Abstract
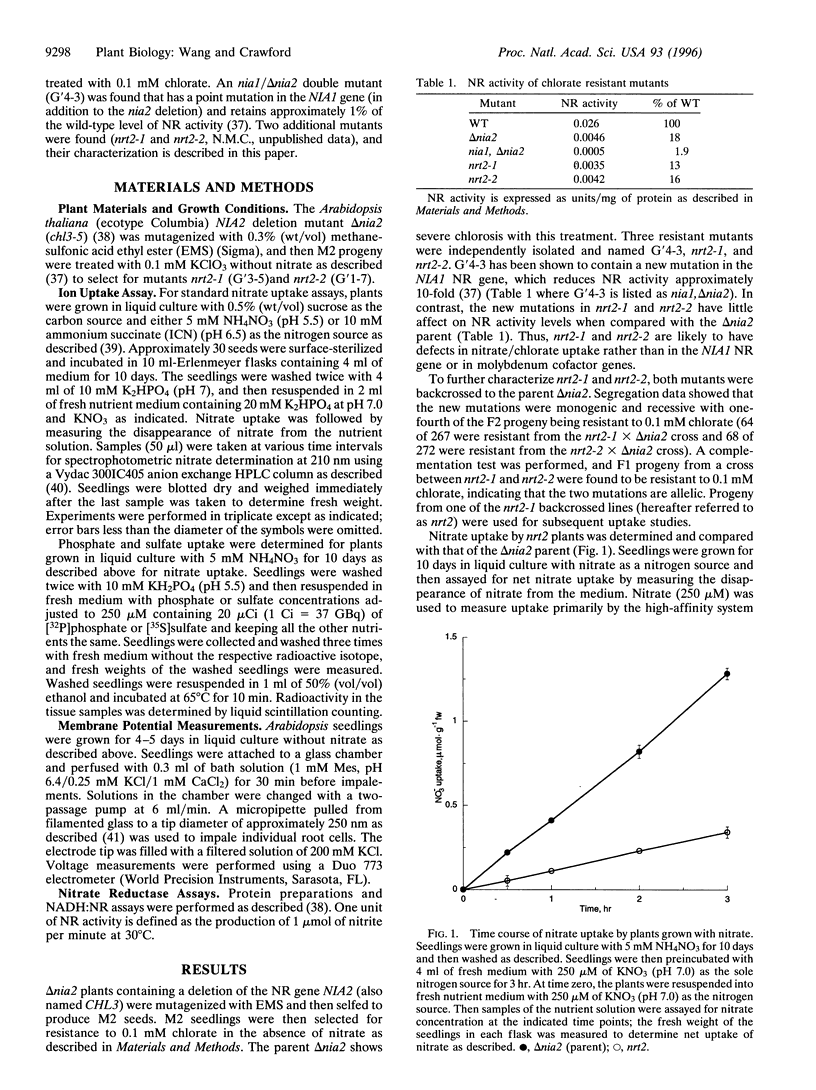
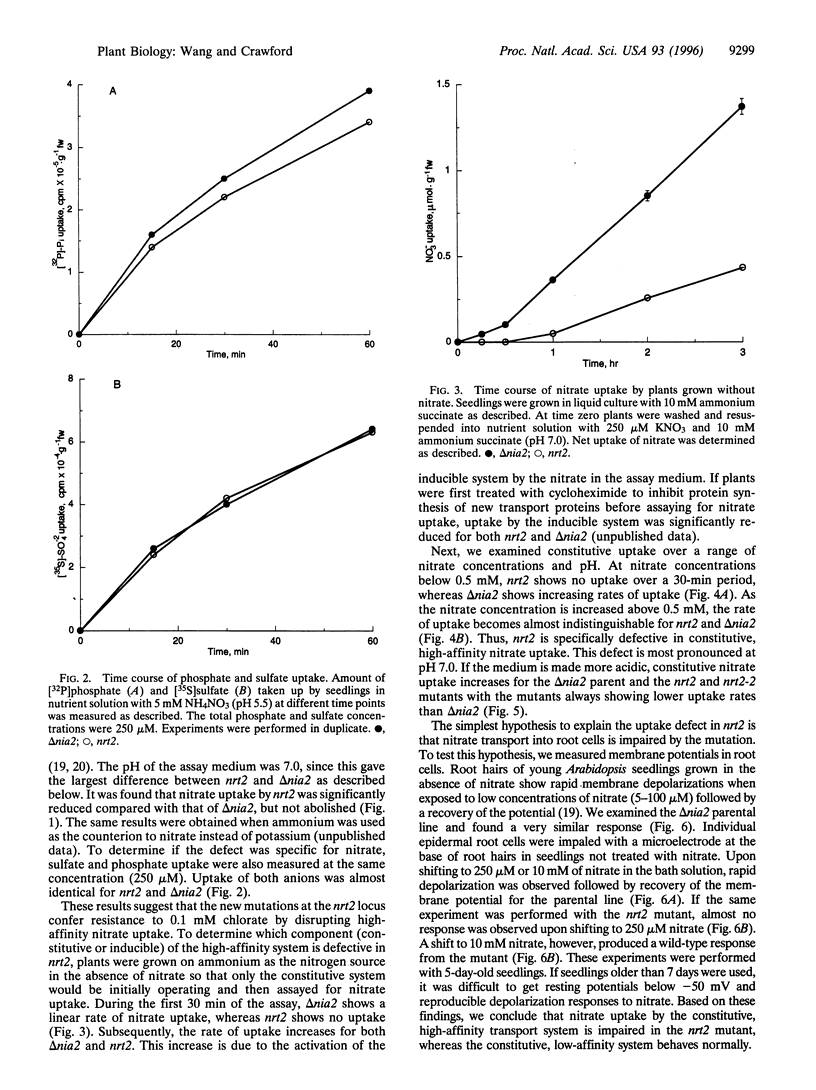
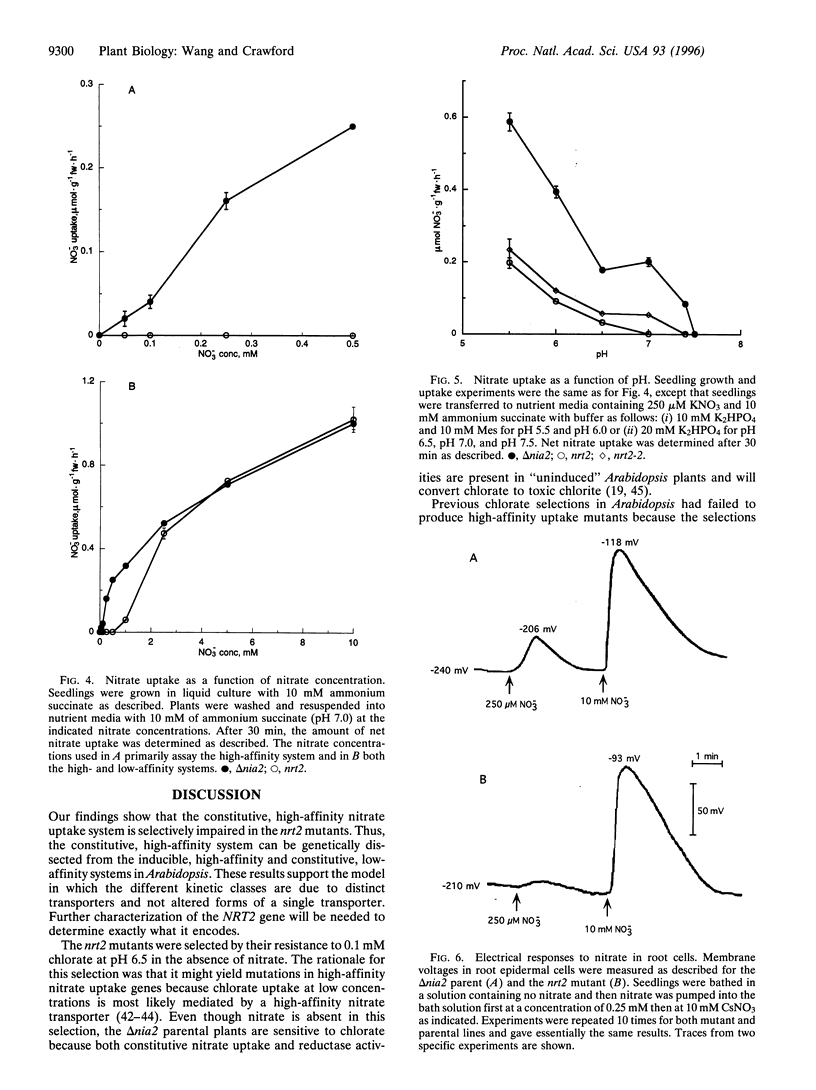
Two mutations have been found in a gene (NRT2) of Arabidopsis thaliana that specifically impair constitutive, high-affinity nitrate uptake. These mutants were selected for resistance to 0.1 mM chlorate in the absence of nitrate. Progency from one of the backcrossed mutants showed no constitutive uptake of nitrate below 0.5 mM at pH 7.0 in liquid culture (that is, within 30 min of initial exposure to nitrate). All other uptake activities measured (high-affinity phosphate and sulfate uptake, inducible high-affinity nitrate uptake, and constitutive low-affinity nitrate uptake) were present or nearly normal in the backcrossed mutant. Electrophysiological analysis of individual root cells showed that the nrt2 mutant showed little response to 0.25 mM of nitrate, whereas NRT2 wild-type cells showed an initial depolarization followed by recovery. At 10 mM of nitrate both the mutant and wild-type cells displayed similar, strong electrical responses. These results indicate that NRT2 is a critical and perhaps necessary gene for constitutive, high-affinity nitrate uptake in Arabidopsis, but not for inducible, high-affinity nor constitutive, low-affinity nitrate uptake. Thus, these systems are genetically distinct.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson B. E., Ward J. M., Schroeder J. I. Evidence for an Extracellular Reception Site for Abscisic Acid in Commelina Guard Cells. Plant Physiol. 1994 Apr;104(4):1177–1183. doi: 10.1104/pp.104.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M., Travis R. L., Huffaker R. C. Comparative kinetics and reciprocal inhibition of nitrate and nitrite uptake in roots of uninduced and induced barley (Hordeum vulgare L.) seedlings. Plant Physiol. 1992;99:1124–1133. doi: 10.1104/pp.99.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N. M., Arst H. N., Jr The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet. 1993;27:115–146. doi: 10.1146/annurev.ge.27.120193.000555. [DOI] [PubMed] [Google Scholar]
- Crawford N. M. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995 Jul;7(7):859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane-Drummond C. E., Glass A. D. Nitrate Uptake into Barley (Hordeum vulgare) Plants : A New Approach Using ClO(3) as an Analog for NO(3). Plant Physiol. 1982 Jul;70(1):50–54. doi: 10.1104/pp.70.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass A. D., Shaff J. E., Kochian L. V. Studies of the Uptake of Nitrate in Barley : IV. Electrophysiology. Plant Physiol. 1992 Jun;99(2):456–463. doi: 10.1104/pp.99.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy M., Zabala G., Filner P. The Kinetics of Chlorate Uptake by XD Tobacco Cells. Plant Physiol. 1988 Mar;86(3):817–821. doi: 10.1104/pp.86.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole D. J., Emran A. M., Fares Y., Drew M. C. Induction of nitrate transport in maize roots, and kinetics of influx, measured with nitrogen-13. Plant Physiol. 1990 Jun;93(2):642–647. doi: 10.1104/pp.93.2.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsande J., Touraine B. N Demand and the Regulation of Nitrate Uptake. Plant Physiol. 1994 May;105(1):3–7. doi: 10.1104/pp.105.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker H. J., Siddiqi M. Y., Glass ADM. Kinetics of NO3- Influx in Spruce. Plant Physiol. 1995 Sep;109(1):319–326. doi: 10.1104/pp.109.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrie S. T., Crawford N. M. A glycine to aspartic acid change in the MoCo domain of nitrate reductase reduces both activity and phosphorylation levels in Arabidopsis. J Biol Chem. 1994 May 20;269(20):14497–14501. [PubMed] [Google Scholar]
- Labrie S. T., Wilkinson J. Q., Crawford N. M. Effect of Chlorate Treatment on Nitrate Reductase and Nitrite Reductase Gene Expression in Arabidopsis thaliana. Plant Physiol. 1991 Nov;97(3):873–879. doi: 10.1104/pp.97.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure P. R., Kochian L. V., Spanswick R. M., Shaff J. E. Evidence for Cotransport of Nitrate and Protons in Maize Roots : II. Measurement of NO(3) and H Fluxes with Ion-Selective Microelectrodes. Plant Physiol. 1990 May;93(1):290–294. doi: 10.1104/pp.93.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure P. R., Kochian L. V., Spanswick R. M., Shaff J. E. Evidence for cotransport of nitrate and protons in maize roots : I. Effects of nitrate on the membrane potential. Plant Physiol. 1990 May;93(1):281–289. doi: 10.1104/pp.93.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharg A. A., Blatt M. R. NO3- transport across the plasma membrane of Arabidopsis thaliana root hairs: kinetic control by pH and membrane voltage. J Membr Biol. 1995 May;145(1):49–66. doi: 10.1007/BF00233306. [DOI] [PubMed] [Google Scholar]
- Quesada A., Galván A., Fernández E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994 Mar;5(3):407–419. doi: 10.1111/j.1365-313x.1994.00407.x. [DOI] [PubMed] [Google Scholar]
- Siddiqi M. Y., Glass A. D., Ruth T. J., Rufty T. W. Studies of the Uptake of Nitrate in Barley: I. Kinetics of NO(3) Influx. Plant Physiol. 1990 Aug;93(4):1426–1432. doi: 10.1104/pp.93.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi M. Y., King B. J., Glass A. D. Effects of nitrite, chlorate, and chlorite on nitrate uptake and nitrate reductase activity. Plant Physiol. 1992 Oct;100(2):644–650. doi: 10.1104/pp.100.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner H. Y., Naider F., Becker J. M. The PTR family: a new group of peptide transporters. Mol Microbiol. 1995 Jun;16(5):825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- Thayer J. R., Huffaker R. C. Determination of nitrate and nitrite by high-pressure liquid chromatography: comparison with other methods for nitrate determination. Anal Biochem. 1980 Feb;102(1):110–119. doi: 10.1016/0003-2697(80)90325-5. [DOI] [PubMed] [Google Scholar]
- Tsay Y. F., Schroeder J. I., Feldmann K. A., Crawford N. M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell. 1993 Mar 12;72(5):705–713. doi: 10.1016/0092-8674(93)90399-b. [DOI] [PubMed] [Google Scholar]
- Ullrich C. I., Novacky A. J. Extra- and Intracellular pH and Membrane Potential Changes Induced by K, Cl, H(2)PO(4), and NO(3) Uptake and Fusicoccin in Root Hairs of Limnobium stoloniferum. Plant Physiol. 1990 Dec;94(4):1561–1567. doi: 10.1104/pp.94.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles S. E., Hawker K. L., Grieve C., Campbell E. I., Montague P., Kinghorn J. R. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):204–208. doi: 10.1073/pnas.88.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J. Q., Crawford N. M. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet. 1993 May;239(1-2):289–297. doi: 10.1007/BF00281630. [DOI] [PubMed] [Google Scholar]
- Wilkinson J. Q., Crawford N. M. Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell. 1991 May;3(5):461–471. doi: 10.1105/tpc.3.5.461. [DOI] [PMC free article] [PubMed] [Google Scholar]



