Abstract
The LEAFY COTYLEDON2 (LEC2) gene plays critically important regulatory roles during both early and late embryonic development. Here, we report the identification of the LEC2 gene from the castor bean plant (Ricinus communis), and characterize the effects of its overexpression on gene regulation and lipid metabolism in transgenic Arabidopsis plants. LEC2 exists as a single-copy gene in castor bean, is expressed predominantly in embryos, and encodes a protein with a conserved B3 domain, but different N- and C-terminal domains to those found in LEC2 from Arabidopsis. Ectopic overexpression of LEC2 from castor bean under the control of the cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis plants induces the accumulation of transcripts that encodes five major transcription factors (the LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON1-LIKE (L1L), FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE 3 (ABI3) transcripts for seed maturation, and WRINKELED1 (WRI1) transcripts for fatty acid biosynthesis), as well as OLEOSIN transcripts for the formation of oil bodies in vegetative tissues. Transgenic Arabidopsis plants that express the LEC2 gene from castor bean show a range of dose-dependent morphological phenotypes and effects on the expression of LEC2-regulated genes during seedling establishment and vegetative growth. Expression of castor bean LEC2 in Arabidopsis increased the expression of fatty acid elongase 1 (FAE1) and induced the accumulation of triacylglycerols, especially those containing the seed-specific fatty acid, eicosenoic acid (20:1Δ11), in vegetative tissues.
Keywords: Castor bean, Eicosenoic acid, LEAFY COTYLEDON2, Seed maturation, Transcription factor, Triacylglycerol
Abbreviations: ABI3-VP1, abscisic acid-insensitive 3-viviparous 1; CaMV, cauliflower mosaic virus; cDNA, complementary DNA; DHA, docosahexaenoic acid; DIG, digoxigenin; FAE1, fatty acid elongase 1; GC, gas chromatography; ORF, open reading frame; RT-PCR, reverse transcription polymerase chain reaction; SSC, sodium chloride-sodium citrate; TAG, triacylglycerol; TF, transcription factor; TLC, thin-layer chromatography
Graphical abstract

Highlights
-
•
Castor bean LEC2 is single copy and shows seed-specific expression.
-
•
Over-expression of castor LEC2 induces genes involved in seed maturation in leaves.
-
•
Castor LEC2 induces the accumulation of triacylglycerols and 20:1 fatty acids in leaves.
-
•
Ectopic expression of castor LEC2 in Arabidopsis affects plant growth.
1. Introduction
Arabidopsis is a model plant of Brassicaceae that has been invaluable for the investigation of oil biosynthesis during seed development. After fertilization of the female gametophyte, initial embryo development is followed by seed maturation, which involves a range of developmental and metabolic changes during seed filling [1]. During seed filling, plant-specific B3 domain transcription factors (TFs) modulate the global transcriptional network that regulates the developmental cues and genes encoding enzymes required for the biosynthesis of fatty acids and the accumulation of triacylglycerols (TAGs) [2–4].
Stone et al. [5] identified LEAFY COTYLEDON2 (LEC2) as a central master regulator containing a B3 domain. This DNA binding region serves critical roles both during embryo development and seed maturation in Arabidopsis. LEC2 controls other master regulators, including LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON1-LIKE (LIL), which contains the B-domain originally identified in the yeast HEME-ACTIVATED PROTEIN 3 (HAP3), and the other B3 domain factors, FUSCA3 (FUS3) and ABSCISIC ACID INSENSITIVE 3 (ABI3). All five of these TFs contribute to a regulatory network required for the maintenance of suspensor morphology, specification of cotyledon identity, progression through the maturation phase for reserving storage proteins and TAGs, and suppression of premature germination [6,7]. LEC2 up-regulates LEC1, FUS3, and ABI3, and then ABI3 and FUS3 positively regulate themselves and each other during seed maturation [2,8].
LEC2 regulates the chemical composition of developing seeds during seed-filling [9]. Mature seeds of a loss-of-function lec2 mutant of Arabidopsis have 15% less protein and 30% less oil, while maintaining higher levels of starch and sucrose than wild-type [10]. Together with FUS3 and ABI3, LEC2 controls seed-specific genes involved in seed maturation [11–14]. LEC2 directly controls the transcription of genes that encode the hydroxysteroid dehydrogenase (HSD1) [14] and the seed-specific OLEOSIN isoforms identified in oil bodies from mature Arabidopsis seeds [15]. LEC2 binds directly to the promoter of HDS1 [14] and the RY element found in the promoters of OLEOSIN genes [15,16]. Baud et al. [17] demonstrated that the WRINKLED1 (WRI1) TF [18] is an essential target of LEC2 for normal regulation of fatty acid metabolism and the accumulation of TAGs in maturing Arabidopsis seeds. In addition to regulating cellular and metabolic processes during embryo maturation, LEC2 promotes the initiation of somatic embryo formation [9] by inducing the expression of genes involved in auxin biosynthesis [3,19]. Accordingly, a lec2-1 mutation impaired in vitro embryogenic responses [20]. Not only can overexpression of LEC2 compensate for the requirement for auxin and trigger somatic embryogenesis in vitro in the absence of exogenous auxin, but analysis of the efficiency in auxin-induced somatic embryogenesis also revealed a negative interaction between auxin treatment and LEC2 overexpression [21]. Microspore embryogenesis in Brassica napus is associated with the induction of LEC2 and reduced abundances of pollen-related transcripts [22,23].
The biotechnological potential of LEC2 has been evaluated by metabolic engineering strategies that aim to express in vegetative tissues those biosynthetic pathways that are usually restricted to seeds. Complex constructs that place co-expression of genes involved in the synthesis of docosahexaenoic acid (DHA) and LEC2 under the control of seed-specific promoters were transiently expressed in Nicotiana bethamiana leaf tissue to validate the potential for metabolic engineering of seed-specific synthetic pathways in vegetative tissue to reduce time-consuming analysis in seeds of transformed plants [24]. Inducible expression of Arabidopsis LEC2, which was regulated by the acetaldehyde-induced Alc gene, increased the abundance of total fatty acids to as much as 6.8% of the dry weight of the leaves of tobacco plants [25]. Ectopic expression of LEC2 in the COMATOSE2 (cts2) Arabidopsis mutant reduced capacity to catabolize fatty acids, resulted in the accumulation of TAGs in senescing tissue [26].
Castor bean (Ricinus communis L.), a member of the Euphorbiaceae family, is an important oil crop. It accumulates large amounts of TAGs, to levels that make up approximately 60% of the fresh weight of mature seeds. The TAGs in castor bean seeds contain 80%–90% ricinoleic acid (12-hydroxy-octadeca-9-enoic acid), which is an unusual fatty acid with numerous industrial applications, including its use in the production of high-quality lubricants, paints, coatings, soaps, medications for treatment of skin infection, and cosmetics [27]. Given its industrial relevance, elucidation of how fatty acid biosynthesis is regulated during the maturation of castor bean seeds would be useful for agro-industrial purposes if it identified ways to produce seeds that accumulate high level of TAGs. However, there is a limitation of using Arabidopsis as a model system for this application: whereas TAGs accumulate in the endosperm of the developing castor bean seed [28], they accumulate in the embryo of the developing Arabidopsis seed. This difference underscores the need to identify a conserved master regulator of TAG accumulation in the seeds of both species. Recently, genomic analyses of members of the Brassicaceae and major crop plants identified orthologs of master regulators of seed filling originally identified in Arabidopsis, such as LEC2, ABI3, and FUS3 [29,30]. Sequence-based quantitative trait loci (QTL) analysis has associated many rice and maize genes that encode products containing B3 domain genes with traits such as grain yield, seed weight and number, and protein content [29]. However, the proposed associations of B3-domain-containing master regulators in crops have yet to be established [4].
In this study, we identified the LEC2 gene from castor bean, which encodes a protein that contains a B3 domain and shows substantial homology to Arabidopsis LEC2 across the full length of its sequence. The castor bean genome contains a single copy of LEC2 which is expressed predominantly in embryo. Depending on the level of expression of castor bean LEC2, ectopic overexpression of the castor bean LEC2 gene in Arabidopsis is associated with phenotypes that range from embryo-like structures in developing seedlings to a bushy dwarf phenotype with reduced apical dominance. In addition, ectopic expression of castor bean LEC2 in Arabidopsis is able to up-regulate the expression of the most important B3-domain-containing TFs required for seed maturation and promote the accumulation of TAGs that contain eicosenoic acid (20:1Δ11), which are normally restricted to seeds, in vegetative tissue.
2. Materials and methods
2.1. Plant materials, growth conditions, and chemical staining
Seeds of the castor-bean variety IT196881 were obtained from the Germplasm Resources Center in the Rural Development Administration, Republic of Korea. Castor bean plants were grown in a greenhouse at temperatures between 18 and 28 °C. Wild-type and transgenic Arabidopsis thaliana (ecotype Columbia-0) were grown in potting soil in controlled-environment growth chambers at 22 °C with a 16-h light/8-h dark photoperiod. Transgenic plant lines transformed with a vector containing the coding sequence of the LEC2 gene from castor bean, which was controlled by the CaMV 35S promoter, were selected based on their resistance to BASTA. Experiments were conducted using members of the T1, T2, T3, and T4 generations of transgenic Arabidopsis seedlings. Seedlings were stained with a 0.1% (wt/vol) solution of Sudan Red 7B as described by Tsukagoshi et al. [31].
2.2. Cloning and vector construction
The LEC2 gene from castor bean was identified by searching the castor bean genomic database (http://castorbean.jcvi.org/index.php) using the BLAST algorithm and previously identified Arabidopsis LEC2 protein sequence [5]. Full-length cDNAs of castor bean LEC2 were amplified by RT-PCR from 1st cDNAs synthesized from RNAs of seeds using following primers: 5′-CACCATGGATTCTTTCCAGAATTCCCAGTTC-3′ (forward) and 5′-TCAAAGAGAAAAATTATACCTATTGAC-3′ (reverse). The amplicons were inserted into pENTR/D-TOPO (Invitrogen) to generate the entry clone pENTR/D-RcLEC2. Using the LR clonase reaction, the RcLEC2 cDNA in the entry vector was inserted downstream of the CaMV 35S promoter in the Gateway vector, pB2GW7 [32]. This plant expression vector was used for transformation into Arabidopsis plants, employing the floral dip method [33].
2.3. Sequence analysis
Nucleotide and deduced amino acid sequences were identified using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Prediction of the open reading frame (ORF) and estimation of the molecular weight of the deduced polypeptide of RcLEC2 were used with the EditSeq program (DNASTAR version 7.2.1). Sequences were compared using the MegAlign program, and default parameters were used to align multiple amino acid sequences with the ClustalW program (DNASTAR Inc., London, UK). A phylogenetic tree was constructed using the Neighbor-Joining algorithm included in the DNASTAR MegAlign (Ver. 7.2.1) software package.
2.4. DNA gel blot analysis
Genomic DNA was extracted from the leaves of castor bean plants by using the hexadecyltrimethylammonium bromide (CTAB) method [34]. Each 10 μg of genomic DNA was digested with the restriction endonucleases EcoRI, XbaI, and EcoRV. The digested genomic DNA was then fractionated by electrophoresis through a 0.8% agarose gel, and transferred to nylon N+ (GE Healthcare) membranes. The probe DNA was labeled with DIG using PCR DIG Probe Synthesis Kit (Roche) and used for hybridization according to the manufacturer's instructions. Hybridization was carried out in a DIG Easy Hyb buffer (Roche) at 42 °C for 24 h. The membranes were washed with 2× SSC, and hybridized signals were detected using a LAS-4000 luminescent image analyzer (FujiFilm). As shown in Fig. 3B, four different sizes of the DNA probes (probe 1 through probe 4) were synthesized using the primers described in Table S1 and the RcLEC2 cDNA as the template DNA.
Fig. 3.
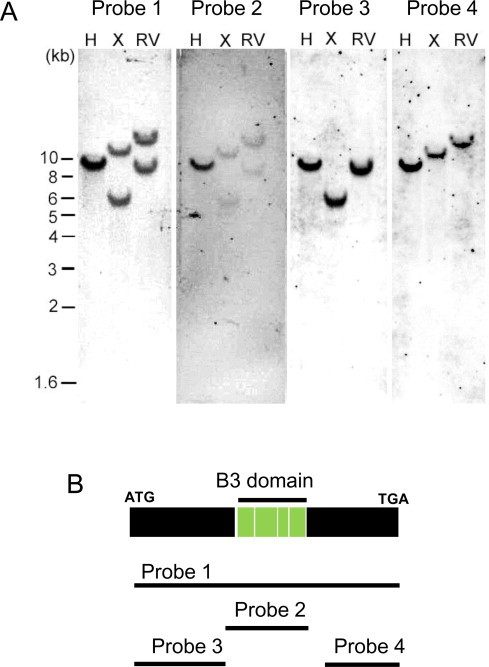
Southern blot analyses of the LEC2 gene from castor bean. (A) Results of hybridization with four LEC2 cDNA fragments hybridized to fragments of the castor bean genome digested with restriction endonucleases. H: HindIII, X: XbaI, RV: EcoRV. (B) RcLEC2 cDNA probes used for Southern blot analysis. Probes 1, 2, 3, and 4 contain the entire RcLEC2 ORF, B3 domain, N-terminal region of the RcLEC2 ORF, and C-terminal region of the RcLEC2 ORF, respectively.
2.5. qRT-PCR and RT-PCR
Total RNA was extracted from various tissues of castor bean and Arabidopsis plants using a plant RNA purification reagent (Invitrogen). A 2 μg of total RNAs extracted from each tissue were used for synthesizing the first-strand cDNAs by PrimeScript II first-strand cDNA synthesis kit (Takara) according to the manufacturer's instructions. RcLEC2 transcript levels were analyzed by quantitative real-time RT-PCR (qRT-PCR) with SYBR Green Premix Ex TaqII (Takara) in 96-well block using the CFX96 Real-Time PCR system (Bio-Rad Laboratories, http://www.bio-rad.com/), as specified by the manufacturer. A castor ACTIN2 gene (RcACT2), Accession No. AY360221 was included in the assays as an internal control to normalize variations in cDNA amounts used. Primers for qRT-PCR, 5′-TGGGACTGAAATTCAAACATGGC-3′ (forward) and 5′-TGGTGCTTTGGCTGTCTTGC-3′ (reverse) for RcLEC2 and 5′-ATTGGCGCTGAGAGATTCCG-3′ (forward) and 5′-T GCAAGGGCGGTGATCTCCTT-3′ (reverse) for RcACT2 were designed using Primer Express Software installed into the system and used for reaction.
RT-PCR reactions for transgenic Arabidopsis plants were carried out with Ex Taq polymerase (Takara) using 2 μl of cDNAs synthesized from total RNA extracted from rosette leaves of Arabidopsis. PCR conditions were as follows: 95 °C for 5 min, 30 cycles that each involved 95 °C for 10 s, 50 °C for 20 s, and 72 °C for 1 min; and 72 °C for an additional 5 min. The gene-specific primers mentioned in Table S2 were used to assess the expression of selected Arabidopsis genes (Figs. 6 and 7).
Fig. 6.
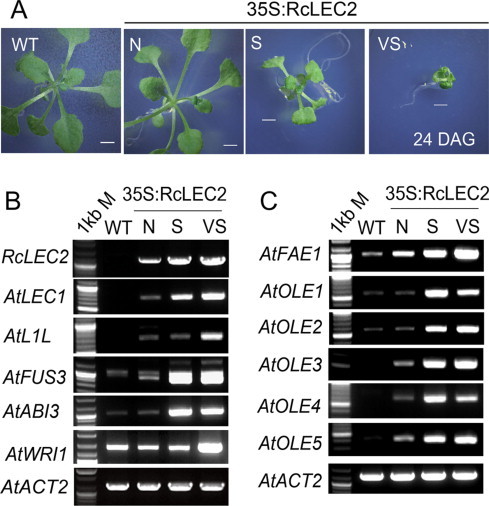
Effect of the expression level of RcLEC2 under the control of CaMV35S promoter in transgenic Arabidopsis plants (35S:RcLEC2) on the severities of their phenotypes 24 days after germination (DAG). (A) Comparison of growth rates between wild-type (WT) and 35S:RcLEC2 transgenic plants. N (normal size), S (small size), and VS (very small size) represent RcLEC2 transgenic plants with different rates of growth. Scale bars represent 1 mm. (B) Expression of RcLEC2 and five seed TF genes regulated by LEC2: LEC1 (At1g21970), L1L (At5g47670), FUS3 (At3g26790), ABI3 (At3g24650), and WRI1 (At3g54320) in rosette leaves of wild-type and transgenic 35S:RcLEC2 Arabidopsis plants. (C) Expression of FAE1 (At4g34520) and five OLEOSIN genes, OLE1 (At3g01570), OLE2 (At3g27660), OLE3 (At4g25140), OLE4 (At5g40420), and OLE5 (At5g51210) in rosette leaves of wild type and transgenic 35S:RcLEC2 Arabidopsis plants. Three biological replicates showed the same expression pattern. 1 kb M indicates 1-kb DNA size marker.
Fig. 7.
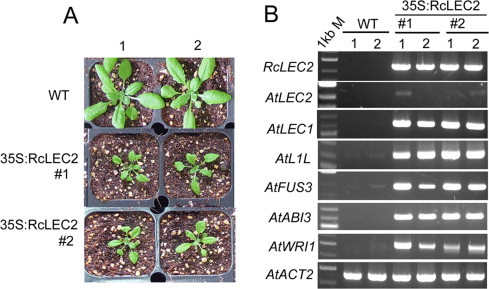
Representatives of 35S:RcLEC2 Arabidopsis transgenic plants and expression of seed master regulator TFs’ genes in rosette leaves. (A) Growth retardation phenotype in T4 generation of 35S:RcLEC2 Arabidopsis transgenic plants compared with wild-type plants on 3 weeks. (B) RT-PCR analyses from leaves of wild-type (WT) and transgenic 35S:RcLEC2 plants (T4 generation). Two different individual plants of WT and two different lines of 35S:RcLEC2 transgenic plants were used for RT-PCR. Expression of the RcLEC2 transgene does not strongly induce expression of Arabidopsis LEC2 (At1g28300) but does induce expression of Arabidopsis LEC1 (At1g21970), L1L (At5g47670), FUS3 (At3g26790), ABI3 (At3g24650), and WRI1 (At3g54320). Three biological replicates showed the same expression pattern. The Arabidopsis ACTIN2 gene, AtACT2, was used as a control. 1 kb M indicates 1-kb DNA size marker.
2.6. Fatty acid analysis
The fatty acid composition of plant tissues was determined by GC after transmethylation at 90 °C for 1 h in 0.5 ml of toluene and 1 ml of methanol containing 5% H2SO4 (v/v). Pentadecanoic acid (15:0) was added to each sample as an internal standard. Following transmethylation, 1 ml of aqueous 0.9% NaCl was added, and fatty acid methyl esters were recovered by three sequential extractions, each with 1 ml of hexane. Fatty acid methyl esters were then analyzed by GC (Shimadzu, Japan) on a 30 m × 0.32 mm (inner diameter) HP-FFAP column (Agilent Technologies), while increasing the oven temperature from 190 to 232 °C at 3 °C min−1.
2.7. Thin-layer chromatography
Leaves were homogenized in 1 ml of 2:1 (v/v) chloroform:methanol and then centrifuged at 20,000×g for 5 min. Supernatants were lyophilized with N2 gas, and the remaining materials were dissolved in 20 μl of chloroform. Lipid was spotted on TLC plates (silica gel G60 plates; EM Separations Technology) and the chromatograms were developed with hexane:diethylether:acetic acid [140:60:2 (by vol.)]. Lipid was visualized by staining with iodine. Relative amounts of TAGs were measured by GC analysis of the total amount of fatty acids extracted from the TAG spot obtained using TLC, and then by comparing the signal from simultaneously loaded glyceryl triheptadecanoate (Sigma T2151), which was used as an internal TAG standard.
3. Results
3.1. Cloning and sequence analysis of LEC2 from castor bean
To investigate the existence of a LEC2 gene(s) in the genome of castor bean, we searched the castor bean genomic database (http://castorbean.jcvi.org) with the BlastP algorithm (http://www.ncbi.nlm.nih.gov/BLAST/), using the protein sequence of Arabidopsis LEC2 (At1g28300) as the probe. The three castor protein sequences with the highest homology to Arabidopsis LEC2 were the B3-domain containing TFs 30190.m010868, 30132.m006860, and 30204.m001803. Of these, the hypothetical protein 30190.m010868 had the lowest P-value (<6.5e−51) and was most homologous to Arabidopsis LEC2, with 45% identity and 58% similarity across the entire amino acid sequence of Arabidopsis LEC2. The two other predicted gene products, 30132.m006860 and 30204.m001803, showed greater homology to Arabidopsis FUS3 (At3g26790) and ABI3 (At3g24650), respectively, than they did to LEC2 (Fig. 1A).
Fig. 1.
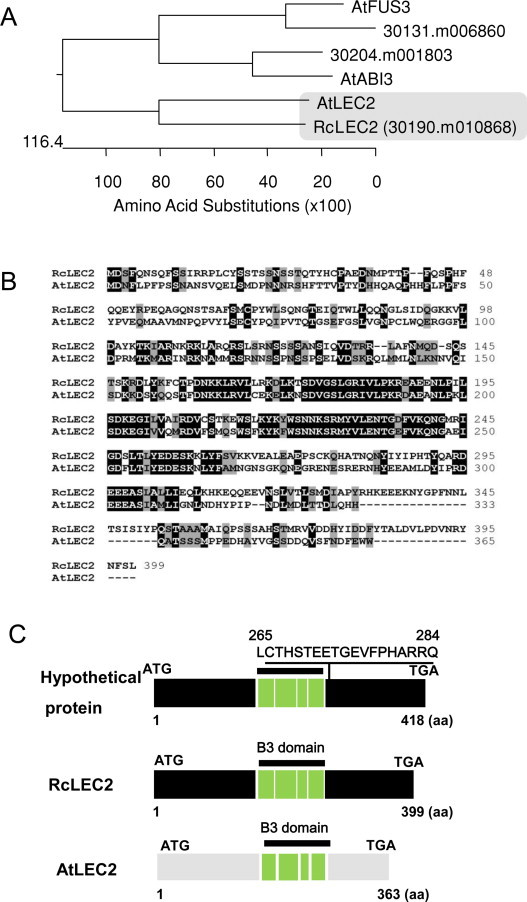
Identification and structure of LEC2 from castor bean (RcLEC2). (A) Phylogenic analysis of deduced amino acids of three Arabidopsis TFs that each contain a B3 domain (LEC2, FUS3, and ABI3) and their most homologous three genes from castor bean, 30190.m010868, 30132.m006860, and 30204.m001803, respectively. Amino acid sequences were aligned using the ClustalW method, and the tree was constructed using the program DNASTAR MegAlign (Ver. 7.2.1). (B) Amino acid alignment of LEC2 proteins from castor bean (RcLEC2) and Arabidopsis (AtLEC2). (C) Structures of cDNAs that encode LEC2 from castor bean and Arabidopsis. Hypothetical protein, XP_002509459, which is encoded by an mRNA transcribed from 30190.m010868 genomic DNA in castor bean. The hypothetical protein XP_002509459 contains a 19-aa sequence (LCTHSTEETGEVFPHARRQ, between aa residues 267 and 284) that this not found in RcLEC2.
The length of the 30190.m010868 gene, including the exons and introns, was predicted to span 2,701 bp (Fig. 2A upper panel). To isolate a full-length 30190.m010868 cDNA, N- and C-terminal primers were designed based on the genomic sequence, and the 1.2-kb cDNA was cloned by PCR amplification from total RNA prepared from developing seeds. Sequencing of the cDNA revealed it to encode a 399-amino acid (aa) protein with a predicted molecular mass of 45.8 kDa and an isolectric point of 7.9 (Fig. 1B and C, middle panel). Perfect alignment of continuous stretches of the cDNA with the nucleotide sequence of 30190.m010868 indicated that the gene comprises six exons and five introns (Fig. 2A, upper panel). Before this study, a hypothetical protein encoded by the 30190.m010868 gene had been registered in GenBank (Genbank: XP_002509459). The hypothetical protein comprises 418 aa, with 19 additional aa on the C-terminal side of the B3 domain, which are not present in the LEC2 protein predicted to be encoded by the cDNA isolated in the present study (Fig. 1C, upper panel). Comparisons of the DNA and protein sequences of 30190.m01868, Arabidopsis LEC2, and our cDNA isolated from castor suggest that the hypothetical protein XP_002509459 may be an alternative splicing product or simply the result of incorrect prediction of the transcript generated from the coding region of 30190.m010868 (Fig. 1C). Herein, we designate our cDNA, which encodes a 399-aa and is derived from the 30190.m010868 genomic sequence of castor bean, as RcLEC2. The RcLEC2 cDNA sequence was registered with NCBI as GenBank: KC146386.
Fig. 2.
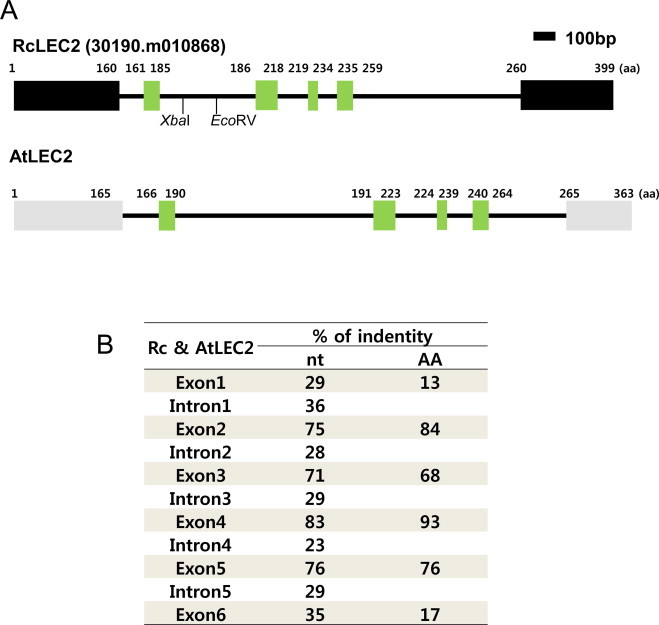
Comparison of LEC2 genes from castor bean and Arabidopsis. (A) Genomic structure of LEC2 genes from castor bean (RcLEC2) and Arabidopsis (AtLEC2). B3 DNA-binding domains are indicated by green rectangles. Sites recognized by the restriction endonucleases XbaI and EcoRV are located in the second intron of the LEC2 gene from castor bean. (B) Nucleotide (nt) and amino acid (aa) sequence identities of the exons and introns of RcLEC2 genes and AtLEC2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Comparison of the genomic structure of the RcLEC2 with that of Arabidopsis (Fig. 2A) showed a high degree of conservation in terms of the number of exons and the nucleotide lengths of each exon. However, the lengths of the second and fifth introns differed between the two genes, with the second intron of the castor bean sequence being shorter and the fifth intron longer than the corresponding introns in the Arabidopsis LEC2 sequence. As shown in Fig. 2B, the nucleotide sequence identities of the introns of the two genes are low, ranging from 23% (intron 4) to 36% (intron 1). The N-terminal (aa 1–160) and C-terminal (aa 260–399) exons of the LEC2 proteins from Arabidopsis and castor bean show low amino acid sequences identities. However, the B3-domains of the two proteins, encoded by the second through fifth exons (aa 161–259) show much higher levels of amino acids sequence identity (Fig. 2B).
3.2. The castor bean genome contains a single copy of LEC2
To investigate the copy number of the LEC2 gene in the castor bean genome, Southern blot analysis was performed after digesting genomic DNA from castor bean with three restriction endonucleases (HindIII, XbaI, and EcoRV). The digested genomic DNA was probed with four types of LEC2-specific probes labeled with digoxigenin (DIG) (Fig. 3B). Whereas the genomic DNA that encodes the LEC2 gene lacks HindIII recognition sites, single recognition sites for each of XbaI and EcoRV were found in the second intron of the LEC2 gene from castor bean (Fig. 2A). Probes 1 and 2, which respectively comprise full-length RcLEC2 cDNA and the part of the cDNA that encodes the B3 domain only (Fig. 3B), hybridized to a single fragment in HindIII-digested genomic DNA and to two fragments in XbaI- and EcoRV-digested DNA, both of which showed an identical hybridization pattern. Probes 3 and 4, which comprised the N- and C-terminal regions of RcLEC2 (Fig. 3B), respectively, hybridized to single fragment in each of HindIII-, XbaI-, and EcoRV-digested samples of genomic DNA. Probes 3 and 4 hybridized to each of the same two fragments that hybridize to probes 1 and 2 in XbaI- and EcoRV-digested genomic DNA (Fig. 3A). Southern blot analysis clearly indicates that castor bean contains a single LEC2 gene.
3.3. Embryo-dominant expression of LEC2 from castor bean
The role of LEC2 as a master regulator of embryogenesis and seed maturation is well documented [4,9]. Arabidopsis LEC2 RNA accumulates predominantly in early- and middle-stage siliques, and either not at all or occasionally at very low levels in vegetative organs [5]. In order to test whether the RcLEC2 gene shows the seed-specific expression pattern as observed in Arabidopsis, qRT-PCR analyses were carried out with total RNA extracted from leaves, stems, flower buds, female flowers, male flowers, developing seeds, and seedlings of castor bean. Of these tissues, transcripts of the LEC2 gene from castor bean were predominantly detected in developing seeds, with very low levels in female flowers and flower buds, and none in leaves, stems, male flowers, and seedlings (Fig. 4A). Significant transcript levels of the RcLEC2 were detected in both embryos and endosperms in developing seeds, but its level was 10-fold higher in embryos than endosperms (Fig. 4B). These results suggest that the RcLEC2 gene in castor bean shows the same expression patterns as that seen for Arabidopsis LEC2, and that the function of the RcLEC2 in castor bean might be identical to that of Arabidopsis during seed development.
Fig. 4.
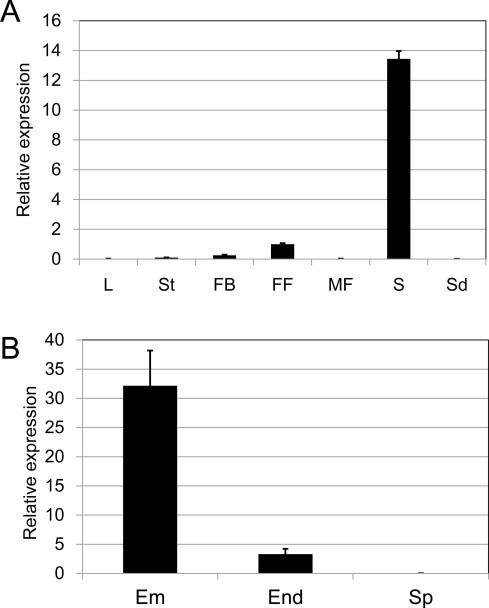
Expression patterns of the LEC2 gene in different tissues of castor bean, determined using qRT-PCR. (A) Seven different tissues were used for analysis (L: leaf, St: stem, FB: flower bud, FF: female flower, MF: male flower, S: developing seed, Sd: seedling). (B) Embryo (Em), endosperm (End), and seed pot (Sp) were dissected from developing seeds. RcLEC2 transcript level was normalized by expression level of castor RcACT2 gene. Three biological replicates showed the same expression profile. Data were averaged with biological triplicates. Data are means ± SD.
3.4. Ectopic overexpression of RcLEC2 in Arabidopsis affects plant development and growth
Ectopic overexpression of Arabidopsis LEC2 induces somatic embryogenesis and affects plant growth by enlarging pistils and root cells [5]. To assess the function of LEC2 from castor bean, the RcLEC2 cDNA was placed under the transcriptional control of the CaMV 35S promoter in Arabidopsis. Thirty-two T1 transgenic plants grown in soil were selected on the basis of their resistance to BASTA. T1 transgenic plants that expressed the RcLEC2 gene exhibited a range of morphological phenotypes. Most of the transgenic plants displayed reduced plant growth rate. The extent of suppression in plant growth was positively correlated with the level of expression of RcLEC2 (Fig. 5A).
Fig. 5.

Phenotypes associated with ectopic overexpression of castor bean LEC2 in Arabidopsis transgenic plants. (A) Series of 8-week-old transgenic 35S:RcLEC2 plants (T1 generation) and levels of RcLEC2 transgene compared with that of wild-type plants (WT). Levels of RcACT2 transcript were used to normalize loading. A bushy phenotype of 12-week-old T1 transgenic plant #2 is shown in the inset. (B) Abnormal seedling phenotype in T2 transgenic plants germinated and grown in solid MS medium. The broader-than-normal hypocotyls of 35S:RcLEC2 seedlings stained strongly with Sudan Red 7B, indicating the accumulation of higher levels of oil than are found in wild-type (WT) seedlings. Scale bar in (B) represents 1 mm. (C) Phenotypes of T2 transgenic plants compared with WT plant. (D) Phenotypes of T3 transgenic plants compared with WT plants.
We did not observe the formation of somatic embryo tissues when RcLEC2 was overexpressed in Arabidopsis, as was previously reported for the overexpression of Arabidopsis LEC2 [5]. However, tiny and the bushy phenotypes of RcLEC2 transgenic Arabidopsis were consistent with previous study [5] (Fig. 5A, inset). Given the diversity of phenotypes caused by ectopic overexpression of LEC2 from castor bean and no observation of somatic embryo formation in these plants during growth in soil, we examined the phenotypes of young T2 seedlings grown in solid MS medium [35]. T2 seeds harvested from a tiny and bushy transgenic plant (T1 generation) were germinated in MS medium, and the development of the seedlings was investigated. Some of the T2 siblings displayed abnormal seedling development, exhibiting growth arrest at the postembryonic stage, swollen hypocotyls, and a failure to develop roots and leaves. The swollen hypocotyls of these transgenic seedlings stained strongly with Sudan Red 7B, indicating the accumulation of higher levels of oil than wild-type seedlings of the same age [31] (Fig. 5B). Ectopic overexpression of LEC2 from castor bean during early seedling development leads to the arrest of postembryonic development and the maintenance of embryo-like features during subsequent development (Fig. 5B). Although most T2 seedlings displayed normal growth and development, transgenic plants were smaller than wild-type plants of comparable age (Fig. 5C). Phenotypes associated with ectopic overexpression of LEC2 from castor bean were transferred to the T3 generation, with similar growth defects at different stages of development to those seen in members of the T2 generation (Fig. 5D).
Overall, the morphological phenotypes associated with the ectopic overexpression RcLEC2 in Arabidopsis were similar to those associated with the ectopic overexpression of Arabidopsis LEC2 in Arabidopsis [5]. In some case, the RcLEC2 protein affected the establishment of Arabidopsis seedlings by increasing the girth of the hypocotyls, and/or delaying and preventing the development of leaves and roots (Fig. 5B). These features resemble those associated with ectopic overexpression of regulators of seed maturation, such as Arabidopsis LEC2 [5], LEC1 [38], and FUS3 [39], as well as null mutation of the PKL gene [37].
3.5. Ectopic expression of LEC2 from castor bean induces the expression of genes that regulate seed maturation and encode oleosin isoforms in vegetative tissues
To investigate the correlation between the retardation of plant growth and level of ectopic expression of RcLEC2 in transgenic Arabidopsis plants, expression of the LEC2 transgene and some of its targets were analyzed in 24-day-old T3 transgenic plants grown in MS medium [2]. T3 transgenic plants were divided into three types depended on their sizes compared with those of wild-type plants: transgenics classified as having a normal size (N), small size (S), or very small size (VS) (Fig. 6A). Interestingly, it seems that the severity of the phenotype depends on the levels of expression of the RcLEC2 gene and its downstream TF genes. Levels of RcLEC2 transcripts in rosette leaves of VS and S transgenic plants were higher than those in N transgenic plants (Fig. 6B). Arabidopsis LEC2 is a master regulator of seed development, which regulates the induction of numerous TFs downstream of pathways that control seedling growth and development [3,5]. In order to study whether 35S:RcLEC2 can regulate downstream TF's in a manner that resembles that of Arabidopsis LEC2, the transcript levels of five TF genes regulated by LEC2 were analyzed. Ectopic expression of RcLEC2 induced expression of the Arabidopsis LEC1, L1L, FUS3, and ABI3 genes, as well as WRI1, a TF that regulates fatty acid biosynthesis in seeds (Fig. 6B). The abundance of these five TFs’ transcripts was positively correlated with the level of the RcLEC2 transcript (Fig. 6B).
Expression of LEC2 from castor bean also induced seed-specific accumulation of the Arabidopsis FAE1 gene [40] and five seed-specific OLEOSIN genes [15] (Fig. 6C). The severity of the phenotypes of the transgenic plants were positively correlated with the abundances of transcripts that encode LEC2 from castor bean and the seed-expressed master TFs LEC1, L1L, FUS3, and ABI3, all of which act downstream of LEC2. Transgenic plants with a normal (N) phenotype had relatively low levels of transcripts encoded by LEC2 and the genes that act downstream of LEC2. However, levels of all five transcripts that encoded downstream TF genes were substantially higher in plants with small (S) and very small (VS) phenotypes than in wild-type plants or plants with normal (N) phenotypes (Fig. 6B). Levels of FAE1 transcripts, which encode an enzyme that produces seed-specific unusual fatty acids found in Arabidopsis TAGs, were much higher in VS plants than in N or S plants (Fig. 6C). In contrast to the abundance of FAE1 transcripts, levels of OLEOSIN transcripts, which are required for the formation of oil bodies, were not always substantially higher in VS plants than in S plants (Fig. 6C).
RT-PCR analysis to detect expression of seed-expressed TFs in rosette leaves of T4 RcLEC2 transgenic Arabidopsis plants (Fig. 7A) shows that LEC2 from castor bean did not highly induce the expression of endogenous Arabidopsis LEC2 than those of downstream four seed master regulators (LEC1, L1L, FUS3, ABI3) and a fatty acid TF(WRI1) (Fig. 7B). This finding suggests that LEC2 may not be subject to strong self-regulation.
3.6. Arabidopsis plants that ectopically overexpress a LEC2 gene from castor bean accumulate TAGs and synthesize an unusual fatty acid in leaves
Given that ectopic overexpression of RcLEC2 in Arabidopsis increased the levels of transcripts that encode three major classes of gene products involved in seed oil accumulation—the WRI1 TF that controls fatty acid synthesis, five OLEOSIN proteins found in oil bodies, and the seed-specific FAE1 protein (Figs. 6 and 7)—we checked whether LEC2 from castor bean can induce TAG accumulation in leaves. Analysis of lipids isolated from the leaves of wild-type and homozygous T4 plants using TLC revealed substantially more TAGs in the leaves of RcLEC2 transgenic plants than TAGs found in wild-type plants (Fig. 8A). We next used gas chromatography (GC) to measure the level of TAGs identified on TLC separation from leaves of comparably aged wild-type plants and RcLEC2 transgenic plants, using glyceryl triheptadecanoate as a TAG standard. Levels of TAGs in the leaves of RcLEC2 transgenic Arabidopsis plants and wild-type Arabidopsis plants were 283 and 102 ng mg−1 fresh weight, respectively (Fig. 8B). This indicates that ectopic overexpression of RcLEC2 increases TAG abundance to levels approximately 2.7-fold higher than the level of TAG in vegetative tissues of wild-type plants.
Fig. 8.
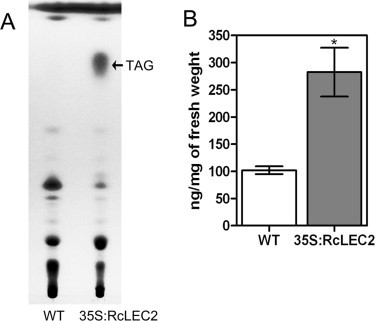
The levels of TAGs in wild-type and 35S:RcLEC2 transgenic Arabidopsis plants. (A) Thin-layer chromatography (TLC) analysis of lipids isolated from leaves of wild-type and 35S:RcLEC2 transgenic Arabidopsis plants (T4 generation). (B) Quantification of TAGs in wild-type and 35S:RcLEC2 T4 transgenic plants. The relative amounts of TAGs represent the sum of total fatty acids extracted from the TAG spot isolated by TLC compared with simultaneous loading of glyceryl triheptadecanoate as a TAG standard during analysis by gas chromatography (GC). Experiments were carried out in triplicate, and data are means ± SD. Asterisks denote statistical differences compared with wild-type plants (t-test, *P < 0.05).
The fatty acid composition of leaves from RcLEC2 transgenic Arabidopsis plants also differed from that of wild-type plants (Table 1). Ectopic overexpression of RcLEC2 in Arabidopsis decreased the levels of certain 16-carbon fatty acids (16:0, 16:1, and 16:3) and increased the amounts of certain 18-carbon fatty acids (18:1 and 18:3, but not 18:2). In addition, the synthesis eicosenoic acid (20:1Δ11), which is not found in wild-type, was induced in leaves of RcLEC2 transgenic Arabidopsis plants. The accumulation of eicosenoic acid to levels equivalent to 2% of the total level of fatty acids coincided with increased FAE1 transcript level driven by ectopic overexpression of RcLEC2 (Table 1, Fig. 6C).
Table 1.
Fatty acid profiles and fatty acid methyl ester (FAME) contents in leaves of wild type plants and 35S:RcLEC2 transgenic plants (T4 generation). Experiments were carried out in triplicate and fatty acid proportions are expressed as mole percent. Data are means ± SD. Asterisk denotes a significant statistical difference compared with wild-type plants (2 way ANOVA, *P < 0.05).
| Fatty acids | 16:0 (mol %) | 16:1t (mol %) | 16:3 (mol %) | 18:0 (mol %) | 18:1 (mol %) | 18:2 (mol %) | 18:3 (mol %) | 20:1 (mol %) |
|---|---|---|---|---|---|---|---|---|
| Wild type Col-0 | 24.8 ± 2.8 | 4.0 ± 0.5 | 8.2 ± 1.2 | 1.9 ± 0.0 | 4.8 ± 0.7 | 18.4 ± 0.1 | 37.7 ± 2.3 | 0.0 ± 0.0 |
| 35S:RcLEC2 | 21.9 ± 2.5 | 2.4 ± 0.5 | 7.6 ± 1.6 | 1.7 ± 0.2 | 6.3 ± 0.7 | 15.0 ± 0.1 | 43.1 ± 1.9* | 2.0 ± 0.7 |
4. Discussion
We searched the genome database for castor bean to identify a putative LEC2 gene from castor bean, and confirmed its identity by analyzing the effects of its ectopic overexpression in Arabidopsis. LEC2 gene has not been identified from any crop plants after first it identified from Arabidopsis [5]. Recently, Peng and Weselake [30] used an in silico genome-wide domain analysis of castor bean and estimated the presence of 21 castor bean proteins with B3 domains of the members of the ABI3-VP1 family, ABI3, FUS3, and LEC2. Among 21 members of the ABI3-VP1 gene family, seven genes each encode a product with a centrally located B3 domain. In this report, we found that castor bean contains a single LEC2 by Southern blot analysis (Fig. 3A).
Cloning of RcLEC2 cDNA enabled us to elucidate the structures of the LEC2 gene from castor bean, which, like LEC2 from Arabidopsis, contains six exons and five introns (Fig. 2A). A centrally located B3 domain is highly conserved in the LEC2 proteins from castor bean and Arabidopsis, whereas the N- and C-terminal regions differ substantially in their amino acid sequences. The 399-amino-acid LEC2 from castor bean has a longer C-terminal region than that found in Arabidopsis LEC2 (Figs. 1C and 2A). qRT-PCR analysis in various tissues indicates that LEC2 from castor bean is expressed predominantly in developing seeds and in embryo (Fig. 4). The conservation of B3 domain structures and similar expression patterns suggests that the LEC2 proteins from castor bean and Arabidopsis serve similar functions in seed development [3,5], despite the low overall similarity of the two protein sequences.
Ectopic overexpression of LEC2 from castor bean in Arabidopsis showed that various phenotypes correlate with the levels of LEC2 transcripts (Fig. 5A) and transcripts that encode TFs that act downstream of LEC2, such as LEC1, L1L, FUS3, and ABI3 (Fig. 6B). Arabidopsis LEC2 gene was not highly induced compared to other downstream seed master regulator TFs by overexpression of RcLEC2 in Arabidopsis leaves indicating that LEC2 may not be subject to strong self-regulation (Fig. 7B). We could not observe the induction of somatic embryogenesis in vegetative tissues, as was seen when Arabidopsis LEC2 was ectopically overexpressed in Arabidopsis [5]. However, ectopic overexpression of LEC2 from castor bean affected seedling growth, most notably by producing embryo-like structures with swollen hypocotyls and an inability to undergo normal shoot and root development (Fig. 5B). This similar phenotype is observed in seedlings of a double knock-out Arabidopsis mutant that lacks the HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2 (HSI2) and HIS2-Like 1(HSL1) genes. These genes encode proteins that each contain a B3 domain, have elevated levels of expression of the seed master regulator genes LEC1, LEC2, FUS3, WRI1, and OleS3 (which encodes an oleosin), and accumulate TAGs in hypocotyls [31]. PICKLE (PKL) is a key regulator responsible for maintaining postembryonic development by suppressing expression of embryonic master TFs, such as LEC1 [36,37]. A null mutation of PKL allows the expression of LEC1 in postembryonic tissues and confers embryo-like properties to primary roots, which differ from the primary roots of wild-type plants insofar as they are more swollen and contain oil bodies [36,37].
Expression of LEC2 from castor bean also induced the accumulation of TAGs and changed the fatty acid composition in vegetative tissues (Fig. 8 and Table 1). The accumulation of TAGs in the transgenic plants coincided with increased levels of the WRI1 TF and five seed-specific OLEOSINs (Fig. 6C). The unusual seed-specific 20:1Δ11 fatty acids identified in vegetative tissue might have been produced by the induction of FAE1 expression (Fig. 6C). These data indicate that expression of LEC2 from castor bean triggered a seed development program that caused the accumulation of TAGs in vegetative tissues of Arabidopsis. Growth defects caused by ectopic overexpression of LEC2 from castor bean in Arabidopsis may result from the initiation of a LEC2-mediated program for seed development in vegetative tissues and/or the production of the unusual fatty acid, eicosenoic acid (20:1Δ11), in vegetative tissue. Epidermal expression of seed-specific FAE1 changed the composition of very-long-chain fatty acids (VLCFAs) in epidermal lipids, and caused trichome cells to die as a consequence of membrane damage [41].
Castor bean is an important oil crop that accumulates large amounts of TAG and ricinoleic acid, an unusual fatty acid normally found exclusively in seeds. Identification of the LEC2 gene from castor bean will help to identify how fatty acid and TAG biosynthesis are coordinated during seed development in castor bean plants.
Acknowledgements
This study was conducted with the support of the Research Program for Agricultural Science & Technology Development (Project No. PJ008556), the National Academy of Agricultural Science, the Rural Development Administration, and the Cooperative Research Program entitled ‘The Next-Generation BioGreen 21 Program’ (SSAC, Project No. PJ009484) of the Rural Development Administration, Republic of Korea.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.11.003.
Appendix. Supplementary materials
This document contains Supplementary Tables S1 and S2.
References
- 1.Baud S., Lepiniec L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 2009;47:448–455. doi: 10.1016/j.plaphy.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Santos-Mendoza M., Dubreucq B., Miquel M., Caboche M., Lepiniec L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005;579:4666–4670. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 3.Stone S.L., Braybrook S.A., Paula S.L., Kwong L.W., Meuser J., Pelletier J., Hsieh T.F., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3251–3256. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos-Mendoza M., Dubreucq B., Baud S., Parcy F., Caboche M., Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- 5.Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki M., Wang H.H., McCarty D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007;143:902–911. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Guo J., Lambert K.N., Lin Y. Developmental control of Arabidopsis seed oil biosynthesis. Planta. 2007;226:773–783. doi: 10.1007/s00425-007-0524-0. [DOI] [PubMed] [Google Scholar]
- 8.To A., Valon C., Savino G., Guilleminot J., Devic M., Giraudat J., Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braybrook S.A., Stone S.L., Park S., Bui A.Q., Le B.H., Fischer R.L., Goldberg R.B., Harada J.J. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angeles-Núñez J.G., Tiessen A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 2011;168:1891–1900. doi: 10.1016/j.jplph.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Roschzttardtz H., Fuentes I., Vásquez M., Corvalán C., León G., Gómez I., Araya A., Holuigue L., Vicente-Carbajosa J., Jordana X. A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol. 2009;150:84–95. doi: 10.1104/pp.109.136531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagaya Y., Okuda R., Ban A., Toyoshima R., Tsutsumida K., Usui H., Yamamoto A., Hattori T. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 2005;46:300–321. doi: 10.1093/pcp/pci031. [DOI] [PubMed] [Google Scholar]
- 13.Wang F., Perry S. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013;161:1251–1264. doi: 10.1104/pp.112.212282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baud S., Dichow N.R., Kelemen Z., d’Andréa S., To A., Berger N., Canonge M., Kronenberger J., Viterbo D., Dubreucq B., Lepiniec L., Chardot T., Miquel M. Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol. 2009;50:1463–1478. doi: 10.1093/pcp/pcp092. [DOI] [PubMed] [Google Scholar]
- 15.Kim H.U., Hsieh K., Ratnayake C., Huang A.H. A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 2002;277:22677–22684. doi: 10.1074/jbc.M109298200. [DOI] [PubMed] [Google Scholar]
- 16.Che N., Yang Y., Li Y., Wang L., Huang P., Gao Y., An C. Efficient LEC2 activation of OLEOSIN expression requires two neighboring RY elements on its promoter. Sci. China C. Life Sci. 2009;52:854–863. doi: 10.1007/s11427-009-0119-z. [DOI] [PubMed] [Google Scholar]
- 17.Baud S., Mendoza M.S., To A., Harscoët E., Lepiniec L., Dubreucq B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007;50:825–838. doi: 10.1111/j.1365-313X.2007.03092.x. [DOI] [PubMed] [Google Scholar]
- 18.Cernac A., Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004;40:575–585. doi: 10.1111/j.1365-313X.2004.02235.x. [DOI] [PubMed] [Google Scholar]
- 19.Braybrook S.A., Harada J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008;13:624–630. doi: 10.1016/j.tplants.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Gaj M.D., Zhang S., Harada J.J., Lemaux P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta. 2005;222:977–988. doi: 10.1007/s00425-005-0041-y. [DOI] [PubMed] [Google Scholar]
- 21.Ledwoń A., Gaj M.D. LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep. 2009;28:1677–1688. doi: 10.1007/s00299-009-0767-2. [DOI] [PubMed] [Google Scholar]
- 22.Malik M.R., Wang F., Dirpaul J.M., Zhou N., Polowick P.L., Ferrie A.M., Krochko J.E. Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol. 2007;144:134–154. doi: 10.1104/pp.106.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik M.R., Wang F., Dirpaul J.M., Zhou N., Hammerlindl J., Keller W., Abrams S.R., Ferrie A.M., Krochko J.E. Isolation of an embryogenic line from non-embryogenic Brassica napus cv. Westar through microspore embryogenesis. J. Exp. Bot. 2008;59:2857–2873. doi: 10.1093/jxb/ern149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrie J.R., Shrestha P., Liu Q., Mansour M.P., Wood C.C., Zhou X.R., Nichols P.D., Green A.G., Singh S.P. Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods. 2010;6:8. doi: 10.1186/1746-4811-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrianov V., Borisjuk N., Pogrebnyak N., Brinker A., Dixon J., Spitsin S., Flynn J., Matyszczuk P., Andryszak K., Laurelli M., Golovkin M., Koprowski H. Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 2010;8:277–287. doi: 10.1111/j.1467-7652.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- 26.Slocombe S.P., Cornah J., Pinfield-Wells H., Soady K., Zhang Q., Gilday A., Dyer J.M., Graham I.A. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 2009;7:694–703. doi: 10.1111/j.1467-7652.2009.00435.x. [DOI] [PubMed] [Google Scholar]
- 27.Caupin H.J. Products from castor oil: past, present, and future. In: Gunstone F.D., Padley F.B., editors. Lipid Technologies and Applications. Marcel Dekker; New York: 1997. pp. 787–795. [Google Scholar]
- 28.Robert L.M., Lord J.M. Protein biosynthetic capacity in the endosperm tissues of ripening castor bean seed. Planta. 1981;152:420–427. doi: 10.1007/BF00385358. [DOI] [PubMed] [Google Scholar]
- 29.Verdier J., Thompson R.D. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008;49:1263–1271. doi: 10.1093/pcp/pcn116. [DOI] [PubMed] [Google Scholar]
- 30.Peng F.Y., Weselake R.J. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 2013;126:1305–1329. doi: 10.1007/s00122-013-2054-4. [DOI] [PubMed] [Google Scholar]
- 31.Tsukagoshi H., Morikami A., Nakamura K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2543–2547. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimi M., Inze D., Depicker A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 33.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Dellaporta S.L., Wood J., Hicks J.B. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1983;1:19–21. [Google Scholar]
- 35.Murashige T., Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 36.Ogas J., Cheng J.C., Sung Z.R., Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- 37.Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junker A., Mönke G., Rutten T., Keilwagen J., Seifert M., Thi T.M., Renou J.P., Balzergue S., Viehöver P., Hähnel U., Ludwig-Müller J., Altschmied L., Conrad U., Weisshaar B., Bäumlein H. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012;71:427–442. doi: 10.1111/j.1365-313X.2012.04999.x. [DOI] [PubMed] [Google Scholar]
- 39.Lumba S., Tsuchiya Y., Delmas F., Hezky J., Provart N.J., Shi Lu Q., McCourt P., Gazzarrini S. The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol. 2012;10:8. doi: 10.1186/1741-7007-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossak M., Smith M., Kunst L. Expression of the FAE1 gene, FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 2001;46:717–725. doi: 10.1023/a:1011603923889. [DOI] [PubMed] [Google Scholar]
- 41.Reina-Pinto J.J., Voisin D., Kurdyukov S., Faust A., Haslam R.P., Michaelson L.V., Efremova N., Franke B., Schreiber L., Napier J.A., Yephremov A. Misexpression of FATTY ACID ELONGATION1 in the Arabidopsis epidermis induces cell death and suggests a critical role for phospholipase A2 in this process. Plant Cell. 2009;21:1252–1272. doi: 10.1105/tpc.109.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains Supplementary Tables S1 and S2.


