Abstract
The eukaryotic stress response involves translational suppression of non-housekeeping proteins and the sequestration of unnecessary mRNA transcripts into stress granules (SGs). This process is dependent on mRNA binding proteins (RBPs) that interact with capped mRNA transcripts through RNA recognition motifs, and exhibit reversible aggregation through hydrophobic poly-glycine domains, some of which are homologous to yeast prion proteins. The activity and aggregation of RBPs appears to be important in the context of unfolded protein diseases. The discovery that mutations in these RBPs can cause familial motor neuron diseases and familial dementias indicates the importance of these genes to neuronal degeneration. Some disorders linked to mutations in RBPs include: amyotrophic lateral sclerosis (ALS), frontotemporal dementia and spinal muscular atrophy (SMA). These RBPs also associate with pathological structures in other neurodegenerative diseases, including Huntington’s chorea, Creutzfeld-Jacob disease, and Alzheimer’s disease. Interestingly, protein levels of RBPs change across the aging spectrum and may be linked to other age-related disorders, such as type 2 diabetes. The link between SG pathways and proteins linked to neurodegenerative diseases suggests a potential role for common pathways in both processes, such as those involved in translational control, and highlights potentially novel targets for therapeutic intervention in neurodegenerative diseases.
Keywords: Alzheimer’s disease, Frontotemporal Dementia, Amyotrophic Lateral Sclerosis, Fragile X Syndrome, Neurofibrillary Tangles, TIA-1, TTP, G3BP, TDP-43, FUS, Tau protein, Prion protein, HuR, Staufen, Pamilio, Dcp1a
II. Introduction
Regulating mRNA translation and protein synthesis allows a cell to rapidly alter the proteome in response to various signals. The discovery of RBPs, RNA granules, and their critical role in determining the fate and activity of mRNA transcripts has brought the importance of translational control into sharp focus. The interaction between RBPs and RNA granules controls the stability and translational activity of mRNAs and plays a critical role in fine-tuning protein expression under both normal conditions and conditions of stress[1].
The RBP family is comprised of about 800 proteins that share conserved domains structures and related functions. These RBPs generally contain two types of conserved domains: glycine rich domains and RNA recognition motifs (RRM). The glycine rich domain is hydrophobic and mediates the reversible aggregation of these proteins. The RRMs also impact on RBP aggregation, but are predominantly important for regulating RNA binding. The RRMs bind to short motifs (<10 bp), but exhibit incomplete sequence binding fidelity that also varies depending on the type of stress presented to the cell [2]. RBPs and RNA granules regulate many aspects of RNA biogenesis including RNA maturation, surveillance, transport, subcellular localization, translation, and RNA degradation. RBPs form dynamic interactions with coding, untranslated, and non–protein-coding RNAs in functional units called ribonucleoprotein (RNP) complexes. The RBPs within RNP complexes can remain stably bound to the RNA throughout its journey from synthesis to degradation, or associate with the RNAs selectively in a temporal and spatial manner [3].
The functions of RBPs can generally be divided into nuclear and cytoplasmic activities. In the nucleus, RBPs regulate mRNA maturation, including splicing, RNA helicase activity, RNA polymerase elongation, and nuclear export. In the cytoplasm, RBPs regulate RNA transport, silencing, translation, and degradation. These cytoplasmic RBPs regulate transcript activity and distribution by forming RNA granules that are macromolecular complexes containing RBPs, translational machinery, and mRNA transcripts consolidated to form granules through protein/protein interactions mediated by the glycine rich domains and protein/mRNA interactions mediated by RRMs. RNA granules vary by molecular composition and function. RNA degradation is mediated by a type of RNA granule, termed Processing-bodies or P-bodies (PBs). Transport granules play important roles in neurons, where they move transcripts from the soma into the dendritic, and possibly axonal, arbors. Stress granules (SGs) are important for the mammalian stress response, sequestering mRNAs and allowing for dynamic sorting of mRNAs for translation, storage, or degradation to allow for cell survival [2].
The pathophysiology of neurodegenerative diseases and many aging processes are characterized by the continual presence of oxidative stress. The stress response in eukaryotic cells involves the activation of defense mechanisms that can promote survival under some conditions, but also can initiate apoptosis under other conditions. The cellular response depends on the type, strength and duration of stress presented; the response is also finely tuned by the response of RBPs and their ability to control translation in both a global and transcript dependent manner. A key component of stress-induced translational suppression, SGs play a dynamic role in mRNA triage by sorting sequestered mRNAs for re-initiation, storage, or degradation, and may be required to allow optimal translation of stress-responsive anti-apoptotic mRNAs [4]. The process of SG formation is initiated by core, nucleating RBPs, including T-cell intracellular antigen 1 (TIA-1), tristetraprolin (TTP), and Ras-GTPase activating protein SH3-domain-binding protein (G3BP), however SGs mature with time to also include microRNAs, translation initiation proteins and other regulatory proteins, such as kinases and GTPases. The core, initiating RBPs each contain prion-like and poly-glycine rich domains, and their aggregation mirrors that of proteins linked to neurodegenerative diseases [5]. Importantly, RBPs including TDP-43, FUS, ataxin-2, hnRNPA1 and B2, optineurin, angiogenin, SMN-1, TIA-1 and TTP have all been shown to associate with SGs, and mutations or malfunctions in some of these RBPs can directly cause neurodegeneration [3] [6]. This review broadly examines the role of RBPs in aging, age-related processes, and diseases, with focus on conveying the therapeutic potential of pathways modulating key players in these processes.
III. RNA granules
RBPs regulate all aspects of RNA biogenesis including RNA maturation, surveillance, transport, subcellular localization, translation, and RNA degradation. RBPs are able to interact with mRNA and other protein components and consolidate to form discrete granules in the cytoplasm, and based upon their composition and function can be identified as ribonucleoprotein particles (RNPs), SGs, or P-bodies. These RNA granules share common protein components, but each kind of RNA granule contains a distinct population of proteins and performs separate functions: (i) RNA transport granules deliver transcripts to dendrites while inhibiting RNA translational activity. RBPs such as FMRP and Pumilio participate in dendritic transcript transport and function as translational repressors. (ii) Stress granules, which will be the main focus of this review, form transiently to reprogram RNA translation under stressful conditions. The primary nucleating SG proteins include TIA-1, TIAR, G3BP, FMRP, and survival of motor neuron protein (SMN), but a number of disease-linked proteins also associate with SGs as they expand; these proteins include fused in sarcoma (FUS), TDP-43, ataxin-2. (iii) Processing-bodies (PBs) are the sites for mRNA degradation, often integrating with miRNA machinery. Dcp1a (decapping enzyme 1a) is a RBP that is classically used to identify PBs. These granules are dynamic and are able to interact with each other and with active polysomes; this regulated trafficking can allow the cell to tailor translation to changes in its environment [1].
a. Ribonucleoprotein particles (RNPs)
Many mRNAs have localization elements within their 3′ or 5′ UTRs; RBPs are able to recognize these elements and direct message localization while suppressing translation until the message is delivered to its destination. These mRNAs are delivered by transport granules composed of RBPs and silenced transcripts. Transcripts may also be stored in the cytoplasm in a repressed state by similar RNA storage granules, which act as a pool of mRNA that, depending on metabolic state and environmental changes, can be directed for activation, repression, or decay [1].
Normal neuron physiology largely depends on the presence of these types of granules, which are the functional units for transport and translation regulation of mRNAs from the soma out into the dendrites to synapses. These RBPs also mediate the process of activity dependent protein synthesis, which is critical in all aspects of biology, but has attracted particularly strong attention at the synapse where it controls synaptic plasticity, habituation and memory. The synaptic function of Fragile X mental retardation protein (FMRP) has been studied extensively and is known to regulate dendritic sprouting.
Neuronal RNA transport granules are distinct from other RNA granules, although they can share some components and get in close contact, likely allowing a flow of mRNAs and proteins. There is also an apparent relationship between neuronal RNA granules and SGs, as both contain polyadenylated mRNAs, ribosomal subunits and a number of common RBPs, including SMN, Staufen, FMRP, Pumilio, TDP-43, and FUS, among others [7].
b. Stress granules
The stress response in eukaryotic cells involves the activation of defense mechanisms that either promote survival or the initiation of apoptosis. The cellular response depends on the type of stress presented. Type I stress, including hypoxia, heat-shock, and arsenite, inhibits translation initiation and induces the formation of SGs as a defense mechanism promoting cell survival [8]. SGs contain non-translating mRNAs, translation initiation components, and many additional proteins effecting mRNA function [4].
In metazoans, five eIF2α kinases monitor environmental stress and directly modulate the translation machinery. These include: (i) PKR (protein kinase R), a double-stranded RNA-dependent kinase that is activated by viral infection, heat and UV irradiation; (ii) PERK (PKR-like endoplasmic reticulum kinase, also known as PEK, or pancreatic eIF2α kinase), a resident endoplasmic reticulum (ER) protein that is activated when unfolded proteins accumulate in the ER lumen; (iii) GCN2 (general control nonderepressible 2), a protein that monitors amino acid levels in the cell and responds to amino acid deprivation; (iv) HRI (heme-regulated initiation factor 2α kinase), a protein that ensures the balanced synthesis of globin chains and heme during erythrocyte maturation and senses oxidative stress produced by arsenite; and (v) Z-DNA kinase, an enzyme involved in the host antiviral response. Stress-induced phosphorylation of eIF2α on Ser51 inhibits global protein translation through depletion of the eIF2–GTP–tRNA-met ternary complex, thus permitting the RBP TIA-1 to bind the 48S complex instead of the ternary complex. This promotes polysome disassembly and the consequent recruitment of mRNAs to SGs [9,10].
These translationally silenced mRNAs are necessary for SG formation and lead to primary nucleation of SGs involving RBPs including TIA-1, TTP, and G3BP, with specific mRNA transcripts. This family of proteins contains prion domains and poly-glycine rich domains, which confer the ability to reversibly aggregate [5]. The primary aggregation of mRNA binding proteins with stalled mRNA transcripts induces protein-protein interactions and protein-mRNA cross-linking by proteins such as poly-A binding protein (PABP-1) that bring other RBPs and non-RBP signaling proteins into the SG (discussed below). This results in the secondary aggregation of proteins with diverse physiological roles that can be modulated by SGs. The composition of SGs includes translation initiation components, small ribosomal subunit (40S), PABP, other RBPs, and mRNAs coding for most cellular proteins except those involved in the stress response. There is also piggy-back recruitment of many important signaling proteins into these granules that may effect cell signaling and survival [11]. However, SG composition may be subtly different according to the nature of the stress stimulus, and it may also change progressively during the response [7].
Once formed, SGs serve as centers of mRNA triage by dynamically sorting sequestered mRNAs for re-initiation, storage, or degradation, and may be required to allow optimal translation of stress-responsive anti-apoptotic mRNAs and thus appear to be protective. Transcripts are also transferred from SGs to PBs to facilitate degradation. SGs have been shown to be positioned adjacent to PBs, with TTP proposed to play an integral role in the shuttling of mRNAs destined for decay from SGs to PBs. Once the stress is removed there is rapid translational recovery in concert with SG disassembly. The mechanism of SG disassembly is poorly understood, but may be dependent upon interactions with heat shock proteins, and active transport by dynein and kinesin motors [12].
SGs have been suggested to suppress apoptosis by suppressing the stress-activated mitogen activated protein kinase MAPK pathway [13]. Additional cell survival mechanisms linked with SG formation are likely to occur, and an emerging example is the sequestration of pro-apoptotic molecules. The TNF receptor associated factor 2 (TRAF2) was the first case reported. TRAF2 facilitates apoptosis by two independent pathways: TNFR activation and caspase activation upon ER-stress induction. TRAF2 interacts with eIF4G and is therefore retained in SGs, thus avoiding apoptosis. More recently, two key molecules that activate the p38/JNK apoptotic pathway, namely RACK1 and ROCK1 were shown to be localized in SGs, thus favoring cell survival [7].
An interesting possibility is that formation of stress granules is required to allow optimal translation of stress responsive mRNAs. The translational arrest that accompanies environmental stress is potentially selective: one study shows that the translation of ~25% of mRNAs is significantly reduced, whereas the translation of another 25% of mRNAs (including transcripts encoding heat-shock proteins) is significantly enhanced. Stress-induced reprogramming of protein expression also entails stabilizing or destabilizing selected groups of mRNAs. Thus, post-transcriptional reprogramming of mRNA translation and decay reconfigures the proteome during adverse environmental conditions [11].
The importance of SGs for cytoprotection is highlighted by the effects of knockout of SG proteins, such as TIA-1, or inhibition of eIF2α phosphorylation, which render cells more vulnerable to acute stresses. Conversely, inhibiting eIF2α dephosphorylation protects against some forms of stress. However, the actual mechanisms by which SGs mediate protection are poorly understood [2].
c. Processing bodies (P-bodies, PBs)
P-bodies are another dynamic structure that contains mRNA decay machinery components. Core components of PBs are translationally inactive mRNA and the decapping factor, which induces mRNA decay and blocks translation by decapping and induction of 5′-3′ mRNA decay. Key to these complexes are Dcp1/2 (decapping enzymes), the Lsm1-7 complex (activators of decapping), Xrn1 (5′-3′ exonuclease), and nonsense mediated decay machinery. P-body assembly factors recruit mRNA into P-bodies through multimerization domains [1].
P-bodies are often observed juxtaposed to SGs but are also present in cells not under stress. Several observations demonstrate that SGs interact with PBs and are likely to exchange mRNAs between them: (i) mammalian PBs and SGs transiently dock with one another during arsenite treatment and can show prolonged docking when TTP is overexpressed and (ii) PBs and SGs share many protein components and the same mRNA species [4]. In neurons, FRAP analysis indicates that the turnover of DCP1a in PBs is dramatically enhanced by synaptic stimulation, indicating that PB dynamics and the release of mRNAs to allow their translation are controlled by neuronal activity, likely playing a role in regulating local protein synthesis at the post-synapse [7].
IV. Stress granules and aging
Throughout the lifetime of an organism, gene expression programs change dynamically. Gene expression patterns vary dramatically in a tissue-specific and age-dependent manner. The gene expression patterns that characterize each tissue at different developmental stages are strongly regulated at the transcriptional level. Gene expression patterns are potently regulated by RBPs, which control post-transcriptional processes such as pre-mRNA splicing, and mRNA cytoplasmic export, turnover, storage, and translation. Unlike transcription factors, much less is known about the role of RBPs in aging and age-related events [14].
RBPs that regulate mRNA turnover and/or translation critically affect subsets of expressed proteins. There has been one study examining the distribution and abundance of four RBPs across the aging spectrum, focusing predominantly on peripheral tissues: HuR, AUF1, TIA-1, and TTP [14]. TIA-1, Each of the RBPs exhibited little loss in abundance with advancing age in vivo. However, in cultured fibroblasts, AUF1 and TIA-1 both decreased greatly at population doublings of 30 or greater, while HuR showed strong decreases only at late stages of senescence and TTP showed strong expression only at late stages of senescence [14].
The aging process is associated with many translational changes that make us susceptible to neurodegenerative disease, metabolic disorders, and cancers where risk is relatively low in younger individuals. Defects in pathways controlling mRNA translation and protein synthesis are now implicated in the pathogenesis of metabolic disorders such as type 2 diabetes at multiple levels, including insulin synthesis and secretion, hepatic glucose uptake and gluconeogenesis, hepatic lipogenesis, and lipid and lipoprotein production [13]. RBPs are also known modulators of cell growth and proliferation, thus their dysfunction is likely to have implications in cancer biogenesis. RBPs hold the fate of many age related processes and their dysfunctions could result in the loss of physiologic function and the onset of diseases associated with advancing age.
There is strong evidence for malfunctions in translational control in metabolic regulation leading to syndromes such as type 2 diabetes. This translational control is important for insulin biosynthesis, hepatic and peripheral insulin sensitivity, and cholesterol homeostasis. For example, Wolcott-Rallison syndrome, a monogenetic form of diabetes, is caused by mutations in PERK, a key kinase regulating eiF2α-phosphorylation, translational, activity, and SG formation. PERK’s function is important for normal islet function and may act as a critical checkpoint for regulating insulin synthesis [1].
In cancer cells, expression of numerous oncoproteins or tumor suppressors is under the control of specific RBPs. Splicing, stability, localization as well as translation of these mRNAs are highly regulated, often in a tissue-specific manner. Many RBPs are aberrantly expressed in cancer cells and thus have a cancer-specific regulatory activity. Deregulation of RBP expression in cancer may have its origin in epigenetic events or on miRNA-dependent controls, although the detailed molecular mechanism is unknown. An additional layer of regulation is provided by signaling: the phosphorylation status of some RBPs is defined by signaling pathways that are deregulated in cancer, and this phosphorylation controls RBP activity and subsequently the expression of its target mRNAs. These signaling pathway alterations occur in different stages of tumor formation and are often correlated with tumor grade [15]. For example, RBP HuR stabilizes the mRNAs coding for cyclins (cyclin D1, E1, A2, B1), favoring cell cycle progression and promoting proliferation of cancer cells. HuR also promotes cancer cell survival by stabilizing transcripts encoding anti-apoptotic factors like Bcl-2, Mcl-1, SIRT1, and p21; mRNAs coding for proteins implicated in invasion and metastasis (MMP-9); or cell migration and adhesion (Urokinase A and uPA receptor) [15].
V. Stress granules and neurodegeneration
RBPs and their aggregation can also be important in the context of unfolded protein diseases. Many aspects of SGs resemble aggresomes and unfolded protein aggregates present in neurodegenerative pathologies. For example, both are mediated by specific prion-like and poly-glycine containing protein-aggregation domains, their dissolution requires molecular chaperones, they contain ubiquitinated proteins, and are enhanced by inhibitors of protein degradation machineries. These commonalities make it tempting to speculate that SGs and intracellular protein aggregates may interact. However, the aggregation processes characterizing the biology of RBPs differ from the conventional models of protein aggregation in that they subserve distinct biological functions and are reversible. In addition, aggresomes tend to form a single large inclusion, originating at the microtubule organizing center, whereas SGs occur as multiple complexes scattered throughout the cytoplasm.
SGs have been shown to be associated with the neurodegenerative disease-linked proteins Huntingtin and PrP prion protein [16,17]. Accumulation of TDP-43 inclusions in ALS and FTLD has been shown to be associated with SGs [18]. Tau inclusions in frontotemporal dementia with parkinsonism-17 (FTDP-17) and Alzheimer’s disease (AD) have been shown to colocalize with SGs, and these structures may increase tau deposition. The colocalization of SGs with neurofibrillary tangles suggests a biological link between the two species, perhaps related to proteins aggregation, and is also consistent with the previous observation that binding of RNA to tau promotes its aggregation [19]. Perhaps the most convincing evidence implicating RBPs in the pathophysiology of neurodegeneration is the discovery that mutations or malfunctions in some of these proteins can directly cause neurological disorders. For instance, impaired expression of FMRP, due to trinucleotide repeat expansions is the cause of fragile X mental retardation syndrome (FXS), which is the most common cause of inherited mental retardation, and with aging also leads to a related neurodegenerative condition. Expanded trinucleotide repeats in several different ataxin genes are the cause of spinocerebellar ataxia. Mutations in survival of motor neuron (SMN1) are linked to spinal muscular atrophy (SMA), and mutations in TDP-43, FUS, ataxin-2 (ATX2), optineurin (OPT) and angiogenenin (ANG) all cause motor neuron diseases including amyotrophic lateral sclerosis (ALS). These disease processes have now been linked to dysfunctional or dysregulation of neuronal RNA granules and SGs [2,20].
i. TDP-43 in ALS
ALS, or Lou Gehrig’s disease, is a progressive fatal neurodegenerative disease that affects motor neurons in the brainstem, spinal cord, and motor cortex. Clinical features involve degeneration of motor neurons producing fasciculation, muscle wasting and weakness, increased spasticity, and hyper-reflexia. With a projected lifetime risk of 1/2000, ALS is considered one of the most common motor neuron diseases. ALS is universally fatal, with a median age of onset of 55 years and a survival of 2–5 years after the onset of symptoms. A common pathological hallmark is the presence of TDP-43+ cytoplasmic inclusions in degenerating neurons that have been shown to colocalize with SGs [18].
TDP-43 is encoded by the TARDBP gene on chromosome 1, 414aa, 43kDa protein. TDP-43 is genetically linked to ALS and FTLD; over 40 TARDBP mutations have been found in ALS and 3 have been found in FTLD patients [13]. TDP-43 is a histopathological marker of ALS and FTLD-U, but also associates with pathology in AD, PD, Huntington’s disease, and hippocampal sclerosis. TDP-43 pathology in the brain and spinal cord is characterized by decreased solubility, ubiquitination, hyper-phosphorylation, and cleavage of TDP-43 into 25 and 35 kDa fragments. Another important feature associated with TDP-43 pathology is translocation from nuclear to cytosolic compartments. Although the significance of cytoplasmic translocation is poorly understood, it seems possible that reductions in nuclear might alter mRNA splicing patterns, which could have long-term consequences for neurons that are deleterious.
TDP-43 plays important roles in neuronal RNA metabolism. It regulates splicing and stability of many RNAs, particularly longer transcripts that encode important proteins for development, synaptic plasticity, and neurodegeneration. The “loss of nuclear function” hypothesis that might be caused by prolonged cytoplasmic translocation presents an attractive mechanism through which TDP-43 might contribute to neurodegeneration. A competing hypothesis (which is not mutually exclusive) is that cytoplasmic aggregation of TDP-43 also contributes to degeneration by interfering with mRNA availability and/or stability, as well as activating processes such as apoptosis. Distinguishing between these two mechanisms is challenging, though, because overexpression TDP-43 is toxic to many cell types, even in absence of aggregation. The mechanism of TDP-43 aggregation remains an additional important line of investigation. The involvement of TDP-43 in RNA granule formation, and in particular, in SG formation provides an appealing mechanism for TDP-43 aggregation. The colocalization of TDP-43 aggregation with TIA1 and eIF3 in ALS and FTLD supports a link between TDP43 aggregation and SGs [13]. TDP-43 contributes to the assembly and maintenance of SGs in response to oxidative stress and specifically regulates G3BP mRNA levels and maturation of G3BP SGs [9]. TDP-43 knockdown delays SG formation by increasing TIA1 levels and decreasing G3BP levels. Familial mutants of TDP-43 led to faster SG localization and larger SGs assembled [13]. However, the mechanisms contributing to persistence of TDP-43 aggregates might evolve and become pleiotropic over time. Proteins such as p62 and heat shock proteins associate with persistent aggregates. In addition, persistent protein aggregates evolve over time, becoming modified by processes such as phosphorylation, ubiquitination, glycosylation, cross-linking and oxidation; these processes render the aggregates increasingly insoluble.
ii. RBPs in Tauopathies
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder, is the most common cause of dementia, and most prevalent form of tauopathy. AD is characterized by progressive cognitive decline and specific pathological changes in the brain. Pathological properties of AD include: extracellular neuritic plaques composed of β-amyloid (Aβ), intracellular neurofibrillary tangles (NFTs) of hyperphosphorylated tau, astrocytic gliosis, reactive microglia and inflammation, and neuronal and synaptic loss. These alterations are accompanied by declining memory, dementia, and neuroinflammation [21]. Our lab has reported that TIA1, TTP, and G3BP are RBPs that accumulate in AD and related tauopathies, and can co-localize with pathogenic tau epitopes.
TIA1, TTP, and G3BP are all evolutionarily conserved RBPs that play a primary role in nucleating SGs. TIA-1 is a RBP of the RNA recognition motif (RRM)/ ribonucleoprotein (RNP) family that has been implicated as effectors of apoptotic cell death [22]. Transcripts targeted by TIA-1 are often C-rich or U-rich and are translationally repressed when they are associated with TIA-1. Translation of mRNAs encoding TNF-α, COX-2, and several other transcripts bearing a TIA-1 motif are specifically regulated by TIA-1. TTP binds mRNAs through two tandem CCCH zinc finger motifs and promotes their decay. TTP target mRNAs typically contain the AU-rich sequence, where TTP can bind and destabilize its targets including TNF-α, GM-CSF, COX-2, IL-3, IL-10, and interferon-γ, indicating its importance in the inflammatory response. G3BP was initially characterized through its interaction with a Ras-GTPase–activating protein (RasGAP p120), and is known to both activate and repress mRNA transcripts in a transcript dependent manner. G3BP is very important in embryonic development and thus knockout is embryonically lethal [23].
The examples of AD and FTLD-17 are particularly striking because in both of these tauopathies, the load of SG positive inclusions that form is large, exhibiting a density that is equal to or greater than the load of NFTs [24]. SG proteins such as TIA-1 and TTP identify most NFTs, but also identify inclusions that appear to lack reactivity with antibodies to phospho-tau (e.g., PHF-1 or CP13) or conformationally altered tau (e.g., Alz-50 or MC1). In contrast, G3BP, identifies neurons that are predominantly negative for pathological tau protein. The abundance of SGs in tauopathies suggests that the load of pathological inclusions is much greater than would be apparent by simply using markers of tau protein.
Tau and SG proteins exhibit an intimate connection. We observed that TIA1 and TTP directly bind to tau protein and may interact with tau to modulate disease progression [24]. Our studies using transfected SH-SY5Y cells demonstrate that SG formation stimulates formation of phosphorylated tau inclusions, and tau appears to stimulate SG formation [24]. The latter result suggests that tau actually contributes to the SG response, and might explain the strong link between tauopathy and SGs. Evaluation of tau biology suggests a mechanism for interaction with SGs. Tau expression is normally restricted to the axon, while the RNA translation machinery is more abundant in the soma and dendrites. However, stress stimulates tau phosphorylation and mislocalization to the soma and dendrite where it can interact with RBPs, as well as RNA associated with SGs [25]. This interaction accelerates SG formation, and may also accelerate tau aggregation [24]. In addition, RNA is a known stimulus for tau aggregation in vitro [19], which makes it possible that the RNA associated with SGs might further promote tau aggregation. Thus, in the case of tauopathies, aggregation of tau protein stimulates SG formation, leading to enhanced SG formation, and SG formation might stimulate tau aggregation.
Interestingly, early tau related deficits develop not from the loss of synapses or neurons, but rather as a result of synaptic abnormalities caused by the accumulation of hyperphosphorylated tau within intact dendritic spines, where it disrupts synaptic function by impairing glutamate receptor trafficking or synaptic anchoring. An interesting possibility is that SGs or other RNPs can impact on the spatial location of hyperphosphorylated tau and help tau to accumulate in the soma and dendritic compartments, where it disrupts trafficking of RNPs, which is necessary for local translation at the synapse and proper function. This would lead to a loss of synapses, and a vicious cycle of tau potentiating an overactive SG response, and SGs further exacerbating tau pathology and trafficking abnormalities.
iii. SMN in Spinal muscular atrophy (SMA)
SMA is a common autosomal recessive neuromuscular disorder associated with the loss of α-motorneurons in the spinal cord and is the leading hereditary cause of infant mortality. SMA is caused by deletions or mutations of the telomeric copy of SMN1 gene and retention of the SMN2 gene. The SMN2 copy, which is present in most SMA patients, produces preferentially exon 7-skipping isoform, SMNΔ7, and inadequate full-length protein that fail to protect motorneurons from the loss of the SMN1 gene. In motorneurons, SMN is localized in granules and can also play a role in RNP complex composition and localization required for axonal projection and maintenance, and SMN can nucleate SGs when overexpressed [26].
Loss of SMN protein is associated with defective RNP assembly and function. Thus, the pathological consequence of SMN loss in SMA is caused by disruption of RNA metabolism events essential for α motorneurons development and survival. In mice the loss of the only SMN gene, SMN1, is embryonically lethal. Introduction of two copies of the human SMN2 gene rescues this embryonic lethal phenotype and results in mice with SMA [3]. Interestingly, the SMN variant preferentially expressed in SMA fails to be recruited to SGs, and SG formation is reduced in SMA patients [7]. Thus, SMN1-mediated assembly or function of RNA granules in motor neurons might influence disease progression through the disruption of protein translation [11].
iv. Regulated protein aggregation
The potential importance of SGs for neurodegenerative disease becomes apparent because the process of SG formation presents a biological pathway that is based on physiological protein aggregation. Such a pathway could be vulnerable to “excessive aggregation” due to mutation in constituent proteins that shift the equilibrium in favor of aggregation or due to nonspecific binding to other protein aggregates that accumulate in neurodegenerative disease. RBPs are a group of proteins that naturally form insoluble aggregates, yet the aggregated material can disperse and resolubilize. Most, if not all, of RBPs linked to neurodegenerative diseases associate with SGs in cell culture. TDP-43, FUS, ataxin-2, SMN, optineurin, and angiogenin have all been shown to co-localize with classic SG markers (TIA-1, TTP, and/or G3BP) in cells undergoing stress. SG proteins such as TIA-1, eIF3, and PABP also co-localize with neuropathology in brain tissue of subjects with AD, FTDP-17, FTLD-TDP, and ALS, or animal models of these diseases In addition, as mentioned above with respect to ALS, AD, SMA, and FXR the dysfunctions of the disease may actually be linked to dysregulation of RNA granules [2].
Recently Wolozin [2], proposed that the aggregation of many pathological, intracellular proteins, including TDP-43, FUS, or tau, proceeds through the SG pathway. Mutations in genes coding for SG associated proteins increase the tendency of these proteins to aggregate, which leads to enhanced SG formation. RBPs exist in equilibrium between dispersed and aggregated. Mutations that favor aggregation will shift this equilibrium in favor of aggregation. Such an equilibrium shift would increase protein aggregation in neurodegenerative diseases by accelerating the process or by favoring the persistence of pathological SGs. Chronic stress might also stimulate formation of persistent pathological SGs. Syndromes such as diabetes, cardiovascular disease or the repeated head trauma experienced by football players each cause prolonged and/or repetitive stress. Such stress would favor persistent SG formation, and secondary modification of these persistent SGs (e.g., by ubiquitination or binding p62) might enhance the persistence. These persistent aggregates could also serve as a nidus for further aggregation of SGs or other aggregation prone proteins, such as tau. The result would be an overactive SG pathway. Over-active stress granule formation could act to sequester functional RBPs away from the nucleus and/or interfere with mRNA transport and synthesis, each of which might potentiate neurodegeneration.
v. Potential for Pharmacological modulation
The RBPs addressed in this review (TIA-1, G3BP, TTP, SMN, and TDP-43) are essential players in RNA metabolism, with the RNPs forming a dynamic regulatory system for all aspects of the life of an mRNA, including nuclear processing, transport, translation, and decay. These proteins are key in the coordination of gene expression of many proteins, and their disruption could impair cell function and interfere with appropriate distribution and translation of mRNA in response to signaling [24].
Whether hyperactive SG formation is good or bad remains to be determined. Acutely SG formation is known to be protective and anti-apoptotic, until the stress is resolved. In neurodegeneration and other aging-associated diseases, there is no resolution and the stress is chronic. At this point SG formation and recruitment of RBPs from their normal functions act as a loss of function situation where RNPs become dysfunctional, and RNA metabolism is no longer in check. Neurons require SGs for an effective stress response, but overactive, overly stable SG complexes could easily interfere with neuronal function by silencing transcripts and sequestering important proteins. Mutations associated with disease-linked RBPs increase the aggregation propensity or cause dysfunctional RNPs, which provides a direct mechanism for overactive SG formation. Chronic stressful diseases or environmental conditions might also stimulate overactive SG formation. For instance, the oxidative stress associated with aging, the trophic stress associated with diabetes, or the cellular stress associated with cancer all enhance SG formation creating the conditions for overactive SG aggregation [2].
The reversibility of the SG pathway also offers novel opportunities to stimulate endogenous biochemical pathways to reverse these pathological stress granules and also perhaps delays the progression of disease [2,24]. The effect of modulating the protein synthesis/SG pathway was recently evaluated in an animal model of Creutzfeld Jacob disease, where pathological misfolding of PrP precipitates neurodegeneration. Mallucci and colleagues forced expression of GADD34 to reduce eIF2α phosphorylation, inhibit SG formation and stimulate protein synthesis [27]. This intervention reduced PrP-induced neurodegeneration. In contrast, salubrinal, which increases eIF2α phosphorylation, increased SG formation, inhibited protein synthesis, and accelerated neurodegeneration [27]. These results suggest that inhibiting the SG pathway and stimulating protein synthesis can inhibit PrP-mediated neurodegeneration. The discovery of over-active SG formation in other diseases raises the possibility that these pathways are overactive in multiple neurodegenerative diseases and other aging processes, and pharmacotherapy that targets SG formation might be protective [2]. Several groups, including our own, are developing non-toxic chemicals that can dampen a system suffering from over-active SG formation. For instance some groups have identified FDA approved chemicals that moderately reduce aggregation of TDP-43, and we are developing a series of novel compounds that strongly reduce TDP-43 aggregation with minimal toxicity. A major challenge in the future will be to define the components of each RNP complex and pinpoint the defects associated with RBP [28] dysfunction. An important issue to resolve is why the absence of certain RBPs or accessory proteins leads to cell-specific defects [3].
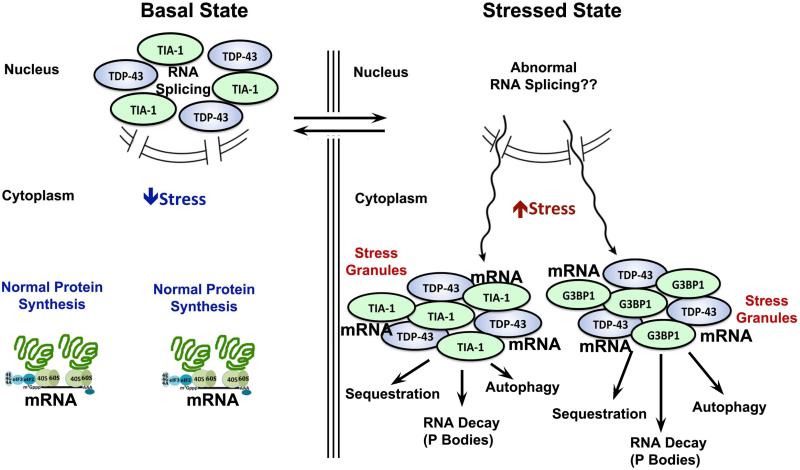
Figure 1.
TIA-1, TDP-43 and many other RBPs reside in the nucleus under basal conditions. Their nuclear functions vary, but RBPs act in part to facilitate RNA transcription and to regulate RNA splicing. Stress causes these nuclear RBPs to leave the nucleus. In the cytoplasm, the RBPs bind stalled, free mRNA and aggregate to form SGs primarily through the glycine rich domains and secondarily by other domains. The process of SG formation is reversible, but prolonged stress leads to maturation and incorporation of other proteins and post-translational modifications. Mature SGs interact with P-bodies, the autophagic and apoptotic systems.
Acknowledgements
This work was supported by grant awards to BW (NIH grants ES020395, NS066108, NS073679, NS060872 and the BrightFocus Foundation).
References
- 1.Adeli K: Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol Endocrinol Metab. 2011;301:E1051–1064. doi: 10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- 2.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, Maclea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013 doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 9.McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 10.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 13.Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P, 3rd, Good SK, Johnson BA, Herz J, Yu G. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda K, Marasa B, Martindale JL, Halushka MK, Gorospe M. Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging (Albany NY) 2009;1:681–698. doi: 10.18632/aging.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurth L. Versatility of RNA-Binding Proteins in Cancer. Comp Funct Genomics. 2012;2012:178525. doi: 10.1155/2012/178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goggin K, Beaudoin S, Grenier C, Brown AA, Roucou X. Prion protein aggresomes are poly(A)+ ribonucleoprotein complexes that induce a PKR-mediated deficient cell stress response. Biochim Biophys Acta. 2008;1783:479–491. doi: 10.1016/j.bbamcr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampers T, Friedhoff P, Biernat J, Mandelkow EM, Mandelkow E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996;399:344–349. doi: 10.1016/s0014-5793(96)01386-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selkoe DJ. Developing preventive therapies for chronic diseases: lessons learned from Alzheimer’s disease. Nutr Rev. 2007;65:S239–243. doi: 10.1111/j.1753-4887.2007.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 22.Beck AR, Medley QG, O’Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Vanderweyde T, Yu H, Varnum M, Liu-Yesucevitz L, Citro A, Ikezu T, Duff K, Wolozin B. Contrasting Pathology of Stress Granule Proteins TIA-1 and G3BP in Tauopathies. J Neurosci. 2012;32:8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoover BR, Reed MN, Su J, Penrod RD, Kotilinek LA, Grant MK, Pitstick R, Carlson GA, Lanier LM, Yuan LL, Ashe KH, Liao D. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, Barrett DA, Tsaytler P, Bertolotti A, Willis AE, Bushell M, Mallucci GR. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd J, Lee P, Feiler M, Zauur N, Liu M, Concannon J, Ebata A, Wolozin B, Glicksman M. A high content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J. Biomol. Screening. 2013 doi: 10.1177/1087057113501553. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]