biomater. 2010;6:137. doi: 10.1016/j.actbio.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdick JA, Vunjak-Novakovic G. Tissue Eng. Part A. 2009;15:205. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]; Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, Patel VN, Lang RA, Mohammadi M. Sci. Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]; He JK, Du YA, Villa-Uribe JL, Hwang CM, Li DC, Khademhosseini A. Adv. Funct. Mater. 2010;20:131. doi: 10.1002/adfm.200901311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Odedra D, Chiu L, Shoichet M, Radisic M. Acta Biomater. 2011;7:3027. doi: 10.1016/j.actbio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JA, Anseth KS. Adv. Mater. 2010;22:61. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan N, Dormer NH, Caldwell KL, Key VH, Berkland CJ, Detamore MS. Tissue Eng. Part A. 2011;17:2845. doi: 10.1089/ten.tea.2011.0135. [DOI] [PubMed] [Google Scholar]
- 33.Rickard DJ, Sullivan TA, Shenker BJ, Leboy PS, Kazhdan I. Dev. Biol. 1994;161:218. doi: 10.1006/dbio.1994.1022. [DOI] [PubMed] [Google Scholar]
- 34.Awad HA, Halvorsen YD, Gimble JM, Guilak F. Tissue Eng. 2003;9:1301. doi: 10.1089/10763270360728215. [DOI] [PubMed] [Google Scholar]; Joyce ME, Roberts AB, Sporn MB, Bolander ME. J. Cell. Biol. 1990;110:2195. doi: 10.1083/jcb.110.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suciati T, Howard D, Barry J, Everitt NM, Shakesheff KM, Rose FRAJ. J. Mater. Sci-Mater. M. 2006;17:1049. doi: 10.1007/s10856-006-0443-9. [DOI] [PubMed] [Google Scholar]
- 36.Fransson J, Hallén D, Florin-Robertsson E. Pharm Res. 1997;14:606. doi: 10.1023/a:1012101027814. [DOI] [PubMed] [Google Scholar]; van de Weert M, Hennink WE, Jiskoot W. Pharm. Res. 2000;17:1159. doi: 10.1023/a:1026498209874. [DOI] [PubMed] [Google Scholar]
- 37.Alsberg E, Anderson KW, Albeiruti A, Franceschi RT, Mooney DJ. J. Dent. Res. 2001;80:2025. doi: 10.1177/00220345010800111501. [DOI] [PubMed] [Google Scholar]
- 38.Moore NM, Lin NJ, Gallant ND, Becker ML. Acta Biomater. 2011;7:2091. doi: 10.1016/j.actbio.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Lai JY, Ma DH, Lai MH, Li YT, Chang RJ, Chen LM. PLoS One. 2013;8:e54058. doi: 10.1371/journal.pone.0054058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat. Mater. 2010;9:518. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haynesworth S, Goshima J, Goldberg V, Caplan A. Bone. 1992;13:81. doi: 10.1016/8756-3282(92)90364-3. [DOI] [PubMed] [Google Scholar]
- 42.Enobakhare BO, Bader DL, Lee DA. Anal. Biochem. 1996;243:189. doi: 10.1006/abio.1996.0502. [DOI] [PubMed] [Google Scholar]