Abstract
We present RNA-MITOMI, a microfluidic platform for integrated synthesis and functional assays across thousands of RNAs. The interaction of a comprehensive library of RNA mutants with stem loop binding protein precisely defined the RNA structural and sequence features that govern affinity. The functional motif reconstructed in a single experiment on our platform uncovers novel binding specificities and enriches interpretation of phylogenetic data.
The ENCODE project has shown that three-quarters of the human genome is capable of being transcribed1. Because only ~1.5% of the genome encodes for protein, non-coding RNAs (ncRNAs) may be a highly abundant output of genetic information. ncRNAs can exert epigenetic, transcriptional, and post-transcriptional regulation of gene networks that affect human health2. Regulatory function is primarily carried out by interactions between ncRNAs and diverse ligands. Yet, it is difficult to pinpoint ncRNA sequence or structural motifs that govern interaction specificity. Moreover, molecular principles that govern the ncRNA interactome are poorly understood3 and pathogenic mutations in ncRNAs that disrupt their regulatory function are difficult to identify4.
In order to address the gap between ncRNA discovery and functional characterization, we developed a general mutagenesis strategy aimed at identifying the contribution of RNA structural and sequence features on affinity for a given ligand (Supplementary Note 1). We applied this strategy to the well-characterized interaction between stem loop binding protein (SLBP) and the 3′ histone mRNA stem loop5 (Supplementary Table 1). After designing library of stem loop mutants per our strategy, we adapted the MITOMI (mechanically induced trapping of molecular interactions)6 microfluidic platform such that the entire RNA library could be simultaneously synthesized and then assayed for ligand binding affinity (Supplementary Note 2).
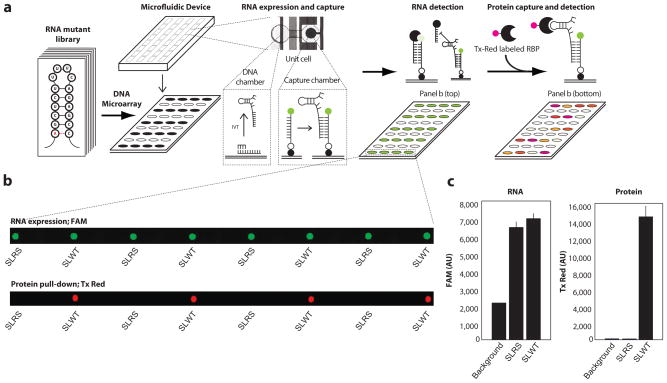
The MITOMI microfluidic chip used in this study has 640 microchambers, each with a volume ~ 1nL. DNA oligos serve as transcription templates for the RNA library. We spotted these oligos as a microarray and overlaid the MITOMI chip onto the array such that each spot was compartmentalized in a unique microchamber of the device. Each microchamber has a back-chamber, which houses the spotted DNA template, and a detection chamber in which the interaction between RNA and ligand can be measured (Fig. 1a). After testing two molecular beacon designs (Supplementary Fig. 1), we chose to immobilize a 5′ biotin and 3′ Fluorescein (FAM) labeled polyT ssDNA capture probe in each detection chamber (Supplementary Fig. 2). We then flowed an in vitro transcription mix onto the chip, filling all back-chambers and transcribing RNA from each DNA spot. The polyA-tailed transcribed RNA molecules hybridized to the surface immobilized capture probe during the IVT reaction. Following RNA synthesis, we used a quencher probe to quantify RNA capture within each detection chamber (Supplementary Fig. 3). Thus, our design allows co-transcriptional RNA folding and quantification of RNA capture without incorporation of fluorophore-modified nucleotides or use of intercalating dyes, which can alter RNA structure or function. Following RNA synthesis, our design enables interrogation of the RNA library against a wide diversity of ligands.
Figure 1. RNA-MITOMI is a microfluidic platform for integrated synthesis and affinity measurement across a programmable RNA library.
(a) An RNA library is encoded on ssDNA oligos, which are spotted as a microarray. The MITOMI microfluidic chip is overlaid onto the microarray and RNA molecules are transcribed from spotted DNA templates in each chamber of the device. RNA is captured prior to incubation with fluorophore tagged ligand. (b) Imaging on orthogonal channels quantifies RNA and protein capture in each cell of the device. (c) The imaged spots shown in B are averaged for SLWT (wild-type stem loop) and SLRS (mutant with no binding affinity to SLBP).
Once we verified RNA capture, we incubated N-terminal glutathione S-transferase (GST)-tagged SLBP with Texas Red dye conjugated GST antibody and flowed the protein across the immobilized RNA library. We quantified both RNA and protein captured in each chamber following equilibration using a microarray scanner (Fig. 1b). The control RNAs (SLWT, wild-type stem loop and SLRS, a compensatory mutant with no binding affinity to SLBP7) had a similar degree of RNA capture, but protein pull-down signal was ~100 fold greater in chambers with SLWT relative to the SLRS negative control (Fig. 1c), demonstrating that the assay had favorable dynamic range and low background signal due to non-specific RNA-protein binding.
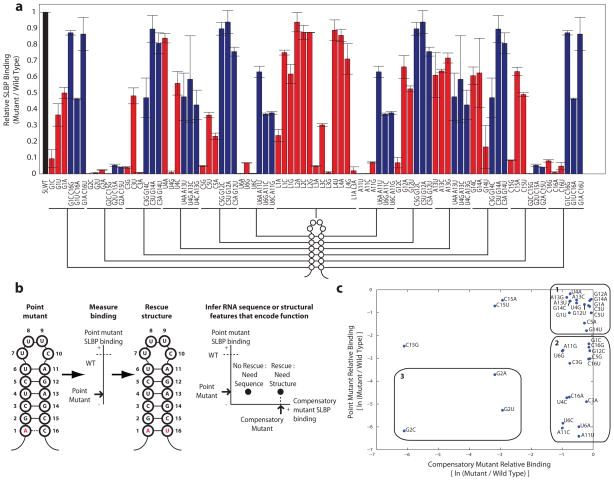
We measured relative SLBP binding across our library of single and double RNA mutants at 3 nM (Fig. 2a) and 30 nM (Supplementary Fig. 4) protein concentrations. We use a scatter plot to display the binding of each point mutant with respect to the relative binding of each double mutant that rescues RNA structure (Fig. 2b and Supplementary Fig. 5). The points on the scatter plot fall into three regimes. Fifteen mutants have little effect on binding, fifteen deleterious mutants are rescued by compensatory mutations that rescue RNA structure, and three mutants cannot be rescued by restoring structure, indicating sequence-based recognition at these positions (Fig. 2c).
Figure 2. SLBP binding landscape across the library of stem loop sequence and structure variants.
(a) SLBP affinity to stem loop point (red) and double compensatory (blue) mutants. Error bars show standard deviation with N=4. (b) Strategy for assigning function to features of stem loop sequence and structure. (c) The scatter plot of relative binding for each point mutant with respect to each double compensatory mutant is separated into three regimes: point mutants with little effect on binding (regime 1), point mutants that are rescued by compensatory mutations (regime 2), and point mutants that cannot be rescued (regime 3).
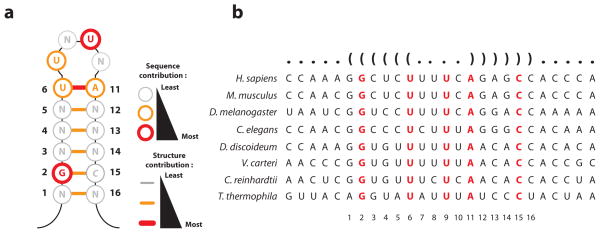
We used the dataset to generate a “functional motif”, which distills sequence and structural requirements for stem loop function (SLBP binding) at single nucleotide resolution (Fig. 3a). We validated the functional motif in two different ways. First, the features identified in the functional motif recapitulated the pattern of phylogenetic conservation of stem loop sequences8, suggesting that SLBP binding is the dominant selective constraint on histone mRNA tail (Fig. 3b). Residues (G2 and U9) and structural features (the U6-A11 base pair) that are critical for SLBP binding are conserved from Tetrahymena to humans, whereas co-variation is observed for base pairs that have less stringent sequence specificity but need to maintain pairing (G1-C16, C3-G14, U4-A13, and C5-G12). As a second validation, we tested nine point mutants with electrophoretic mobility shift analysis (EMSA). The shift data agreed with binding affinity measurements obtained with RNA-MITOMI (Supplementary Fig. 6), confirming that measurements on MITOMI can be recapitulated with conventional biochemical assays. Concordance of the functional motif with the evolutionary conservation of histone 3′ sequences also suggests that MITOMI measurements can identify features that are functionally relevant in vivo.
Figure 3. Reconstruction and validation of the functional stem loop motif.
(a) The functional motif distills stem loop sequence and structural features that are required for function (SLBP binding). (b) Phylogenetic analysis shows sequence conservation at features that are required for SLBP binding as well as co-variation without sequence preservation at base pairs for which structure, but not sequence, is important.
Our results show that SLBP recognizes much of the stem via RNA secondary structure rather than sequence. For nine of the twelve bases in the stem, function is not dependent upon sequence, as individual base pair substitutions at these positions are tolerated. In some cases, base substitutions are functionally neutral only if structure is preserved. In particular, Watson-Crick base pairing is required at the U6 -A11 position at the top of the stem (Supplementary Fig. 7). Nucleotide specific contacts are also important for SLBP binding. Consistent with prior studies7, two residues in the loop (U7 and, especially, U9) as well as G2 are required for binding (Supplementary Fig. 8).
Extending the results from prior studies9,7, our panel of mutants also identified non-canonical base pairs that are tolerated. Base pair substitutions in the position (C15) opposing G2 are tolerated, as mutations that allow wobble base-pairing (C15U) or prevent Watson-Crick base pairing (C15A) had nearly wild type binding. Like the C15A mutation, C5A establishes a functionally tolerated GA “base pair.” In addition, G12A and G1A mutations created a functionally tolerated AC pairs and G14C created a tolerated CC (Supplementary Table 1). Yet, these non-canonical pairs can be deleterious if combined (Supplementary Fig. 9), suggesting limited tolerance for structure perturbation within the stem. Furthermore, some of these non-canonical pairs are not tolerated at other positions in the stem, such as GA created by U4G and C16A as well as CC created by G1C, which may indicate higher sensitivity for structural deformation.
The functional motif reconstructed by RNA-MITOMI can enrich interpretation of phylogenetic data, as sequence co-variation patterns can be viewed through a functional lens. For example, only the G in the G2C15 pair of histone mRNA stem loop is required for SLBP affinity and non-canonical base-pairs resulting from mutations of C15 are functionally tolerated. With this in mind, our results suggest that there could be a number of undiscovered stem loop variants in histone genes that bind SLBP with similar affinity to the wild-type stem loop. The fact that none of these variants have been observed suggests that they have been selected against. This may be explained by additional functions carried out by the stem loop, including the formation of a SLBP-3′hExo ternary complex10, 11 or interactions with other factors that play a role in histone mRNA metabolism, which contribute to the selective pressure on the stem loop sequence.
Systematic mutagenesis and functional interrogation using the strategy and technology presented in this study offer a novel way to gain insights into the relationship between RNA sequence, structure, and function. Our approach is complementary with sequencing-based methods for RNA interactomics12,13,14,15, which can identify thousands of putative RNA-ligand interactions. These RNAs can be synthesized, mutated, and assayed on RNA-MITOMI in order to explore the underlying molecular principles of these interactions and consequences of mutations on function 4. Furthermore, motifs may be recombined and assayed to support engineering of ncRNAs with novel functions2, 16.
Online methods
Microarray preparation
Each RNA mutant was encoded on a DNA oligo, which were ordered in antisense orientation relative to the mutant sequence in Supplementary Table 1. Equimolar (9 μM) amounts of antisense oligo and sense T7 promoter oligo were annealed for 1 minute at 95°C and allowed them to cool at room temperature. After annealing, the oligos were diluted 1000-fold and re-suspended in a final concentration of 10 mg/mL BSA (Sigma, 100 mg/mL stock) for array spotting. The annealed oligos were arrayed in a 384 well plate and loaded into a custom microarry spotter with 36-pin printhead (Parallel Synthesis 32 pin microarray print head) and 3 pins (Parallel Synthesis silicon pins with 75x75μm tips). Oligos were arrayed from 384 well plate to an epoxy slide (25x75mm VEPO-25 Vantage expoxy slides) slide, resulting in an array orientation shown in Supplementary Table 2. Common T7 promoter (AATTTAATACGACTCACTATAGG) and RNA capture tail (CCCCAAAAAAAAAAAAAAAAAAAAAAAA) sequences were used for all oligos listed in Supplementary Table 1. We chose an un-structured polyA sequence to reduce the likelihood of intra-molecular interactions between the stem-loop and tail, increasing reliability RNA capture. We tested nine RNA species that lack the polyA tail using EMSA (Supplementary Fig. 6) and observed concordant results with RNA-MITOMI, suggesting that the polyA tail does not affect the RNA-SLBP interaction.
MITOMI chip fabrication
MITOMI devices were fabricated as described previously6. An image of the chip CAD drawing as well as instructions for ordering are shown in Supplementary Fig. 10.
Surface chemistry
After four hours of thermal bonding between the slide and MITOMI chip at 80°C, the slide surface was derivatized using biotinylated BSA (bBSA) (ThermoFisher, 2 mg/mL), neutravidin (ThermoFisher, 1 mg/mL), and HEPES buffer (pH 8.0) as described previously6. Derivitization resulted in a region of exposed avidin underneath each button valve. After testing molecular beacon designs (Supplementary Fig. 1), we chose a ssDNA probe (IDT ; 5Biosg//iSp18/TTTTTTTTTTTTTTTTTTTTTTTTGGGGAA/36-FAM/), which was flowed for 30 minutes after derivtization and was pulled-down specifically to the regions with exposed avidin underneath each button valve. After capture probe pull-down and 10 minutes of buffer wash, the button valve was depressed. An in vitro transcription mix (MEGAshortscript IVT kit, Ambion) was prepared (3 μL water, 2 μL IVT buffer, 8 μL NTP mix, 4 μL enzyme, 1 μL superasin (Ambion), 2 μL BSA), flowed across the chip for 30 minutes, and finally flowed into the cDNA-containing back-chambers of each unit cell by releasing the neck valve. The neck valve was then depressed again and transcription proceeded in the back-chambers at 37°C for 30 minutes while the chip was flushed with buffer (HBS-EP binding buffer, GE Healthcare). After flushing was complete, the button and neck valves were released and sandwich was depressed in order to allow RNA to be simultaneously transcribed and to hybridize with capture probe in each unit cell for an additional hour at 37°C. Once IVT was complete a quencher probe (IDT;/5IAbFQ/TTCCCCAAAAAAAAAAAAAAA) was used to confirm transcription and pull-down of RNA. The FAM dye on each capture probe slide was imaged using a Tecan LS Reloaded scanner at 488 nm excitation and FAM filter set.
SLBP expression and purification
SF9 insect cells were grown in suspension culture in SF900-II media supplement with 10% FBS to a density of 2 X 106 cells/mL. Cells were infected with baculovirus produced using standard protocol from Invitrogen Bac-to-Bac system. Infection was allowed to proceed for 3 days. Cells were pelleted an lysed in buffer containing 50 mM Tris-HCl (pH 8.5), 5 mM 2-mercaptoethanol, 1 mM PMSF, 1% Nonidet P-40. Clarified lysate was then bound to Ni-NTA resin and recombinant protein was eluted into buffer containing 20 mM Tris-HCl (pH 8.5), 100 mM KCl, 100 mM imidazol, 5 mM 2-mercaptoethanol, 10% glycerol. Protein aliquots were stored at −80°C.
SLBP preparation
A 3 μL aliquot of purified N-terminal GST tagged SLBP at 1.8 μg/μL (~ 31 μM) was thawed and 1 μL of SLBP was mixed with 5 μL of 1.0 μg/μL Texas-Red conjugated anti-GST antibody (Abcam ab34733) in 1000 μL of 1X buffer (HBS-EP, GE Healthcare). The mix was incubated for 1 hour at room temperature. Following incubation, 88 μL of protein-antibody complex was mixed with 1 μL of 100 mg/mL BSA (Sigma), 1 μL of 1 mg/mL salmon sperm DNA (Ambion), and 10 μL of 10 mg/mL yeast tRNA (Ambion) for a maximum assayed protein concentration of ~ 30 nM. SLBP concentration was titrated by adjusting the abundance of protein-antibody complex in the final preparation. The mix was flowed across the chip for 30 minutes in order to ensure equilibration between RNA and protein prior to trapping the bound SLBP with the button valve and imaging of a Tecan LS Reloaded scanner at 532 nm with the Texas Red filter set.
Array data analysis
Scanned files were analyzed using GenePix v6.0 software to determine FAM and Texas Red intensity under the button valve. The Texas Red intensity value for the wild-type stem loop was used to normalize all other variants (Supplimentary Table 1), resulting in normalized binding data. The natural log of relative intensity values is used for the scatter plot comparisons between point and compensatory mutants, as relative binding are displayed as negative values relative to 0 (wild-type relative affinity).
Mobility Shift Experiments
5 femtomoles of uniformly labeled RNA was incubated on ice with varying amounts of recombinant SLBP protein in 10 mM HEPES (pH 7.6), 50 mM KCl, 0.1mM EDTA, 10% glycerol, 1 μg/μL yeast tRNA, 0.1 μg/μL BSA on ice for 10 minutes. Reactions were directly loaded onto 8% native polyacrylamide gel (acrylamide:bisacrylamide was 29:1 in TBE buffer) without loading dyes. Gels were dried and visualized by autoradiography. Images were processed using the gel analysis route in ImageJ software.
Supplementary Material
Acknowledgments
We thank R. C. Spitale and C. Chu for helpful discussions. We thank J.C. Liang, R. J. Bloom, and C. D. Smolke for helpful discussions and assistance with Biacore. We thank A. Clore of Integrated DNA Technologies for assistance with probe designs. L.M. was funded by the Department of Defense (DoD) National Defense Science & Engineering Graduate Fellowship (NDSEG). Our work was supported by NIH R01-HG004361 (to H.Y.C.) and R01GM29832 (to W.F.M.). H.Y.C. is an Early Career Scientist and S.R.Q. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
L.M., S.R.Q., and H.Y.C. conceived the study. L.M., M.M., S.M.L., and R.V.S. performed experiments. L.M., M.M., S.M.L., W.F.M., S.R.Q., and H.Y.C. discussed the results and analyzed the data. L.M. and H.Y.C. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Djebali S, et al. Nature. 2012;488:101–108. [Google Scholar]
- 2.Martin L, Chang HY. J Clin Invest. 2012;122:1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Rinn JL. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper GM, Shendure J. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 5.Marzluff WF, Wagner EJ, Duronio RJ. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maerkl SJ, Quake SR. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 7.Battle DJ, Doudna JA. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davila Lopez M, Samuelsson T. RNA. 2007;14:1–10. doi: 10.1261/rna.782308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams AS, Marzluff WF. Nucleic Acids Research. 1995;23:654–662. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF. Molecular Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 11.Yang XC, Purdy M, Marzluff WF, Dominski Z. Journal of Biological Chemistry. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- 12.Koenig J, Zarnack K, Luscombe NM, Ule J. Nat Rev Genet. 2012;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, et al. Molecular Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granneman S, Kudla G, Petfalski E, Tollervey D. Proceedings of the National Academy of Sciences. 2009;106:9613. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Molecular Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang JC, Bloom RJ, Smolke CD. Molecular Cell. 2011;43:915–926. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.