Abstract
The mouse germ-line presents a unique opportunity to study epigenetic reprogramming in vivo1. Recently we showed that genome-wide active DNA demethylation in primordial germ cells (PGCs) is linked to changes in nuclear architecture and extensive loss of histone modifications brought about by widespread histone replacement2. Notably, this chromatin remodelling follows the onset of genome-wide DNA demethylation, which raises a possibility that DNA demethylation may be linked to a DNA repair process2. Here we show the activation of components of the base excision repair (BER) pathway and the presence of single-strand DNA (ssDNA) breaks at the time of genome-wide DNA demethylation in PGCs. We found high levels of expression of critical BER components as well as chromatin-bound XRCC1 together with nuclear poly-ADP-ribosylation (PAR), specifically at the time when PGCs are undergoing DNA demethylation. A similar wave of genome-wide DNA demethylation occurs in the zygote affecting only the paternal genome3-5 where we observed a strikingly similar activation of BER components. Notably, maternally inherited Stella promotes this epigenetic asymmetry, since in zygotes lacking this protein, DNA demethylation is detected in both pronuclei. Crucially, zygotes lacking Stella exhibit aberrant targeting of active BER to both pronuclei. Finally we demonstrate that small molecule inhibitors of diverse BER components administered during in vitro fertilisation interfere with the progress of DNA demethylation.
Our combined observations demonstrate that DNA repair through BER represents a core component of genome-wide DNA demethylation and provides a vital mechanistic link to the extensive chromatin remodelling in developing PGCs2.
The establishment of the founder population of PGCs at embryonic day 7.25 (E7.25) is accompanied by the initiation of epigenetic modifications and re-expression of pluripotency related genes1. At E10.5, PGCs enter into the genital ridges and exhibit widespread epigenetic reprogramming at E11.5, including erasure of genomic imprints, genome-wide DNA demethylation and large-scale chromatin remodelling2,6. This is followed by meiotic and mitotic arrest in female and male embryos, respectively (Supplementary Fig. 1). We recently showed that extensive chromatin remodelling occurs in PGCs at E11.5, which follows the onset of genome-wide DNA demethylation. This lead us to propose that the chromatin changes could be a consequence of DNA repair process that might be linked to the genome-wide DNA demethylation process2.
DNA repair driven DNA demethylation would involve replacement of a methylcytosine (5mC) containing nucleotide by an unmethylated cytosine7-9. Importantly, epigenetic changes in PGCs occur in the G2 phase of the cell cycle, and are thus independent of DNA replication2. Hence, the likely mechanisms for the replacement of 5mC are the nucleotide excision repair (NER), and base excision repair (BER) pathways. We first examined NER by looking for the expression of ERCC1 (Excision Repair Cross-complementing rodent repair deficiency, Complementation group 1) and XPA (Xeroderma pigmentosum, complementation group A), which are core NER components. We only detected a weak signal for ERCC1 in PGCs and neighbouring somatic cells at the time of epigenetic reprogramming, while we saw a dose dependent increase and nuclear localisation of ERCC1 in our control UV-irradiated primary embryonic fibroblasts (PEFs; Supplementary Fig. 2b). Although we detected XPA in both somatic cells and PGCs (Supplementary Fig. 3a), this factor should be chromatin bound and insoluble upon activation of NER10. Pre-extraction of soluble proteins prior to fixation resulted in a complete loss of XPA signal from both PGCs and neighbouring somatic cells (Supplementary Fig. 3b), indicating that XPA and consequently the NER pathway are largely inactive during the reprogramming process in germ cells. Next, we examined XRCC1 (X-ray Repair Complementing defective repair in Chinese hamster cells 1) expression, which is a core component of the BER pathway11. Indeed, we detected a strong signal in nuclei of PGCs between E10.5-E12.5 (Supplementary Fig. 4). Notably, we also detected the presence of high amounts of PARP1 (Poly (ADP-ribose) polymerase family, member 1) and APE1 (Apurinic/apyrimidinic endonuclease) in PGCs compared to their levels in the surrounding somatic cells (Supplementary Fig. 4).
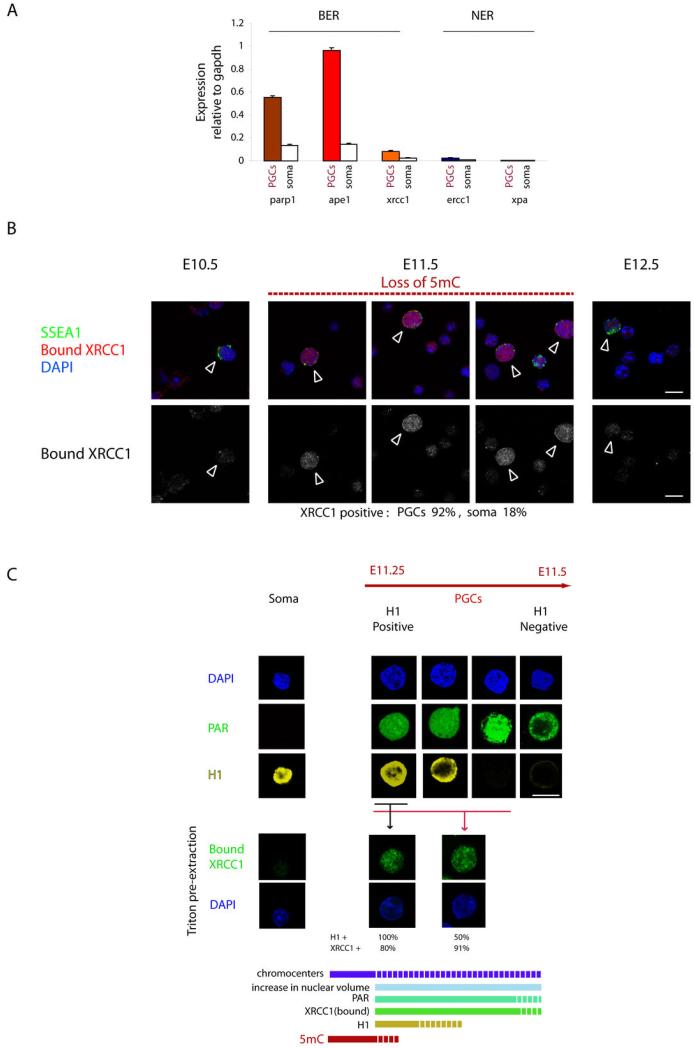
To obtain a quantitative measure of expression of BER versus NER components in PGCs, we performed qPCR analysis on their expression. We observed a striking up regulation of BER component transcripts, parp1, ape1 and xrcc1 in E11.5 PGCs, which was not seen in the neighbouring somatic cells. Also, the levels of expression detected far exceed those observed in embryonic stem cells (ES), suggesting their likely importance in germ cell reprogramming. By contrast, we observed little expression of NER components, ercc1 and xpa in PGCs. (Fig. 1a and Supplementary Fig. 5). The observations on the transcription of BER components therefore complement our immunofluorescence analysis described above. Thus, the detection of large amounts of XRCC1, PARP1 and APE1, which are all key components of the BER apparatus11, suggested that BER might be involved in DNA demethylation in PGCs
Figure 1. Activation of BER during DNA demethylation in gonadal PGCs.
a. qPCR analysis of PGC and neighbouring somatic cells at E11.5; note high expression of BER but not NER factors specifically in PGCs b. Chromatin bound XRCC1 detected by immunofluorescence in gonadal PGCs (arrowheads) at E11.5 at the time of DNA demethylation.. Single cell suspensions of embryonic genital ridges were pre-extracted with detergent prior to fixation (see Materials and Methods). c. Kinetics of BER activation with respect to chromatin changes in PGCs compared with neighbouring somatic cells at E11.25-E11.5 (Samples were prepared as described: see Supplementary Fig.1c). Immunofluorescence analysis of single cell suspension (upper panel), or after pre-extraction to detect chromatin bound XRCC1 (bottom panel). Note a transient subpopulation of H1 positive PGCs, which also display PAR and bound XRCC1 signals. There is than a progressive loss of H1 during between E11.25-E11.5, while the XRCC1 and of PAR signals persist in PGC population lacking H1. The kinetics of the process is depicted at the bottom with respect to the loss of 5mC. (Scale bar 10μM).
We next sought additional evidence for BER, specifically to distinguish between the inactive and active ongoing DNA repair in PGCs. It is known that XRCC1 is a soluble nuclear factor that is not associated with DNA in the absence of DNA damage, but which binds to DNA when ssDNA breaks occur so as to provide a scaffold for the assembly of other BER components11. To distinguish between soluble and chromatin bound XRCC1 in gonadal PGCs, we pre-extracted soluble proteins with detergents from isolated PGCs prior to fixation12. Whereas we observed an overall enrichment of XRCC1 in PGCs during E10.5-E12.5, we found a striking enhancement in chromatin bound XRCC1 specifically in PGCs at E11.5, which coincides precisely with the stage at which genome-wide DNA demethylation occurs (Fig. 1b). The presence of chromatin bound XRCC1 provides strong evidence for the existence of ssDNA breaks in PGCs at the time of DNA demethylation13. An additional marker of active BER is formation of the PAR polymer, a product of activated PARP1 enzyme11,14. Notably, we also detected high levels of PAR specifically in E11.5 PGCs (Fig. 1c). The presence of ssDNA breaks and active BER specifically in PGCs during ongoing epigenetic reprogramming are consistent with our previous proposal that DNA demethylation may be linked to DNA repair pathway2.
We have recently established detailed kinetics of chromatin changes during reprogramming in germ cells, which showed that this includes transient loss of the linker histone H1, loss of chromocenters and changes in nuclear size2. We have moreover shown that the onset of DNA demethylation precedes loss of H1 and changes in histone modifications2. To further investigate the link between DNA repair and widespread chromatin remodelling in PGCs, we examined the chronology of these processes. In PGCs spanning a period of approximately 6h, between E11.25-E11.5, we detected increasing levels of PAR prior to the loss of signal for histone H1 (Fig. 1c). Since histone H1 is a well described target for PARP1 ribosylation14,15, and PARP1 itself has been shown to displace H116, it is likely that high levels of PARP1-mediated PAR synthesis in the nuclei of PGCs might be directly involved in H1 displacement. Additionally, we observed a correlation between high nuclear PAR signals and the disappearance of chromocenters in PGCs (Supplementary Fig 5b)2. This is in agreement with a proposed role for PARP1 in the regulation of higher-order chromatin structure14,17,18 and further points towards an involvement of DNA repair in large-scale chromatin remodeling observed in PGCs2.
Concomitantly with the appearance of PAR signal in the nuclei of PGCs, we also detected high amounts of chromatin bound XRCC1 – documenting the presence of ssDNA breaks in nuclei of PGCs undergoing DNA demethylation process (Fig. 1c)13. The chromatin bound XRCC1 was detectable in PGCs before the loss of H1, which is in agreement with our previous data showing that 5mC is lost prior to the complete disappearance of H1 staining2. Thus, we note the existence of a highly transient population of PGCs in which we detect PAR signal, together with chromatin bound XRCC1 and H1, in the course of epigenetic reprogramming in PGCs (see Fig 1c). Notably, both PAR signal and bound XRCC1 persisted thereafter in a population of PGCs undergoing chromatin remodelling following the loss of H1 signal (Fig. 1c).
Next, we wished to establish whether there is a direct mechanistic link between BER and DNA demethylation. However, since PGCs are difficult to culture and manipulate in vitro, we examined mouse zygotes where there is also active genome-wide DNA demethylation that specifically affects the paternal but not the maternal pronucleus present in the same cell (Supplementary Fig. 6)3-5. This wave of globally active DNA demethylation occurs 4-5 hours following fertilisation (Supplementary Fig. 6)5, and while the kinetics of the process is well established, the mechanism is unknown. Based on our work on PGCs described above, we anticipated that activation of DNA repair markers might also be linked to DNA demethylation in the zygote. First, similarly to our observations on PGCs, we found negligible levels of ERCC1 and chromatin bound XPA throughout zygotic development (Supplementary Figs. 7 and. 8). In contrast, we found high levels of PARP1 and APE enzymes in zygotic pronuclei as well as accumulation of PAR (Supplementary Fig 9 and 10). Additionally, we observed high levels of XRCC1, albeit in both parental pronuclei (Fig. 2a). However, following pre-extraction of soluble proteins, we strikingly detected bound XRCC1 protein specifically only in the male pronucleus (Fig. 2b). This provides evidence for the existence of ssDNA breaks localised to the paternal genome but not to the maternal pronucleus in the same cell. The asymmetric chromatin bound XRCC1 in the male genome is detectable from early pronuclear stage (PN) 3 that coincides with the onset of DNA demethylation (Fig. 2b)5. We note that while ssDNA breaks are specifically confined to the male pronucleus, we did detect high levels of PAR in both the male and female pronuclei (Supplementary Fig. 9b). It is possible that the PAR in the paternal and maternal pronuclei are of different types having diverse biological roles, since the biological function of PAR polymers depends on the length (number of units) and the type of polymer branching19,20, which are indistinguishable by antibody staining. Indeed, the observed asymmetry in PARP1 protein that is detected primarily in the paternal pronucleus (Supplementray Fig 10), suggests that the PAR polymers present in maternal pronucleus might be a product of a different enzyme of the PARP family.
Figure 2. Chromatin bound XRCC1 in zygotes depicting ssDNA breaks in male pronucleus.
a. Detection of total XRCC1 in male and female pronuclei. b. Chromatin bound XRCC1 specifically in the male pronucleus visible from PN3. ♀ – maternal pronucleus, ♂ – paternal pronucleus, pb – polar body. (Scale bar 10μM).
To establish that the activation of BER pathway was specifically associated with DNA demethylation, we wanted to exclude other possible triggers, including protamine-histone exchange and DNA replication. As previously reported, protamine-histone exchange occurs during PN121 (Supplementary Fig 11), which is significantly before the onset of DNA demethylation or the detection of active BER. We also observed high levels of APE1 and chromatin bound XRCC1 in the presence of aphidicolin, an inhibitor of replicative DNA polymerase, indicating that the process occurs independently of DNA replication (Supplementary Fig. 12)22, as suggested previously4,3.
We decided to seek additional evidence for the link between DNA demethylation and BER. In this context, we note that the epigenetic asymmetry in DNA demethylation in zygotes has been shown to be promoted by the maternal inheritance of the Stella protein23; absence of Stella results in aberrantly targeted DNA demethylation to both the maternal and paternal pronuclei. Using this as a genetic model, we found that in zygotes from Stellla null females, the activation of BER components is detectable in both parental pronuclei. The chromatin bound XRCC1 (presence of ssDNA breaks) was clearly detectable in both pronuclei of Stella depleted zygotes in contrast to the wild type controls where the chromatin bound XRCC1 was predominantly confined to the paternal pronucleus (Supplementary Fig. 13). This genetic model provides further evidence that activation of BER is linked to DNA demethylation.
To further verify the mechanistic link between DNA demethylation and BER, we used small-molecule inhibitors of key BER components, which we predicted should impede the progression of DNA demethylation. Indeed, the presence of either PARP inhibitor 3-aminobenzamide (3AB)24, ABT-888 or the APE inhibitor CRT0044876 (APE1-i)25 in the fertilisation medium obstructed the progress of DNA demethylation, yielding zygotes with significantly higher levels of DNA methylation in the paternal pronucleus as judged by 5mC staining (Fig. 3, Supplementary Fig. 15), without affecting DNA methylation levels in the maternal pronucleus (data not shown). This effect on DNA methylation was further confirmed by bisulphite sequencing of the repetitive Line1 elements (Fig.3, Supplementary Figs. 14 and 15). The use of Ape1 inhibitor causes persistence of abasic sites in the template DNA that prevents amplification of the affected molecules, which may explain an apparent lack of changes in Line1 methylation in our bisulphite analysis. Notably, the amplified sequences in these samples showed signs of amplification skewing (data not shown). It should be pointed out that the inhibitors did not affect development of the zygotes, as judged by the rate of fertilisation and progression through the pronuclear stages (PN1-5).
Figure 3. Inhibition of BER and the consequences for DNA demethylation in zygotes.
a,c Small molecule PARP inhibitor 3AB and, APE1 inhibitor (CRT0044876) impede progression of DNA demethylation as detected by 5mC staining. b,d. Quantification of 5mC staining. The values represent a ratio between 5mC signal from paternal pronuclei relative to the signal from maternal pronuclei. Statistical analysis was by Student’s t-test. e. Bisulphite analysis of Line1 repetitive elements. The values show the percentage of methylated CpGs. Each line represents a unique DNA clone. Filled and open circles represent methylated and unmethylated CpGs, respectively. Scale bar 10μM. ♀ – maternal pronucleus, ♂ – paternal pronucleus, pb – polar body.
We demonstrate that the genome-wide active DNA demethylation process is mechanistically linked to the appearance of ssDNA breaks and the activity of the BER repair pathway, which, in turn, probably triggers extensive chromatin remodelling and histone exchange in PGCs2. This provides the foundation for detailed investigations, including how 5mC might be excised. While methylcytosine-specific DNA glycosylases (ROS1 and DME) exist in plants, enzymes with similar activity remain unknown in animals7,8, Alternatively, deamination of 5mC resulting in T-G mismatches could trigger a response from glycosylases such as TDG or MBD426 thus leading to a loss of 5mC. However, while MBD4 (but not TDG) is detectable in PGCs, both MBD4 and TDG are absent from mouse zygotes (Supplementary Figs 16 and 17). Furthermore, transcription analysis by qPCR of known candidates of deamination revealed that aid, was undetectable in PGCs, while apobec1 was down regulated prior to reprogramming (Supplementary Fig 18). In addition, we exclude the proposed deamination driven by Dnmt3a or Dnmt3b27, because PGCs lack both these enzymes at this time. Thus, the combined evidence for 5mC deamination is weak in our system. However, 5mC could be subject to another modification that might be recognized by an abundant glycosylase with a different specificity (Fig. 4). One possibility is the modification of 5mC to 5-hydroxymethylcytosine (5hmC)28,29. In support of this possibility, we detected significant expression of tet1, a gene that encodes a key enzyme for this modification29, specifically in PGC and not in the neighbouring somatic cells at E11.5 (Supplementary Fig. 18). Strikingly, we also note that the increase in tet1 expression coincides with significant transcription of BER components in E11.5 PGCs (see Supplementary Figs 5 and 18). Thus, we believe that our systematic approach may not only provides significant insights into the relationship between the key components of a dynamic epigenetic reprogramming event in germ cells, but it also offers one of the best opportunities for unravelling all aspects of an important yet elusive mechanism of active DNA demethylation.
Figure 4. Molecular pathways for DNA demethylation and chromatin remodelling in mouse germ cells.
The appearance of ssDNA breaks involves the BER pathway components (APE1, XRCC1 and PARP1) in the process of DNA demethylation. Several molecular pathways can lead to the activation of BER in the course of DNA demethylation.. 5mC could be subject to a chemical modification such as cytosine deamination although we did not detect expression of known candidate enzyme. Alternatively, 5mC may be converted to 5hmC by tet1 or other such enzymes that are expressed in PGCs, which would trigger a response from glycosylases. Alternatively, 5mC specific glycosylases, which have been identified in plants may have equivalents in mammals but none have yet been found. Note the temporal connections between DNA demethylation, appearance of ssDNA breaks and the activation of BER and the chromatin remodelling observed in PGCs.
Methods
Embryo collection and PGC preparation
PGCs were isolated from outbred MF1 mice. Noon of the day of the vaginal plug was designated as E0.5. For the FACS sorting the dissected urogenital ridges from embryos carrying Oct4-GFP transgene6 were trypsinized and treated with hyaluronidase prior to the FACS sorting using a MoFlo 3 laser Cytometer (Beckman Coulter). The purity of PGCs was examined independently using OCT4 staining and was always in excess of 98%. Stella ko mice were published previously30.
Immunofluorescence staining
The FACS sorted PGCs, somatic cells or single cell suspension obtained by trypsinisation of urogenital ridges were allowed to attach to the poly-L-lysine (Sigma-Aldrich) treated slides. The cells were briefly washed in PBS and fixed in 4% paraformaldehyde (PFA, prepared in PBS) for 15 min at room temperature. The cells were permeabilised for 30 min using PBS, 1% BSA, 0.1% Triton X-100 followed by antibody staining that was carried out in the same buffer at 4°C overnight. The slides were subsequently washed three times in PBS, 1%BSA, 0.1% Triton X-100 (5 min each wash) and incubated with Alexa fluorophore conjugated secondary antibodies (Molecular Probes) for 1 hr at room temperature in the dark, washed in PBS, 1% BSA, 0.1% Triton X-100 for 5 min and twice in PBS for 5 min. Finally, the slides were mounted in Vectashield containing DAPI (Vector Laboratories) and imaged using BioRad Radiance 2100 or Zeiss Meta 510 confocal microscopes.
For Triton pre-extraction cells were allowed to attach to poly-L-lysine treated (Sigma-Aldrich) slides and treated with ice-cold permeabilisation solution (50mM NaCl, 3mM MgCl2, 0.5% Triton X-100, 300mM sucrose in 25mM Hepes pH7.4) for 10 min on ice. The slides were then washed twice with PBS and fixed for 10 min using 4% PFA in PBS. Cells were stained as described above.
For immunofluorescence staining of zygotes the zona pellucida was removed using the Acid Tyrode’s solution (Sigma-Aldrich), the zygotes were washed in M2 medium (Sigma-Aldrich), rinsed in PBS, 1% BSA and fixed in 4% PFA in PBS. Following three 10 min washes in PBS, 1% BSA the zygotes were permeabilised in PBS, 1% BSA, 0.1% Triton X-100 for 30 min and incubated with primary antibodies at 4°C overnight in the same buffer. The zygotes were washed three times in PBS, 1%BSA, 0.1% Triton X-100 for 10 min, incubated with the Alexa fluorophore conjugated secondary antibodies (Molecular Probes) for 1 hr at room temperature in the dark. The zygotes were then washed in PBS 1% BSA, 0.1% Triton X-100 for 10 min, twice in PBS, 1% BSA for 10 min, stained with DAPI and mounted in Vectashield (Vector Laboratories) using imaging spacers. The zygotes were imaged as Z-series confocal sections using Zeiss Meta510 confocal microscope.
For Triton pre-extraction zygotes were taken with an intact zona pellucida. Zygotes were incubated in ice-cold permeabilisation solution for 10 min on ice. The zygotes were then washed three times with permeabilisation solution without Triton X-100 and fixed for 20 min using 4% PFA in PBS at room temperature. Zygotes were stained as described above. In order to prevent the zona pellucida from collapsing under osmotic stress upon transfer to the mounting media, zygotes were washed in PBS, 1% BSA with increasing concentrations of glycerol before mounting.
In vitro fertilisation of mouse oocytes
The procedure was carried out as in Nagy et al31. F1 mice were used as source of gametes. The sperm capacitation as well as fertilisation was carried out in the HTF fertilisation medium (Lonza) supplemented with BSA (4mg/ml). The zygotes were collected 7-8hrs following fertilisation.
In order to inhibit replication 2mg/ml aphidicolin (Sigma-Aldrich) was included in the fertilisation medium. For small molecule inhibitors of BER, oocytes were pre-incubated in the fertilisation medium containing either 5mM 3-aminobenzamide (Sigma-Aldrich), 38μM ABT-888 (Alexis Biochemicals) or 100μM CRT0044876 (Calbiochem) for at least 40 min prior to the addition of sperm. All compounds were solubilised in pure DMSO and used at 0.1% DMSO final concentration.
5mC staining of mouse zygotes
Following the removal of zona pellucida using Acid Tyrode’s solution, mouse zygotes were fixed in 4% PFA in PBS for 20 min and washed three times in PBS, 1% BSA for 10 min. Zygotes were then permeabilised in PBS, 1% BSA, 0.5% Triton X-100 for 30min, washed three times in PBS, 1%BSA for 10 min and treated with RNAseA (10mg/ml) for 1hr at 37°C. Following three subsequent 10 min washes in PBS, 1% BSA, samples were rinsed in PBS and treated with 4M HCl for 20min at 37°C. Zygotes were then rinsed in PBS, washed three times in PBS, 1% BSA for 10 min, incubated in PBS, 1% BSA, 0.1% Triton X-100 for 30 min and incubated in the same buffer with 5mC antibody at 4°C overnight. Zygotes were subsequently washed three times in PBS, 1% BSA, 0.1% Triton X-100 for 10 min and incubated with Alexa fluorophore conjugated secondary antibodies (Molecular Probes) for 1 hr at room temperature in the dark. Zygotes were then washed once in PBS, 1%BSA, 0.1% Triton X-100 for 10 min and twice in PBS, 1% BSA for 10 min, followed by propidium iodide (PI) staining (0.25mg/ml) for 25 min. The final wash was carried out in PBS, 1% BSA for 20 min to remove excess of PI, the zygotes mounted in Vectashield (Vector laboratories) using imaging spacers and imaged as above.
Images were analyzed using Volocity 4.3.1. Maternal and paternal pronuclei were identified by size and proximity to the polar body. For each pronucleus the center z-section was identified using PI staining. The total intensity of 5mC staining was calculated using the center section of each pronucleus. The total intensity values were normalized to background values by subtracting the average background pixel intensity multiplied by the total pixel size of each pronucleus. The level of paternal methylation was reported as a ratio of paternal over maternal methylation signal.
Antibodies
XRCC1 (Serotec) 1:200, APE1 (Abcam) 1:5000, ERCC1 (NeoMarkers) 1:100, PAR (Trevigen) 1:300, PARP1 (Alexis Biochemicals) 1:100, Oct4 (BD Biosciences) 1:200, SSEA-1 (kind gift of P.Beverly) 1:2, PGC7/Stella (kind gift of T.Nakano) 1:2000, H1 (Abcam) 1:200; 5mC antibody (kind gift of A.Nivelau) 1:50; XPA (Bethyl Laboratories) 1:200, TDG (Santa Cruz) 1:50; MBD4 (Santa Cruz) 1:50
Bisulphite sequencing
Zygotes obtained by IVF were treated with acid Tyrode’s (Sigma Aldrich) to remove zona pellucida and snap frozen in liquid nitrogen. Bisulphite sequencing was subsequently carried out as using agarose bead modification as described in32. The following primers were used for the amplification of Line1 elements : F1: gttagagaatttgatagtttttggaatagg, R1: ccaaaacaaaacctttctcaaacactatat, R2: tcaaacactatattactttaacaattccca. The semi-nested approach was used: 1st PCR (F1,R1 primers), 2nd PCR (F1,R2 primers). PCR conditions: 95°C 5min,(95°C 1min, 56°C 1min, 72°C 45sec) 35×, 72°C 5min, 4°C.
Single cell amplification and quantitative PCRs
PGCs of different developmental stages were purified from genital ridges of mice carrying Oct4-GFP transgene using the FACS sort as explained above. For the ES cell control, ES cells carrying the same Oct4-GFP transgene were purified using identical FACS procedure. The sorted cells were snap frozen in liquid nitrogen and the RNA subsequently purified using RNeasy micro kit (Qiagen). 1ng of total RNA was amplified using the single cell amplification protocol33. The amplified product was diluted 40x and used for quantitative PCR analysis (ABI 7000 instrument). The list of primers used is available upon request.
Acknowledgments
The authors would like to thank Y. Galanty, S. Polo and members of Surani lab for stimulating discussions. S.J. Jeffries is a NIH-Cambridge Health Science Scholar. This work was funded by grants from the Wellcome Trust to M.A.S.
References
- 1.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452(7189):877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 4.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 5.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 6.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1-2):15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 8.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 9.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009 doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Svetlova M, et al. Reduced extractability of the XPA DNA repair protein in ultraviolet light-irradiated mammalian cells. FEBS Lett. 1999;463(1-2):49–52. doi: 10.1016/s0014-5793(99)01592-6. [DOI] [PubMed] [Google Scholar]
- 11.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenk R, Ransom L, Kaufmann Y, Penman S. A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell. 1977;10(1):67–78. doi: 10.1016/0092-8674(77)90141-6. [DOI] [PubMed] [Google Scholar]
- 13.Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst) 2007;6(2):254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41(1):60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Huletsky A, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264(15):8878–8886. [PubMed] [Google Scholar]
- 16.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319(5864):819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 17.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 18.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16(9):923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 19.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 20.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35(21):e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodman TC, Pruslin FH, Hoffmann HP, Allfrey VG. Turnover of basic chromosomal proteins in fertilized eggs: a cytoimmunochemical study of events in vivo. J Cell Biol. 1981;90(2):351–361. doi: 10.1083/jcb.90.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howlett SK. The Effect of Inhibiting DNA-Replication in the One-Cell Mouse Embryo. Rouxs Archives of Developmental Biology. 1986;195(8):499–505. doi: 10.1007/BF00375890. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9(1):64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 24.Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185(3):775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhusudan S, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33(15):4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 28.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payer B, et al. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13(23):2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 31.Nagy A. Manipulating the mouse embryo: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2003. [Google Scholar]
- 32.Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24(24):5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2(3):739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]