Abstract
Rationale: Emphysema occurs in distinct pathologic patterns, but little is known about the epidemiologic associations of these patterns. Standard quantitative measures of emphysema from computed tomography (CT) do not distinguish between distinct patterns of parenchymal destruction.
Objectives: To study the epidemiologic associations of distinct emphysema patterns with measures of lung-related physiology, function, and health care use in smokers.
Methods: Using a local histogram-based assessment of lung density, we quantified distinct patterns of low attenuation in 9,313 smokers in the COPDGene Study. To determine if such patterns provide novel insights into chronic obstructive pulmonary disease epidemiology, we tested for their association with measures of physiology, function, and health care use.
Measurements and Main Results: Compared with percentage of low-attenuation area less than −950 Hounsfield units (%LAA-950), local histogram-based measures of distinct CT low-attenuation patterns are more predictive of measures of lung function, dyspnea, quality of life, and health care use. These patterns are strongly associated with a wide array of measures of respiratory physiology and function, and most of these associations remain highly significant (P < 0.005) after adjusting for %LAA-950. In smokers without evidence of chronic obstructive pulmonary disease, the mild centrilobular disease pattern is associated with lower FEV1 and worse functional status (P < 0.005).
Conclusions: Measures of distinct CT emphysema patterns provide novel information about the relationship between emphysema and key measures of physiology, physical function, and health care use. Measures of mild emphysema in smokers with preserved lung function can be extracted from CT scans and are significantly associated with functional measures.
Keywords: emphysema, chronic obstructive pulmonary disease, spiral computed tomography, epidemiology
At a Glance Commentary
Scientific Knowledge on the Subject
Emphysema occurs in distinct pathologic patterns, but it is technically challenging to efficiently extract information on these patterns from computed tomography (CT) scans. Consequently, little is known about the clinical associations of these patterns.
What This Study Adds to the Field
This study demonstrates that quantitative measures of distinct emphysema patterns from CT provide novel information relevant to chronic obstructive pulmonary disease epidemiology and phenotyping that is distinct from the information provided by a standard quantitative CT emphysema measure.
Emphysema, defined as alveolar destruction and airspace enlargement distal to the terminal bronchiole, is a characteristic pathologic process of chronic obstructive pulmonary disease (COPD) (1). Quantitative measures of emphysema can be assessed in vivo with computed tomography (CT), and semiautomated processes for extracting this information have enabled efficient quantitative characterization of emphysema in large-scale studies. Numerous studies have demonstrated association between quantitative measures of emphysema severity and physiologic parameters of lung function (2–5), functional status and dyspnea (6–9), genetic polymorphisms (10–18), and clinical outcomes (19–21).
The standard approach to CT-based quantification of emphysema is to define a threshold in the lung density histogram that differentiates emphysematous from preserved lung (22). One limitation of this approach is that, by representing the burden of emphysema as a single number, information present in the local patterns of emphysematous involvement is lost. Three distinct patterns of emphysema based on distribution within the secondary lobule (centrilobular, panlobular [PL], and pleural-based [PB]) have been well-described in pathologic studies, and previous work has demonstrated that these emphysematous patterns can be accurately detected on CT (23–25). More sophisticated quantitative emphysema detection methods have been shown to be capable of accurately classifying discrete lung regions into distinct emphysema patterns from CT scans, but these methods are difficult to apply to large cohorts because of the high computational cost of these algorithms (26–29).
We have characterized the percentage of five CT emphysema patterns (mild centrilobular, moderate centrilobular, severe centrilobular, PL, and PB) and the percentage of nonemphysematous (NE) lung present in the CT scans of 9,313 smokers using a tissue classification scheme based on the local histogram of lung density (30). We hypothesized the following: (1) the local histogram-based emphysema (LHE) patterns identified by this method would be associated with COPD-related physiologic and functional measures; (2) these associations would persist after adjustment for a standard measure of quantitative emphysema on CT, the percent of emphysema defined by lung tissue with an attenuation value less than −950 Hounsfield units (%LAA-950); and (3) LHE patterns would be more predictive of COPD-related physiologic and functional measures than %LAA-950. Some of the results of these studies have been previously reported in the form of an abstract (31).
Methods
Study Population
Between 2007 and 2011, 10,192 non-Hispanic white (n = 6,784) and African-American (n = 3,408) smokers were enrolled into the COPDGene Study, a previously described (32) multicenter study designed to investigate the genetic and epidemiologic associations of COPD. Volumetric CT scans of the chest were obtained at full inflation and relaxed exhalation. Spirometry was performed with an NDD EasyOne TM Spirometer (Zurich, Switzerland) in accordance with American Thoracic Society/European Respiratory Society recommendations (33). Dyspnea and lung disease–specific quality of life measures were obtained through the use of previously validated questionnaire items (34, 35). Participants with a history of active lung disease other than asthma, emphysema, or COPD were excluded from COPDGene. The COPDGene study was approved by the institutional review boards of all participating centers.
Local Histogram-based Quantitative Emphysema Measures
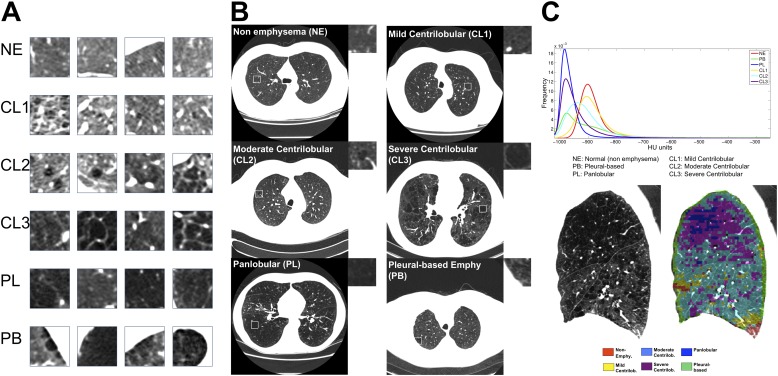
Characterization of the emphysema pattern was performed using local histogram information (30). To train the algorithm, 267 CT scans at full inspiration from the COPDGene cohort were processed to yield 1,337 regions of interest (ROIs) of size 24.18 × 24.18 mm2. Each of these ROIs were labeled by a physician with expertise in chest radiology, and these labels were used to learn a classifier that would classify new ROIs into one of six categories (NE, centrilobular emphysema by increasing amount of severity [CL1, CL2, CL3], PL emphysema, and PB emphysema). Figure 1 shows representative ROIs with the corresponding axial CT slice.
Figure 1.
Prototypic lung computed tomography (CT) patches for each local histogram emphysema pattern as regions of interest (A) and in context (B). Representative regions of interest local histograms (C, top) and local histogram emphysema classification results for a sagittal slice corresponding to a COPDGene subject with severe emphysema (C, bottom). CL1 = mild centrilobular; CL2 = moderate centrilobular; CL3 = severe centrilobular; NE = nonemphysematous; PB = pleural-based; PL = panlobular.
Detailed definitions for each emphysema type are listed in Table E1 in the online supplement. Briefly, areas of lung with normal-appearing architecture were labeled NE. Centrilobular emphysema labels were applied to regions with emphysematous destruction within the secondary pulmonary lobule with overall preservation of architecture (CL1), more confluent emphysema but preservation of the bronchovascular bundle (CL2), or confluent emphysema with obliteration of the bronchovascular bundle but preservation of septa (CL3). Regions with complete effacement of the parenchyma including the septa and bronchovascular bundles were labeled as PL. Regions of emphysema abutting a pleural surface were labeled as PB.
Using the expert-provided labels from the training set as a reference, the LHE method was applied to the remaining scans in COPDGene to generate for each scan six continuous measures representing the lung volume percentage that was classified into each of the six patterns. The LHE approach exploits the difference that is observed between the local histogram for each emphysema pattern ROI illustrated in Figure 1. The local histogram for each ROIs was computed using a kernel density estimator (36). A k nearest neighbors (kNN) classifier assigned each ROI local histogram to the emphysema patterns based on a majority consensus from the k nearest training samples to the ROI local histogram. The distance between the test ROI local histogram and the training local histograms was computed using the L1 norm. For this initial kNN classification, PB emphysema training samples were excluded and each ROI was assigned a label for one of the following types: NE, CL1, CL2, CL3, or PL.
Given that PB emphysema is defined as emphysema occurring near the chest wall, we classified a sample as PB if it abutted the chest wall and the corresponding label was classified as one of the emphysema patterns: CL1, CL2, CL3, or PL. An ROI was defined to be corresponding to the chest wall if the percentage of high-density pixels (defined as >0 HU) was greater than 0.25. This threshold was obtained based on the PB training datasets. The output of the classification method for a sagittal slice is illustrated in Figure 1C.
Detailed information about the computational approach is included in the online supplement along with information about the validity and repeatability of the local histogram method (see Tables E3 and E4 and Figure E3).
Threshold-based Emphysema Measures
Threshold-based measures of %LAA-950 and %LAA-910 were calculated for each lung CT scan by quantifying the percentage of the overall lung density histogram below the −950 or −910 Hounsfield unit threshold, respectively.
Statistical Analysis
Bar plots were used to visualize the median percentage of each emphysema pattern by Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage and by discretized %LAA-950. Nonlinearities in the relation between emphysema patterns were visualized with scatterplot matrices. LHE patterns were related to COPD-related variables of interest (e.g., spirometric measures, functional status, and so forth) through separate regression models. A full list of COPD-related measures tested for association is presented in Tables E6–E8. In these models, the COPD-related variable of interest was the dependent variable and each LHE pattern was included as an independent variable in a separate regression equation. Linear and logistic regressions were used for continuous and binary outcomes, respectively. For ordered categorical outcomes, linear regression was used and proportional odds logistic regression was also conducted to confirm the validity of associations. Associations were tested first in a base model that adjusted for age, sex, race, and pack-years of smoking exposure. These associations were subsequently tested in a model adjusting for %LAA-950 in addition to the base variables.
To evaluate the relative contributions of %LAA-950 and the LHE patterns in predicting key COPD-related measures, multivariate models were used to relate %LAA-950 and the five pathologic LHE patterns (leaving out the NE pattern) to four COPD-related measures (FEV1, the Modified Medical Research Council [MMRC] dyspnea score [37], 6-minute walk distance [6MWD], and number of COPD exacerbations over the past year). For each COPD-related measure, three models were used to relate %LAA-950, LHE patterns, and both %LAA-950 and LHE patterns, respectively, to the measure. Linear regression was used for all four outcomes, and for MMRC score and number of exacerbations ordinal logistic regression was also performed. The goodness of fit of these models was assessed by the F statistic and Akaike information criterion. Both measures account for the differing degrees of freedom of the LHE and %LAA-950 models. All analyses were conducted using R 2.14 (38). This same process was also performed to compare the predictive performance of %LAA-910 with the LHE measures.
Results
Of the 10,192 smokers enrolled in COPDGene, complete quantitative emphysema and covariate data were available for 9,313 individuals. The clinical characteristics of these individuals are shown in Table 1. Of the 9,313 individuals, 44% were control subjects with normal spirometry (smoking control subjects); 8% were GOLD stage 1; 36% were GOLD stages 2–4; and 12% were not classifiable by GOLD criteria because of reduced FEV1 percent predicted with an FEV1/FVC ratio above 0.7. The relative amounts of each LHE pattern are presented by GOLD stage. The most common patterns are the NE, mild centrilobular, and moderate centrilobular patterns. The PB pattern is less common, and the severe centrilobular and PL patterns are present only in severely affected GOLD 2–4 subjects. To focus only on individuals without spirometrically diagnosed COPD or with definite COPD, we excluded from further analysis those individuals who were GOLD stage 1 or who were unclassifiable by GOLD criteria.
TABLE 1.
SUBJECT CHARACTERISTICS
| All Subjects | GOLD U* | GOLD 0† | GOLD 1 | GOLD 2–4 | |
|---|---|---|---|---|---|
| N |
9,313 |
1,113 |
4,069 |
746 |
3,385 |
| Age, mean (SD) |
60 (9) |
57 (8) |
57 (8) |
62 (9) |
63 (9) |
| Sex, % female |
46 |
54 |
47 |
42 |
44 |
| Race, % African-American |
32 |
42 |
40 |
22 |
22 |
| Pack-years, median (IQR) |
39 (27) |
38 (26) |
34 (23) |
40 (26) |
47 (32) |
| FEV1, % of predicted, mean (SD) |
77 (25) |
71 (8) |
98 (12) |
91 (9) |
51 (18) |
| Emphysema at −950 HU, median (IQR) |
2 (6) |
1 (1) |
1 (2) |
3 (6) |
8 (18) |
| NE, median (IQR) |
64 (43) |
72 (25) |
74 (22) |
60 (36) |
32 (48) |
| CL1, median (IQR) |
24 (16) |
23 (18) |
21 (15) |
26 (15) |
26 (16) |
| CL2, median (IQR) |
5 (16) |
2 (4) |
2 (4) |
7 (13) |
22 (33) |
| CL3, median (IQR) |
0 (1) |
0 (0) |
0 (0) |
0 (1) |
1 (7) |
| PL, median (IQR) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (3) |
| PB, median (IQR) | 2 (4) | 2 (2) | 2 (2) | 3 (4) | 5 (7) |
Definition of abbreviations: CL1 = mild centrilobular; CL2 = moderate centrilobular; CL3 = severe centrilobular; GOLD = Global Initiative for Chronic Obstructive Lung Disease; IQR = interquartile range; NE = nonemphysematous; PB = pleural-based; PL = panlobular.
Quantitative emphysema values are % of computed tomography lung volume.
GOLD U: GOLD “unclassified” subjects (i.e., those with FEV1% predicted < 80% but FEV1/FVC > 0.7).
GOLD 0: control smokers.
The Relation of LHE Patterns to GOLD Stage and %LAA-950
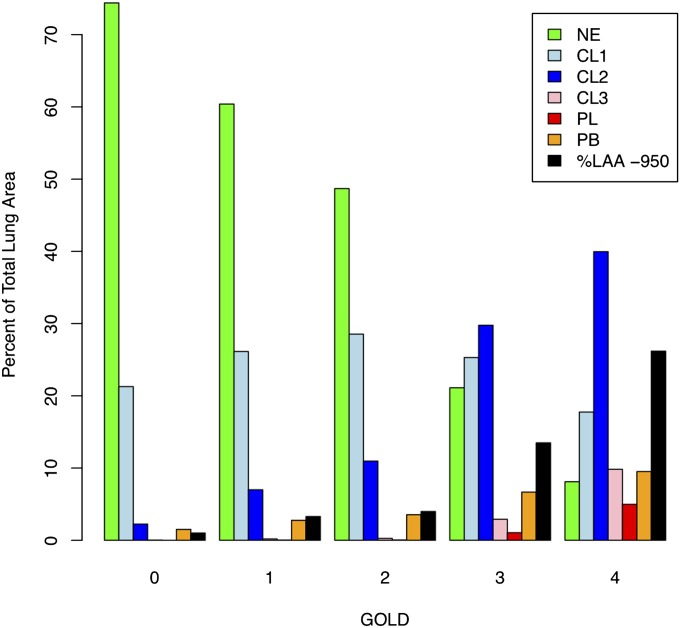
The median percentage of each LHE pattern by GOLD stage is shown in Figure 2. As expected, the amount of LHE increases with increasing GOLD stage, except for the NE and mild centrilobular patterns. The median amount of the mild centrilobular pattern is highest in GOLD 2 subjects, and it is present to a significant degree even in smoking control subjects.
Figure 2.
Median percentage of lung tissue with an attenuation value less than −950 Hounsfield units (%LAA-950) and each local histogram-based emphysema pattern stratified by Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage. CL1 = mild centrilobular; CL2 = moderate centrilobular; CL3 = severe centrilobular; GOLD 0 = control smokers; NE = nonemphysematous; PB = pleural-based; PL = panlobular.
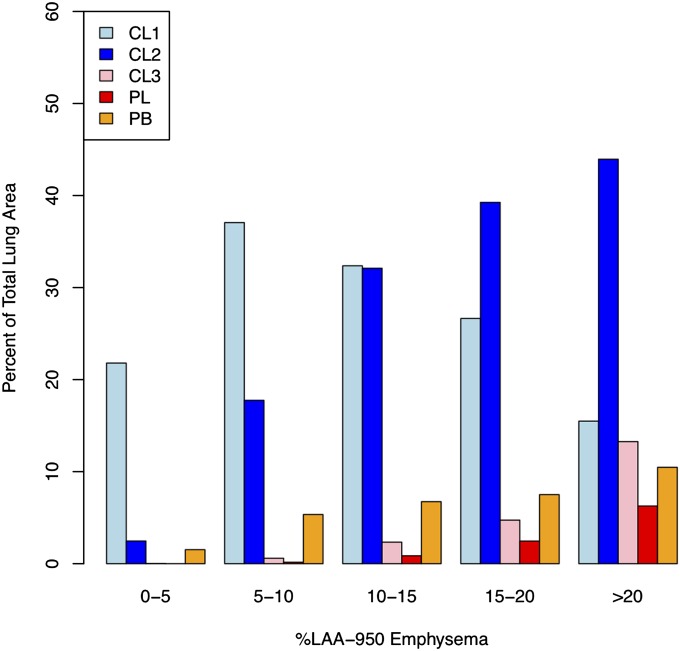
The relationship between LHE patterns and %LAA-950 is shown in Figure 3. The relative abundance of LHE patterns differs by the overall burden of %LAA-950. In individuals whose %LAA-950 emphysema was less than 10%, the mild centrilobular pattern is the predominant class of diseased lung. At greater than or equal to 10% LAA-950 emphysema, the moderate centrilobular pattern predominates, with an increasing amount of severe centrilobular, PL, and PB types as %LAA-950 emphysema increases.
Figure 3.
Median percentage of each pathologic local histogram-based emphysema pattern within strata of lung attenuation less than −950 Hounsfield units (%LAA-950). CL1 = mild centrilobular; CL2 = moderate centrilobular; CL3 = severe centrilobular; PB = pleural-based; PL = panlobular.
Relation of LHE Patterns to Each Other and to %LAA-950
The scatterplot matrix in Figure E5 demonstrates the highly nonlinear bivariate relation between some LHE patterns and %LAA-950. This nonlinearity is most pronounced for the mild centrilobular and PB pattern. There are two distinct populations of subjects with moderate levels of the mild centrilobular pattern: those with relatively low amounts of emphysema by standard quantitative measurement (i.e., %LAA-950), and those with a significant burden of %LAA-950. In contrast, the severe centrilobular and PL patterns show a roughly linear relation with %LAA-950. The Spearman correlation between %LAA-950 and the moderate centrilobular, severe centrilobular, and PL patterns ranges from 0.82–0.96 (see Table E5). PB emphysema also shows a strong correlation with %LAA-950 of 0.89, although visual inspection clearly demonstrates a nonlinear relationship.
Association of LHE Patterns with Physiologic, Functional Measures, and Health Care Use
All LHE patterns were strongly associated with multiple measures of respiratory physiology, functional status, and health care use (see Tables E6–E8). Most of these associations remained significant after adjustment for %LAA-950, suggesting that LHE patterns provide independent information from that provided by %LAA-950.
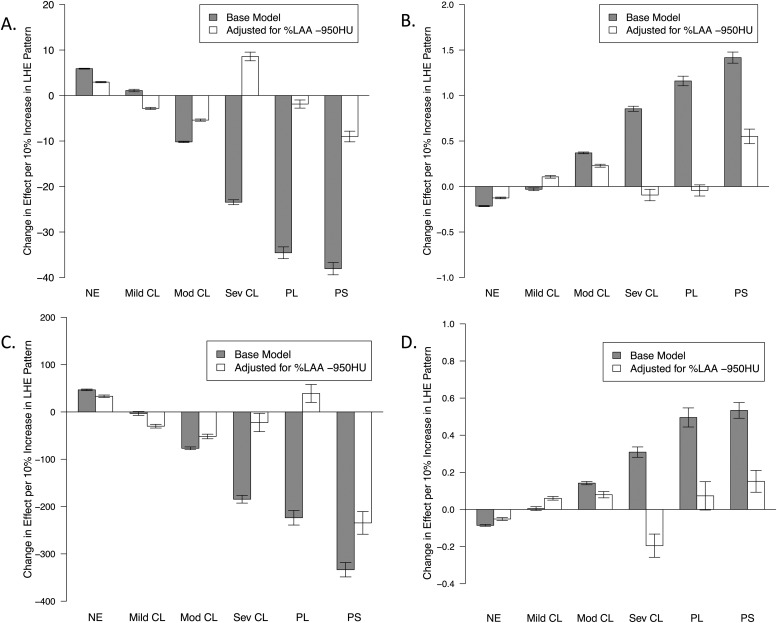
The estimated impact for each 10% increase in each LHE pattern is shown in Figure 4 for key COPD-related measures of airflow obstruction; dyspnea (MMRC score); functional status (6MWD); and exacerbation frequency. Before adjustment for %LAA-950, the effect of each emphysema type increases with the severity of LHE pattern (i.e., normal < mild centrilobular < moderate centrilobular < severe centrilobular < PL). The PB emphysema pattern was generally associated with the largest relative impact, a pattern that persisted after adjustment for %LAA-950. After %LAA-950 adjustment, the effect of moderate centrilobular emphysema was attenuated but remained highly significant (P < 10−6 for all three outcomes), and the effects for the mild centrilobular, severe centrilobular, and PL patterns were highly attenuated or, in some cases, reversed.
Figure 4.
Estimated effect sizes from separate regression models relating each local histogram emphysema pattern to chronic obstructive pulmonary disease (COPD)–related measures of disease severity. Barplots show the estimated effect of a 10% increase in each local histogram-based emphysema pattern on FEV1 % of predicted (A), Modified Medical Research Council (MMRC) dyspnea score (B), 6-minute walk distance (C), and number of COPD exacerbations in the past year (D). CL1 = mild centrilobular; CL2 = moderate centrilobular; CL3 = severe centrilobular; NE = nonemphysematous pattern; PB = pleural-based; PL = panlobular.
Relative Predictive Power of LHE Patterns and %LAA-950
To compare the predictive ability of LHE patterns versus %LAA-950 for key COPD-related physiologic, functional, and health care use measures, we examined the model fit and percentage of variance explained by models incorporating each measure as a predictor. We constructed three sets of regression models for each outcome relating (1) LHE patterns, (2) %LAA-950, and (3) both LHE and %LAA-950 to the following outcomes: FEV1% of predicted, 6MWD, MMRC dyspnea score, and number of exacerbations over the past year. For each outcome, the model incorporating LHE patterns provides a significantly better fit to the observed data than the model using %LAA-950 alone (P < 5 × 10−6 in all cases) and explains an additional 1–4% of phenotypic variance (see Table E9). For FEV1, 6MWD, and MMRC, the best model was that which included both LHE and %LAA-950 (P < 5 × 10−6 for all). For the number of exacerbations over the past year there was no significant difference between the LHE model and the model including both LHE and %LAA-950 (P = 0.92). Comparison of the LHE patterns against %LAA-910 also showed the LHE patterns were more predictive of these four COPD-related outcomes (see Table E12), and the relative performance of the LHE measures was insensitive to the exclusion of GOLD 1 subjects from analysis (see Table E13).
Mild Centrilobular Pattern as an Early Disease Indicator in Smokers with Preserved Lung Function
Given the relatively large burden of the mild centrilobular pattern in control smokers, we conducted association analyses limited to GOLD 0 smokers to test the association between LHE patterns and physiologic and functional measures in “healthy” smokers (see Table E14). In control smokers, the mild centrilobular pattern is significantly associated with lower FEV1, more dyspnea, shorter 6MWD, and a higher likelihood of steroid inhaler use (P < 5 × 10−6, 0.003, < 5 × 10−6, and 0.0004, respectively).
Discussion
In this paper we describe the epidemiologic associations between distinct CT local attenuation patterns and physiologic, functional, and health care use measures in a large cohort of smokers. We demonstrate that these patterns are strongly associated with a wide range of COPD-related physiologic, functional, and health care use measures, and we show that, compared with %LAA-950, these patterns are more predictive of FEV1, MMRC dyspnea score, 6MWD, and annual number of exacerbations. Thus, compared with a standard measure of quantitative emphysema, these distinct CT patterns provide novel, independent information relevant to COPD epidemiology and phenotyping.
There have been several investigations focused on the detection and classification of parenchymal disease on CT scan. Although collectively these methods may be referred to as textural analysis, each varies in its approach and clinical application. Chabat and colleagues (29) used a 13-feature vector and (29) Bayesian classifier to discriminate parenchymal disease type in 33 subjects. More recently Park and coworkers (28) applied a texture-based classifier to the CT scans of 39 subjects with emphysema and demonstrated that disease type may be a stronger predictor of lung function than %LAA. Uppaluri, Xu, and Hoffman were able to demonstrate in multiple publications that the adaptive multiple feature method technique could detect distinct patterns of emphysema on CT with reasonable agreement with human observers and excellent reproducibility (27, 39, 40). Sørenson and coworkers (26) investigated multiple texture-based techniques using CT data from 39 smokers and determined that a method using local binary patterns combined with intensity histograms provided the best performance in a cross-validation study. Ginsburg and coworkers (41) trained a multiple regression classifier with run and gap-length textural features to detect emphysema patterns in 105 CT scans from smokers and nonsmokers. The emphysema classification approach applied in this paper uses local lung density histogram information estimated by means of a kernel density estimator to classify parenchymal regions according to their emphysema status using a kNN classifier. This more parsimonious technique allows for a high degree of computational efficiency and scalability to large data sets. It has been previously shown that local lung density is the most discriminative feature in terms of classification performance when compared with local binary patterns with and without local density information (30).
This paper presents the first large-scale epidemiologic investigation of distinct emphysema patterns with physiologic and functional measures in smokers representing the full spectrum of smoking-related emphysema. The emphysema patterns identified by the local histogram method are based on the three main classes of emphysema as defined by pathology (i.e., centrilobular, PL, and paraseptal). The high prevalence of the mild centrilobular pattern in control smokers coupled with the association of this pattern with worse lung function and functional status suggests that radiographic evidence of clinically relevant emphysematous destruction may be detectable even in control smokers. The moderate centrilobular pattern shows strong associations and large effects with many clinically relevant measures, and these associations persist after adjustment for %LAA-950 indicating that this pattern provides a strong signal for emphysematous lung destruction across GOLD stages. The severe centrilobular and PL patterns are less prevalent than the mild and moderate centrilobular patterns, and they are highly correlated with %LAA-950, suggesting that the information captured by these patterns is largely captured by %LAA-950. The PB pattern detects emphysema associated with a pleural surface, and interestingly this pattern shows the strongest effects for many of the measures studied. Although this suggests that PB emphysema may be a marker of severe disease, it should also be noted that the prevalence of this pattern is low compared with the mild and moderate centrilobular patterns. It is more accurate to describe this pattern as PB rather than paraseptal emphysema, because the pattern captured by the algorithm includes emphysema that may be a contiguous extension of centrilobular and PL emphysema. The automatic detection of paraseptal emphysema poses unique challenges and requires information about relative position within the lung in addition to textural information. Additional work needs to be done to improve the automatic identification of this emphysema type.
Our findings shed new light on traditional densitometric measures, such as %LAA-950. LHE patterns vary greatly by overall level of %LAA-950 quantified emphysema, suggesting that there is a great deal of diversity in emphysema pattern that is not captured by %LAA-950. In addition to providing better understanding of densitometric measures, LHE patterns provide phenotypic information above and beyond that provided by %LAA-950 emphysema. When the predictive power of LHE and %LAA-950 emphysema were compared in multivariate models for FEV1, MMRC score, 6MWD, and exacerbation frequency, the LHE patterns explained a larger proportion of variance than %LAA-950 emphysema, although both emphysema quantification measures provided some degree of independent information.
One of the limitations of this study is that it does not assess the correlation between local histogram emphysema patterns and pathology. Although LHE patterns are based on an expert’s visual assessment, the relationship between local histogram measures, visual emphysema assessments, and pathologic emphysema requires further investigation. We present extensive data in the online supplement on the repeatability and visual confirmation of these patterns. Such confirmation is important for the interpretation and validation of emphysema patterns; however, the validity of the clinical associations observed in this paper does not depend on the correlation of these measures with pathology or visual assessments. There are limitations to the use of visual emphysema in particular as a gold standard for quantitative emphysema assessment, because the interobserver agreement in visual assessment of emphysema patterns has been shown to be fair to moderate (39, 42).
Many ROIs do not perfectly overlap a secondary lobule and may capture heterogeneous areas of emphysematous involvement. However, the variability of the LHE percentages for two different spacing schemes for the ROI center was less than 0.5% in median value (see Figure E4). Despite this limitation, strong associations with COPD-associated measures were observed with LHE patterns, suggesting that clustering of emphysema types in the lung may have minimized the effect of ROI heterogeneity. This observation is in line with theoretical work suggesting that a complex interplay of forces, including mechanical forces, promotes the local propagation of emphysema (43).
Our LHE classification is based on distinct emphysema patterns defined according to the Fleischner Society (44). However, it is clear that in our data certain patterns, such as the CL3 and PL, are highly correlated with each other and with %LAA-950, raising the question of how truly distinct are these patterns. Additional work remains to be done integrating information from data-driven emphysema quantification algorithms and expert-derived emphysema classification patterns. Data-driven classification algorithms, such as the LHE approach, can offer a different perspective on fundamental questions, such as the number of different subtypes of emphysema.
Using a novel classification technique based on local density histograms, we quantified the amount of distinct emphysema patterns in CT scans from 9,313 smokers in the COPDGene study. The associations between these patterns and various physiologic, functional, and health care use measures suggest that information relating to distinct CT emphysema patterns provides novel insight into COPD epidemiology and can be more predictive of COPD-related measures than %LAA-950.
Acknowledgments
COPDGene Investigators—Core Units:
Administrative Core: James Crapo, M.D. (PI), Edwin Silverman, M.D., Ph.D. (PI), Barry Make, M.D., Elizabeth Regan, M.D., Ph.D., Rochelle Lantz, Lori Stepp, and Sandra Melanson
Genetic Analysis Core: Terri Beaty, Ph.D., Barbara Klanderman, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Peter Castaldi, M.D., M.Sc., Merry-Lynn McDonald, Ph.D., Jing Zhou, M.D., Ph.D., Manuel Mattheissen, M.D., Ph.D., Emily Wan, M.D., Megan Hardin, M.D., Jacqueline Hetmanski, M.S., Margaret Parker, M.S., and Tanda Murray, M.S.
Imaging Core: David Lynch, M.B., Joyce Schroeder, M.D., John Newell, Jr., M.D., John Reilly, M.D., Harvey Coxson, Ph.D., Philip Judy, Ph.D., Eric Hoffman, Ph.D., George Washko, M.D., Raúl San José Estépar, Ph.D., James Ross, M.Sc., Mustafa Al Qaisi, M.D., Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, and Douglas Stinson
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, Ph.D.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, Ph.D., Stacey Meyerer, Shivam Chandan, and Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D., Andre Williams, Ph.D., Carla Wilson, M.S., Anna Forssen, M.S., Amber Powell, and Joe Piccoli
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, M.P.H., Ph.D., Marci Sontag, Ph.D., Jennifer Black-Shinn, M.P.H., Gregory Kinney, M.P.H., Ph.Dc., Sharon Lutz, M.P.H., Ph.D.
COPDGene Investigators – Clinical Centers:
Ann Arbor VA, Ann Arbor, MI: Jeffrey Curtis, M.D., and Ella Kazerooni, M.D.
Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S., Philip Alapat, M.D., Venkata Bandi, M.D., Kalpalatha Guntupalli, M.D., Elizabeth Guy, M.D., Antara Mallampalli, M.D., Charles Trinh, M.D., Mustafa Atik, M.D., Hasan Al-Azzawi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., and Anne Marie Marciel, M.D.
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H., Craig Hersh, M.D., M.P.H., George Washko, M.D., Francine Jacobson, M.D., M.P.H., Hiroto Hatabu, M.D., Ph.D., Peter Clarke, M.D., Ritu Gill, M.D., Andetta Hunsaker, M.D., Beatrice Trotman-Dickenson, M.B.B.S., and Rachna Madan, M.D.
Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H., Byron Thomashow, M.D., John Austin, M.D., and Belinda D’Souza, M.D.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D., Lacey Washington, M.D., and H. Page McAdams, M.D.
Fallon Clinic, Worcester, MA: Richard Rosiello, M.D., Timothy Bresnahan, M.D., Joseph Bradley, M.D., Sharon Kuong, M.D., Steven Meller, M.D., and Suzanne Roland, M.D.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H., and Joseph Tashjian, M.D.
Johns Hopkins University, Baltimore, MD: Robert Wise, M.D., Nadia Hansel, M.D., M.P.H., Robert Brown, M.D., Gregory Diette, M.D., and Karen Horton, M.D.
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, M.D., Janos Porszasz, M.D., Ph.D., Hans Fischer, M.D., Ph.D., Matt Budoff, M.D., and Mehdi Rambod, M.D.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, M.D., Charles Trinh, M.D., Hirani Kamal, M.D., Roham Darvishi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., and Anne Marie Marciel, M.D.
Minneapolis VA, Minneapolis, MN: Dennis Niewoehner, M.D., Quentin Anderson, M.D., Kathryn Rice, M.D., and Audrey Caine, M.D.
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S., Gloria Westney, M.D., M.S., and Eugene Berkowitz, M.D., Ph.D.
National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D., David Lynch, M.B., Joyce Schroeder, M.D., Valerie Hale, M.D., John Armstrong, II, M.D., Debra Dyer, M.D., Jonathan Chung, M.D., and Christian Cox, M.D.
Temple University, Philadelphia, PA: Gerard Criner, M.D., Victor Kim, M.D., Nathaniel Marchetti, D.O., Aditi Satti, M.D., A. James Mamary, M.D., Robert Steiner, M.D., Chandra Dass, M.D., and Libby Cone, M.D.
University of Alabama, Birmingham, AL: William Bailey, M.D., Mark Dransfield, M.D., Michael Wells, M.D., Surya Bhatt, M.D., Hrudaya Nath, M.D., and Satinder Singh, M.D.
University of California, San Diego, CA: Joe Ramsdell, M.D., and Paul Friedman, M.D.
University of Iowa, Iowa City, IA: Alejandro Cornellas, M.D., John Newell, Jr., M.D., and Edwin J.R. van Beek, M.D., Ph.D.
University of Michigan, Ann Arbor, MI: Fernando Martinez, M.D., MeiLan Han, M.D., and Ella Kazerooni, M.D.
University of Minnesota, Minneapolis, MN: Christine Wendt, M.D., and Tadashi Allen, M.D.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D., Joel Weissfeld, M.D., M.P.H., Carl Fuhrman, M.D., Jessica Bon, M.D., and Danielle Hooper, M.D.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D., Sandra Adams, M.D., Carlos Orozco, M.D., Mario Ruiz, M.D., Amy Mumbower, M.D., Ariel Kruger, M.D., Carlos Restrepo, M.D., and Michael Lane, M.D.
Footnotes
Supported by NIH grants P01HL105339 (E.K.S.), K08HL102265 (P.J.C.), K25HL104085 and R01HL116931 (R.S.J.E.), K23HL089353 and HL107246-03 (G.R.W.), and TEC2010-21619-C04-02 (C.S.M., CICYT, Spain). The project described was supported by Award Number R01HL089897 (J.D.C.) and Award Number R01HL089856 (E.K.S.) from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the NIH. The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, and Sunovion.
Author Contributions: Conception and design, P.J.C., R.S.J.E., E.K.S., and G.R.W. Acquisition, analysis, and/or interpretation, P.J.C., R.S.J.E., N.L., J.D.C., D.A.L., E.K.S., and G.R.W. Drafting the manuscript for important intellectual content, P.J.C., R.S.J.E., C.S.M., C.P.H., N.L., J.D.C., D.A.L., E.K.S., and G.R.W.
Originally Published in Press as DOI: 10.1164/rccm.201305-0873OC on August 27, 2013
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ciba Guest Symposium Report. Definitions and classifications of chronic obstructive pulmonary emphysema and related conditions. Thorax. 1959;14:286–299. [Google Scholar]
- 2.Washko GR, Criner GJ, Mohsenifar Z, Sciurba FC, Sharafkhaneh A, Make BJ, Hoffman EA, Reilly JJ. Computed tomographic-based quantification of emphysema and correlation to pulmonary function and mechanics. COPD. 2008;5:177–186. doi: 10.1080/15412550802093025. [DOI] [PubMed] [Google Scholar]
- 3.Mohamed Hoesein FA, de Hoop B, Zanen P, Gietema H, Kruitwagen CL, van Ginneken B, Isgum I, Mol C, van Klaveren RJ, Dijkstra AE, et al. CT-quantified emphysema in male heavy smokers: association with lung function decline. Thorax. 2011;66:782–787. doi: 10.1136/thx.2010.145995. [DOI] [PubMed] [Google Scholar]
- 4.Marsh S, Aldington S, Williams MV, Nowitz M, Kingzett-Taylor A, Weatherall M, Shirtcliffe P, Pritchard A, Beasley R. Physiological associations of computerized tomography lung density: a factor analysis. Int J Chron Obstruct Pulmon Dis. 2006;1:181–187. doi: 10.2147/copd.2006.1.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould GA, Redpath AT, Ryan M, Warren PM, Best JJ, Flenley DC, MacNee W. Lung CT density correlates with measurements of airflow limitation and the diffusing capacity. Eur Respir J. 1991;4:141–146. [PubMed] [Google Scholar]
- 6.Lee YK, Oh YM, Lee JH, Kim EK, Lee JH, Kim N, Seo JB, Lee SD KOLD Study Group. Quantitative assessment of emphysema, air trapping, and airway thickening on computed tomography. Lung. 2008;186:157–165. doi: 10.1007/s00408-008-9071-0. [DOI] [PubMed] [Google Scholar]
- 7.Grydeland TB, Dirksen A, Coxson HO, Eagan TM, Thorsen E, Pillai SG, Sharma S, Eide GE, Gulsvik A, Bakke PS. Quantitative computed tomography measures of emphysema and airway wall thickness are related to respiratory symptoms. Am J Respir Crit Care Med. 2010;181:353–359. doi: 10.1164/rccm.200907-1008OC. [DOI] [PubMed] [Google Scholar]
- 8.Diaz AA, Bartholmai B, San José Estépar R, Ross J, Matsuoka S, Yamashiro T, Hatabu H, Reilly JJ, Silverman EK, Washko GR. Relationship of emphysema and airway disease assessed by CT to exercise capacity in COPD. Respir Med. 2010;104:1145–1151. doi: 10.1016/j.rmed.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambod M, Porszasz J, Make BJ, Crapo JD, Casaburi R. Six minute walk distance predictors, including computed tomography measures, in the COPDGene(R) cohort. Chest. 2012;141:867–875. doi: 10.1378/chest.11-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai SG, Kong X, Edwards LD, Cho MH, Anderson WH, Coxson HO, Lomas DA, Silverman EK ECLIPSE and ICGN Investigators. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:1498–1505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sørheim IC, DeMeo DL, Washko G, Litonjua A, Sparrow D, Bowler R, Bakke P, Pillai SG, Coxson HO, Lomas DA, et al. International COPD Genetics Network Investigators. Polymorphisms in the superoxide dismutase-3 gene are associated with emphysema in COPD. COPD. 2010;7:262–268. doi: 10.3109/15412555.2010.496821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, et al. ICGN Investigators. Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. Am J Respir Cell Mol Biol. 2009;41:324–331. doi: 10.1165/rcmb.2008-0427OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, et al. ECLIPSE Study NETT Investigators. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183:43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambrechts D, Buysschaert I, Zanen P, Coolen J, Lays N, Cuppens H, Groen HJ, Dewever W, van Klaveren RJ, Verschakelen J, et al. The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am J Respir Crit Care Med. 2010;181:486–493. doi: 10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 15.Ito I, Nagai S, Handa T, Muro S, Hirai T, Tsukino M, Mishima M. Matrix metalloproteinase-9 promoter polymorphism associated with upper lung dominant emphysema. Am J Respir Crit Care Med. 2005;172:1378–1382. doi: 10.1164/rccm.200506-953OC. [DOI] [PubMed] [Google Scholar]
- 16.DeMeo DL, Hersh CP, Hoffman EA, Litonjua AA, Lazarus R, Sparrow D, Benditt JO, Criner G, Make B, Martinez FJ, et al. Genetic determinants of emphysema distribution in the national emphysema treatment trial. Am J Respir Crit Care Med. 2007;176:42–48. doi: 10.1164/rccm.200612-1797OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minematsu N, Nakamura H, Iwata M, Tateno H, Nakajima T, Takahashi S, Fujishima S, Yamaguchi K. Association of CYP2A6 deletion polymorphism with smoking habit and development of pulmonary emphysema. Thorax. 2003;58:623–628. doi: 10.1136/thorax.58.7.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakao S, Tatsumi K, Igari H, Watanabe R, Shino Y, Shirasawa H, Kuriyama T. Association of tumor necrosis factor-alpha gene promoter polymorphism with low attenuation areas on high-resolution CT in patients with COPD. Chest. 2002;122:416–420. doi: 10.1378/chest.122.2.416. [DOI] [PubMed] [Google Scholar]
- 19.Stolk J, Putter H, Bakker EM, Shaker SB, Parr DG, Piitulainen E, Russi EW, Grebski E, Dirksen A, Stockley RA, et al. Progression parameters for emphysema: a clinical investigation. Respir Med. 2007;101:1924–1930. doi: 10.1016/j.rmed.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Zulueta JJ, Wisnivesky JP, Henschke CI, Yip R, Farooqi AO, McCauley DI, Chen M, Libby DM, Smith JP, Pasmantier MW, et al. Emphysema scores predict death from chronic obstructive pulmonary disease and lung cancer. Chest. 2012;141:1216–1223. doi: 10.1378/chest.11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haruna A, Muro S, Nakano Y, Ohara T, Hoshino Y, Ogawa E, Hirai T, Niimi A, Nishimura K, Chin K, et al. CT scan findings of emphysema predict mortality in COPD. Chest. 2010;138:635–640. doi: 10.1378/chest.09-2836. [DOI] [PubMed] [Google Scholar]
- 22.Müller NL, Staples CA, Miller RR, Abboud RT. “Density mask.” An objective method to quantitate emphysema using computed tomography. Chest. 1988;94:782–787. doi: 10.1378/chest.94.4.782. [DOI] [PubMed] [Google Scholar]
- 23.Hruban RH, Meziane MA, Zerhouni EA, Khouri NF, Fishman EK, Wheeler PS, Dumler JS, Hutchins GM. High resolution computed tomography of inflation-fixed lungs. Pathologic-radiologic correlation of centrilobular emphysema. Am Rev Respir Dis. 1987;136:935–940. doi: 10.1164/ajrccm/136.4.935. [DOI] [PubMed] [Google Scholar]
- 24.Spouge D, Mayo JR, Cardoso W, Müller NL. Panacinar emphysema: CT and pathologic findings. J Comput Assist Tomogr. 1993;17:710–713. [PubMed] [Google Scholar]
- 25.Webb WR. Thin-section CT of the secondary pulmonary lobule: anatomy and the image—the 2004 Fleischner lecture. Radiology. 2006;239:322–338. doi: 10.1148/radiol.2392041968. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen L, Shaker SB, de Bruijne M. Quantitative analysis of pulmonary emphysema using local binary patterns. IEEE Trans Med Imaging. 2010;29:559–569. doi: 10.1109/TMI.2009.2038575. [DOI] [PubMed] [Google Scholar]
- 27.Uppaluri R, Mitsa T, Sonka M, Hoffman EA, McLennan G. Quantification of pulmonary emphysema from lung computed tomography images. Am J Respir Crit Care Med. 1997;156:248–254. doi: 10.1164/ajrccm.156.1.9606093. [DOI] [PubMed] [Google Scholar]
- 28.Park YS, Seo JB, Kim N, Chae EJ, Oh YM, Lee SD, Lee Y, Kang SH. Texture-based quantification of pulmonary emphysema on high-resolution computed tomography: comparison with density-based quantification and correlation with pulmonary function test. Invest Radiol. 2008;43:395–402. doi: 10.1097/RLI.0b013e31816901c7. [DOI] [PubMed] [Google Scholar]
- 29.Chabat F, Yang GZ, Hansell DM. Obstructive lung diseases: texture classification for differentiation at CT. Radiology. 2003;228:871–877. doi: 10.1148/radiol.2283020505. [DOI] [PubMed] [Google Scholar]
- 30.Mendoza CS, Washko GR, Ross J, Diaz AA, Lynch DA, Crapo JD, Silverman EK, Begona A, Serrano C, Estepar RSJ.Emphysema quantification in a multi-scanner HRCT cohort using local intensity distributions. Proc IEEE Int Symp Biomed Imaging2012:474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castaldi PJ, Estepar RS, Mendoza CS, Hersh CP, Laird N, Crapo J, Lynch DA, Silverman EK, Washko GR.Distinct quantitative CT emphysema patterns are associated with physiology and function in smokers. Am J Respir Crit Care Med2013;188:A3728. [DOI] [PMC free article] [PubMed]
- 32.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 34.Jones PW, Quirk FH, Baveystock CM, Littlejohns P. A self-complete measure of health status for chronic airflow limitation. The St. George’s Respiratory Questionnaire. Am Rev Respir Dis. 1992;145:1321–1327. doi: 10.1164/ajrccm/145.6.1321. [DOI] [PubMed] [Google Scholar]
- 35.Bestall JC, Paul EA, Garrod R, Garnham R, Jones PW, Wedzicha JA. Usefulness of the Medical Research Council (MRC) dyspnoea scale as a measure of disability in patients with chronic obstructive pulmonary disease. Thorax. 1999;54:581–586. doi: 10.1136/thx.54.7.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botev ZI, Grotowski JF, Kroese DP. Kernel density estimation via diffusion. Ann Stat. 2010;38:2916–2957. [Google Scholar]
- 37.Fletcher CM. Standardized questionaries on respiratory symptoms. Br Med J. 1960;2:1665. [Google Scholar]
- 38.R Development Core Team R: a language and environment for statistical computing. 2011. Available from: http://www.R-project.org
- 39.Uppaluri R, Hoffman EA, Sonka M, Hartley PG, Hunninghake GW, McLennan G. Computer recognition of regional lung disease patterns. Am J Respir Crit Care Med. 1999;160:648–654. doi: 10.1164/ajrccm.160.2.9804094. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Sonka M, McLennan G, Guo J, Hoffman EA. MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE Trans Med Imaging. 2006;25:464–475. doi: 10.1109/TMI.2006.870889. [DOI] [PubMed] [Google Scholar]
- 41.Ginsburg SB, Lynch DA, Bowler RP, Schroeder JD. Automated texture-based quantification of centrilobular nodularity and centrilobular emphysema in chest CT images. Acad Radiol. 2012;19:1241–1251. doi: 10.1016/j.acra.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr RG, Berkowitz EA, Bigazzi F, Bode F, Bon J, Bowler RP, Chiles C, Crapo JD, Criner GJ, Curtis JL, et al. COPDGene CT Workshop Group. A combined pulmonary-radiology workshop for visual evaluation of COPD: study design, chest CT findings and concordance with quantitative evaluation. COPD. 2012;9:151–159. doi: 10.3109/15412555.2012.654923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suki B, Lutchen KR, Ingenito EP. On the progressive nature of emphysema: roles of proteases, inflammation, and mechanical forces. Am J Respir Crit Care Med. 2003;168:516–521. doi: 10.1164/rccm.200208-908PP. [DOI] [PubMed] [Google Scholar]
- 44.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246:697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]