Abstract
N-myristoylation is the irreversible attachment of a C14-fatty acid, myristic acid, to the N-terminal glycine of a protein via formation of an amide bond. This modification is catalyzed by myristoyl-CoA : protein N-myristoyltransferase (NMT), an enzyme ubiquitous in eukaryotes that is up-regulated in several cancers.
Here we report a sensitive fluorescence-based assay to study the enzymatic activity of human NMT1 and NMT2, based on detection of coenzyme A by 7-diethylamino-3-(4-maleimido-phenyl)-4-methylcoumarin. We also describe expression and characterization of NMT1 and NMT2, and assay validation with small molecule inhibitors. This assay should be broadly applicable to NMTs from a range of organisms.
Keywords: N-myristoyltransferase (NMT), fluorescence, 7-diethylamino-3-(4-maleimido-phenyl)-4-methylcoumarin (CPM), coenzyme A, screening
Myristoyl-coenzyme A : protein N-myristoyltransferase (N-myristoyltransferase, NMT) is a ubiquitous enzyme in eukaryotes that catalyzes co- and post-translational transfer of a C14 saturated fatty acid (myristic acid) from myristoyl-coenzyme A (myristoyl-CoA) to the N-terminal glycine residue of target proteins [1] (Fig. 1A). NMT was first identified in yeast [2], and subsequently characterized in fungi, parasitic protozoa, insects, plants, humans and other mammals. N-myristoylation of proteins can promote reversible protein-protein interactions, enhance interactions of the protein with the membrane and change protein stability [1]. The role of myristoylation is still not entirely understood, and not all myristoylated proteins have been experimentally determined [3; 4]. However, in humans, protein myristoylation is connected with several diseases including cancer [5], genetic disorders [6] and infection [7]. In Homo sapiens, NMT is encoded by two distinct genes, Nmt1 and Nmt2, and RNA interference experiments suggest that Nmt1 knockdown inhibits tumor growth, making NMT1 a potential target for the development of novel anti-cancer therapies [8]. The host myristoylation is essential for the formation of HIV viral capsids, suggesting potential for mammalian NMT inhibitors as anti-viral agents [9]. Furthermore, NMT is established as a promising anti-fungal [10] and anti-parasitic drug target, for example in African sleeping sickness [11; 12], malaria [13] and leishmaniasis [14; 15]. An effective in vitro enzyme assay is required for drug discovery against NMT; the few assays reported in detail to date are based on detection of a radioisotopic label in a myristoylated peptide or protein [13; 16]. Such assays allow sensitive measurements, but are discontinuous, expensive, and require handling and disposal of radioactive materials.
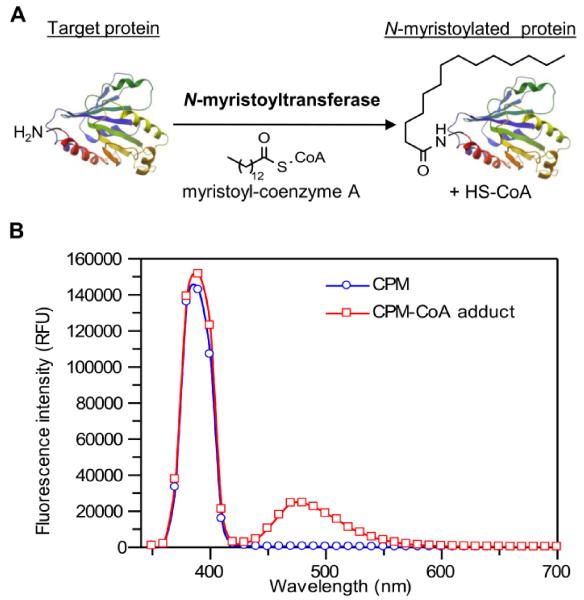
Fig. 1.
(A) NMT-catalysed N-myristoylation of target proteins. (B) Changes in fluorescence emission spectra upon formation of the CPM-coenzyme A adduct in assay buffer at 25 °C with excitation at 390 nm (RFU: relative fluorescence units).
Herein we report the development of a robust fluorogenic assay for NMT activity suitable for continuous reaction monitoring and end-point assays. This assay monitors coenzyme A (CoA) production in real time using a pro-fluorescent probe, 7-diethylamino-3-(4-maleimido-phenyl)-4-methylcoumarin (CPM), a commercially available coumarin derivative containing a thiol-reactive maleimide [17] (Fig. 1B). The maleimide quenches coumarin fluorescence, but the in situ reaction with CoA thiol generated during myristoylation results in release of fluorescence.
The proteins used for this study are the catalytic domains of human NMT isoforms 1 and 2. These proteins share 83% sequence identity over 388 amino acids and lack the long N-terminal extensions involved in NMT subcellular targeting [18] (Supplementary data, Fig S1). Details of gene constructs, protein production and purification may be found in Supplementary Data. The assay was developed in 96-well black polypropylene microplates (Greiner Bio-One, UK) and reagent solutions were prepared in a buffer containing 20 mM potassium phosphate (pH 7.9-8.0) with 0.5 mM EDTA, 0.1% (v/v) Triton® X-100 and a final concentration of 2.7% (v/v) DMSO. Thiol-containing reagents should be avoided as they react with CPM and interfere with the assay. Fluorescence readings were obtained on a SpectraMax M2e microplate reader (Molecular Devices, Canada) or an EnVision Xcite reader (Perkin Elmer, UK). The peptide H-Gly-Ser-Asn-Lys-Ser-Lys-Pro-Lys-NH2 (Hs pp60src(2-9)), derived from the N-terminal sequence of myristoylated Homo sapiens proto-oncogene tyrosine kinase pp60src [19], was used as substrate for both human NMTs.
10 μL of a 10% DMSO/water (v/v) solution, 25 μL of myristoyl-CoA solution, 50 μL of NMT (final concentration: 6.3 nM) and 10 μL of CPM solution (final concentration: 8 μM) were combined in an assay well. The enzymatic reaction was started by adding 15 μL of peptide substrate solution and fluorescence intensity was monitored over 30 minutes at 1 minute intervals (excitation 380 nm, emission at 470 nm) at 25 °C. The initial velocity was calculated over the first 4 minutes of the experiment after subtraction of the background fluorescence (measured in the absence of enzyme). The Michaelis-Menten (Km) constant of Hs pp60src(2-9) was determined using a saturating concentration of the co-substrate, myristoyl-CoA (30 μM). Under these conditions, the peptide Hs pp60src(2-9) displayed a Km of 2.76 ± 0.21 μM and 2.77 ± 0.14 μM for NMT1 and NMT2 respectively. Subsequently, the Km of myristoyl-Co A was determined in the presence of a saturating concentration (30 μM) of Hs pp60src(2-9) substrate. This experiment led to Km values of 8.24 ± 0.62 μM and 7.24 ± 0.79 μM for NMT1 and NMT2 respectively, which are in good agreement with previously reported data (7.6 μM for NMT1 using pp60v-src(2-17) as substrate) [20].
While continuous assays are generally preferred for analytical purposes, they are time-consuming and unsuitable for the screening of large compound libraries; we therefore investigated the feasibility of an endpoint assay. A suitable enzyme concentration (6.3 nM) was selected to give a linear reaction rate over 30 minutes at 25 °C. If necessary, this concentration can be reduced down to 2.1 nM which in these conditions gives a reasonable signal to background ratio of ~ 2. Then, we examined the possibility of quenching the reaction by acidifying the reaction mixture since the rate of the reaction between the coenzyme A and CPM is strongly pH-dependent [21] (Supplementary data, Fig S2). Screening a range of conditions, we found that addition of 60 μL of 0.1 M sodium acetate buffer pH 4.75 (‘quenching solution’) 30 minutes into the assay immediately decreased the pH of the reaction solution to pH 4.9-5.1. At this pH, quenching the fluorogenic reaction and giving a signal that remains stable over 8 hours (Supplementary data, Fig. S3).
To validate the assay, two known NMT inhibitors were evaluated against NMT1 and NMT2 (Fig. 2): 1, a pseudo-peptidic NMT inhibitor [22] and 2, a small molecule that has been recently described as an inhibitor with low-nM affinity for Trypanosoma brucei NMT and Homo sapiens NMT [11]. Inhibition assays were carried out using the endpoint method, with final concentrations of 4 μM Hs pp60src(2-9) and 4 μM myristoyl-CoA. Briefly, the inhibitor in 10% DMSO/water (10 μL), myristoyl-CoA (25 μL) and NMT (50 μL) solutions were combined in a 96-well plate as described above, and the enzymatic reaction was started by adding 25 μL of a solution containing 17.6 μM Hs pp60src(2-9) and 35.2 μM CPM in assay buffer. The reaction was stopped after 30 minutes at 25 °C by adding 60 μL quenching solution. Positive controls excluded inhibitor, negative controls excluded both NMT and inhibitor. Under these assay conditions, the average Z′ value was between 0.7 - 0.9. The apparent Km of the peptide was determined to be 2.66 ± 0.20 μM for NMT1 and 3.25 ± 0.22 μM for NMT2.
Fig. 2.
(A) Chemical structure of inhibitors 1 and 2. (B) Inhibition assay: IC50 values were determined in the presence of 4 μM Hs pp60src(2-9) and 4 μM myristoyl-CoA. Data points represent means of duplicates.
As expected, 1 behaved as an inhibitor of both human NMTs with IC50 values of 0.35 μM and 0.51 μM against NMT1 and NMT2 respectively, conforming to previously reported data (IC50 = 0.50 ± 0.37 μM against NMT1) from a radioactive HPLC-based assay [22]. Similarly, 2 was tested against both human NMTs and led to IC50 values of 13.7 nM (NMT1) and 14.4 nM (NMT2), similar to the result previously obtained against NMT1 with a scintillation proximity assay (4 nM) [11].
In summary, this fluorogenic assay constitutes an attractive alternative to radioactive assays in the search for NMT inhibitors. Moreover, the possibility of using it in either continuous or endpoint mode makes it suitable both for kinetic/mechanistic studies and for high-throughput screening. Whilst strongly nucleophilic reagents or inhibitors should be avoided as they lead to increased background fluorescence, thiols in the enzyme are tolerated. Although this assay was developed on human NMT1 and NMT2 it depends only on the generation of CoA-SH, and could be readily adapted to the study of parasitic or fungal NMT activity.
Supplementary Material
Acknowledgements
This work was supported by the MRC (grant G0900278), Wellcome Trust (grant 087792) and the BBSRC (David Phillips Research Fellowship to EWT, grant BB/D02014X/1). ET and EWT thank CR-UK for a research studentship grant (C29637/A10711), TOO thanks Imperial College for a Scholarship, and RS thanks the EU for the award of an Intra-European Fellowship (PIEF-GA-2010-273868). The authors thank Tony Holder (MRC NIMR) and Deborah Smith (University of York) and their co-workers for helpful discussions.
References
- [1].Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol. 2009;3:19–35. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, Gordon JI. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc Natl Acad Sci U S A. 1987;84:2708–12. doi: 10.1073/pnas.84.9.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. N-Myristoyl transferase-mediated protein labelling in vivo. Org Biomol Chem. 2008;6:2308–15. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- [4].Heal WP, Wickramasinghe SR, Bowyer PW, Holder AA, Smith DF, Leatherbarrow RJ, Tate EW. Site-specific N-terminal labelling of proteins in vitro and in vivo using N-myristoyl transferase and bioorthogonal ligation chemistry. Chemical Communincations. 2008;4:480–2. doi: 10.1039/b716115h. [DOI] [PubMed] [Google Scholar]
- [5].Felsted RL, Glover CJ, Hartman K. Protein N-myristoylation as a chemotherapeutic target for cancer. J Natl Cancer Inst. 1995;87:1571–3. doi: 10.1093/jnci/87.21.1571. [DOI] [PubMed] [Google Scholar]
- [6].Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, Lipzen A, Zampino G, Mazzanti L, Digilio MC, Martinelli S, Flex E, Lepri F, Bartholdi D, Kutsche K, Ferrero GB, Anichini C, Selicorni A, Rossi C, Tenconi R, Zenker M, Merlo D, Dallapiccola B, Iyengar R, Bazzicalupo P, Gelb BD, Tartaglia M. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nature Genetics. 2009;41:1022–6. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Provitera P, El-Maghrabi R, Scarlata S. The effect of HIV-1 Gag myristoylation on membrane binding. Biophys Chem. 2006;119:23–32. doi: 10.1016/j.bpc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- [8].Ducker CE, Upson JJ, French KJ, Smith CD. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol Cancer Res. 2005;3:463–76. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Seaton KE, Smith CD. N-Myristoyltransferase isozymes exhibit differential specificity for human immunodeficiency virus type 1 Gag and Nef. J Gen Virol. 2008;89:288–96. doi: 10.1099/vir.0.83412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sheng C, Xu H, Wang W, Cao Y, Dong G, Wang S, Che X, Ji H, Miao Z, Yao J, Zhang W. Design, synthesis and antifungal activity of isosteric analogues of benzoheterocyclic N-myristoyltransferase inhibitors. European Journal of Medicinal Chemistry. 2010;45:3531–40. doi: 10.1016/j.ejmech.2010.03.007. [DOI] [PubMed] [Google Scholar]
- [11].Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, Price HP, Guther ML, Torrie LS, Robinson DA, Hallyburton I, Mpamhanga CP, Brannigan JA, Wilkinson AJ, Hodgkinson M, Hui R, Qiu W, Raimi OG, van Aalten DM, Brenk R, Gilbert IH, Read KD, Fairlamb AH, Ferguson MA, Smith DF, Wyatt PG. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464:728–32. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Panethymitaki C, Bowyer PW, Price HP, Leatherbarrow RJ, Brown KA, Smith DF. Characterization and selective inhibition of myristoyl-CoA:protein N-myristoyltransferase from Trypanosoma brucei and Leishmania major. Biochem J. 2006;396:277–85. doi: 10.1042/BJ20051886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bowyer PW, Gunaratne RS, Grainger M, Withers-Martinez C, Wickramsinghe SR, Tate EW, Leatherbarrow RJ, Brown KA, Holder AA, Smith DF. Molecules incorporating a benzothiazole core scaffold inhibit the N-myristoyltransferase of Plasmodium falciparum. Biochem J. 2007;408:173–80. doi: 10.1042/BJ20070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bowyer PW, Tate EW, Leatherbarrow RJ, Holder AA, Smith DF, Brown KA. N-myristoyltransferase: a prospective drug target for protozoan parasites. Chemmedchem. 2008;3:402–408. doi: 10.1002/cmdc.200700301. [DOI] [PubMed] [Google Scholar]
- [15].Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, Price HP, Meier F, Leatherbarrow RJ, Tate EW, Smith DF, Wilkinson AJ. N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol. 2010;396:985–99. doi: 10.1016/j.jmb.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McIlhinney RA, McGlone K. A simplified assay for the enzyme responsible for the attachment of myristic acid to the N-terminal glycine residue of proteins, myristoyl-CoA: glycylpeptide N-myristoyltransferase. Biochem J. 1989;263:387–91. doi: 10.1042/bj2630387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chung CC, Ohwaki K, Schneeweis JE, Stec E, Varnerin JP, Goudreau PN, Chang A, Cassaday J, Yang L, Yamakawa T, Kornienko O, Hodder P, Inglese J, Ferrer M, Strulovici B, Kusunoki J, Tota MR, Takagi T. A fluorescence-based thiol quantification assay for ultra-high-throughput screening for inhibitors of coenzyme A production. Assay Drug Dev Technol. 2008;6:361–74. doi: 10.1089/adt.2007.105. [DOI] [PubMed] [Google Scholar]
- [18].Glover CJ, Hartman KD, Felsted RL. Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J Biol Chem. 1997;272:28680–9. doi: 10.1074/jbc.272.45.28680. [DOI] [PubMed] [Google Scholar]
- [19].Lacal PM, Pennington CY, Lacal JC. Transforming activity of ras proteins translocated to the plasma membrane by a myristoylation sequence from the src gene product. Oncogene. 1988;2:533–7. [PubMed] [Google Scholar]
- [20].McIlhinney RA, Patel PB, McGlone K. Characterization of a polyhistidine-tagged form of human myristoyl-CoA: protein N-myristoyltransferase produced in Escherichia coli. Eur J Biochem. 1994;222:137–46. doi: 10.1111/j.1432-1033.1994.tb18851.x. [DOI] [PubMed] [Google Scholar]
- [21].Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–58. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- [22].Nagarajan SR, Devadas B, Zupec ME, Freeman SK, Brown DL, Lu HF, Mehta PP, Kishore NS, McWherter CA, Getman DP, Gordon JI, Sikorski JA. Conformationally constrained [p-(omega-aminoalkyl)phenacetyl]-L-seryl-L-lysyl dipeptide amides as potent peptidomimetic inhibitors of Candida albicans and human myristoyl-CoA:protein N-myristoyl transferase. J Med Chem. 1997;40:1422–38. doi: 10.1021/jm9608671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.