Abstract
Rationale: Gene promoter methylation detected in sputum predicts lung cancer risk in smokers. Compared with non-Hispanic whites (NHW), Hispanics have a lower age-standardized incidence for lung cancer.
Objectives: This study compared the methylation prevalence in sputum of NHWs with Hispanics using the Lovelace Smokers cohort (n = 1998) and evaluated the effect of Native American ancestry (NAA) and diet on biomarkers for lung cancer risk.
Methods: Genetic ancestry was estimated using 48 ancestry markers. Diet was assessed by the Harvard University Dietary Assessment questionnaire. Methylation of 12 genes was measured in sputum using methylation-specific polymerase chain reaction. The association between NAA and risk for methylation was assessed using generalized estimating equations. The ethnic difference in the association between pack-years and risk for lung cancer was assessed in the New Mexico lung cancer study.
Measurements and Main Results: Overall Hispanics had a significantly increased risk for methylation across the 12 genes analyzed (odds ratio, 1.18; P = 0.007). However, the risk was reduced by 32% (P = 0.032) in Hispanics with high versus low NAA. In the New Mexico lung cancer study, Hispanic non–small cell lung cancer cases have significantly lower pack-years than NHW counterparts (P = 0.007). Furthermore, compared with NHW smokers, Hispanic smokers had a more rapidly increasing risk for lung cancer as a function of pack-years (P = 0.058).
Conclusions: NAA may be an important risk modifier for methylation in Hispanic smokers. Smoking intensity may have a greater impact on risk for lung cancer in Hispanics compared with NHWs.
Keywords: ethnicity, sputum, diet, risk, lung cancer
At a Glance Commentary
Scientific Knowledge on the Subject
The detection of promoter methylation in exfoliated lung cells provides an assessment of the extent of field cancerization and is a validated biomarker for predicting lung cancer risk in smokers. Compared with non-Hispanic whites, New Mexican Hispanics have lower age-standardized incidence rates for lung cancer and lower prevalence of chronic obstructive pulmonary disease that may be linked in part to lower cigarette smoking, Native American ancestry, and lifestyle.
What This Study Adds to the Field
Although Native American genetic ancestry in Hispanics is protective against acquiring gene methylation, Hispanics on average still have a significantly increased risk for promoter methylation. Using a lung cancer case-control study, we also provide the first evidence that Hispanics may actually be more susceptible for cigarette smoking–induced lung cancer. In addition, the higher risk for methylation in Hispanics may in part be explained by lower folate intake and multivitamin use in this ethnic group.
Lung cancer is the leading cause of cancer death in men and women in the United States mainly because of a lack of established early screening strategies. Silencing of tumor-suppressor genes through promoter methylation is a major and causal epigenetic event that occurs during lung cancer initiation and progression (1). The detection of promoter methylation in exfoliated lung cells collected in sputum samples provides an assessment of the extent of field cancerization in the lungs and is a validated biomarker for identifying smokers at higher risk for lung cancer incidence (2, 3). Several studies conducted by our group and using the Lovelace Smokers cohort (LSC) identified sex, dietary factors, and variants in genes involved in DNA replication, DNA damage response, and one-carbon metabolism and transsulfuration pathways as important factors affecting this epigenetic alteration in the lungs of smokers (4–7).
Hispanics are one of the fastest growing populations in the United States, accounting for 16.3% (50.5 million per 310 million) of the US population in 2010 (8). New Mexico has the highest percentage (45%) of Hispanics of any state, with 83% of these native-born and 17% foreign-born (9). New Mexican Hispanics display relatively higher European and Native American ancestry and lower African ancestry compared with Hispanics in Puerto Rico and the Southeastern United States (10). Although Hispanics in the United States have worse average socioeconomic indicators, Hispanics tend to have comparable or even better health outcomes than non-Hispanic whites (NHW) (11). This “Hispanic paradox” has been applied to smoking-induced pulmonary diseases, such as chronic obstructive pulmonary disease (COPD), with an observed 50% lower prevalence of COPD in Hispanics (12). Hispanics also have 40–50% lower age-standardized incidence rates for lung cancer compared with NHWs in New Mexico and nationwide (8, 13). These observations have been suggested to be mainly caused by the traditionally lower rates of cigarette smoking and fewer cigarettes per day consumed by smokers (14). However, among smokers who smoke less than 20 cigarettes per day, Hispanics still have a risk of lung cancer that is approximately one-half to one-third that of NHWs (15). In addition, New Mexican Hispanic smokers have 50% reduced odds of COPD and reduced pulmonary function decline compared with NHWs even after controlling for smoking history (12). Thus, these studies suggest that factors other than tobacco smoke may contribute to the observed lower incidence and/or prevalence of pulmonary diseases in Hispanic smokers. Because Hispanics have a significant amount of Native American ancestry (NAA) and Native Americans have the lowest risk for lung cancer among New Mexican populations (13), NAA may contribute to the lower incidence and/or prevalence for smoking-induced pulmonary diseases in Hispanics. This premise was strongly supported by studies demonstrating that higher NAA is associated with better pulmonary function and lower odds of COPD in Hispanics (12, 16).
Accumulating evidence from our group and others suggests that subjects with different ethnic backgrounds may have different susceptibility for acquiring gene methylation during carcinogenesis. NHW patients with lung cancer have significantly higher prevalence of O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in tumors (31–45%) than East Asians (2–16%) (17). Our recent study suggested that the ethnic difference of MGMT methylation in lung cancer may be largely accounted for by a promoter-enhancer single-nucleotide polymorphism (rs16906252) that regulates MGMT gene expression and has a large difference in allele frequency between NHWs and East Asians (15–20% vs. 0%) (18; S. Leng unpublished data). Furthermore, global methylation levels in blood leukocytes and tumors were also affected by ethnicity (19, 20). Based on the fact that Hispanics have lower prevalence for smoking-induced pulmonary diseases and that gene methylation detected in sputum is an intermediate biomarker for lung cancer, we tested the hypothesis that NAA would modify the risk for acquiring gene methylation in the lungs of Hispanic smokers from the LSC.
Methods
Lovelace Smokers Cohort
The LSC was established in 2001 to conduct longitudinal studies on biomarkers of respiratory carcinogenesis and diseases in biologic specimens from smokers at risk for lung cancer (7). Enrollment was restricted to current and former smokers age 40–74 years with a minimum of 10 pack-years of smoking. A detailed questionnaire written in English was used to collect information on demographics; medical, smoking, and exposure history; socioeconomic status; and quality of life. Participants completed the adult version of the Harvard University Dietary Assessment questionnaire, a self-administered instrument that includes approximately 150 food items (4). Sputum was collected by induction and stored in Saccomanno fixative. All participants signed a consent form, and the Western Institutional Review Board approved this project.
New Mexico Lung Cancer Study
The New Mexico lung cancer study is a population-based case-control study that has been enrolling lung cancer cases through the Veterans Hospital and the University of New Mexico Hospital since 2004 (21). Control subjects with no history of any cancer were recruited from the Veterans Smokers Cohort and the LSC and were frequency matched to cases by age group (by 5-yr intervals), sex, smoking status (current vs. former), and cohort. A total of 435 incident non–small cell lung cancer (NSCLC) cases and 474 control subjects who are either NHW or Hispanic smokers were identified and included in data analysis with demographics described in Table E1 in the online supplement.
Gene Promoter Methylation
Twelve genes were selected for methylation analysis in cytologically adequate sputum samples based on our previous studies establishing their association with risk for lung cancer and their specificity to methylation in lung epithelial cells (1–3). Given the low percentage (<3%) of lung epithelial cells in sputum samples that also varied significantly between individuals, a two-stage nested methylation-specific PCR (MSP) was used to detect methylated alleles (1–3). The gene names and primer sequences for MSP are listed in Table E2. Methylation status for each individual gene was scored as 0 (unmethylated) or 1 (methylated). Our assay can reproducibly detect one methylated allele in a background of 10,000 unmethylated alleles (2).
Genotyping of Ancestry Informative Markers and Estimation of Genetic Ancestry
Illumina GoldenGate Genotyping assay (Illumina, Inc., San Diego, CA) was used to genotype 1,998 LSC subjects for 48 ancestry informative markers that have been used to establish precise estimates of genetic ancestry from African, European, Native American, and East Asian for each individual (12, 22; F. Gilliland, unpublished data). Genetic ancestry was analyzed using the Bayesian Markov Chain Monte Carlo algorithm implemented in STRUCTURE 2.3.3 (23) under the admixture model. Self-identified ethnicity was used as prior population information to enhance the detection of population admixture (23). Publicly available genotype data from three parental populations (24) was used to seed the STRUCTURE runs (Table 1).
TABLE 1.
GENETIC ANCESTRY ESTIMATES FOR THE LSC SUBJECTS AND THE PARENTAL POPULATIONS USING 48 ANCESTRY INFORMATIVE MARKERS
| Self-identified Ethnicity | n | Genetic Ancestry (Mean Proportion ± SD) |
||
|---|---|---|---|---|
| NAA | European | African | ||
| LSC subjects |
|
|
|
|
| NHW |
1,526 |
0.03 ± 0.03 |
0.97 ± 0.03 |
0.00 ± 0.02 |
| Hispanic |
418 |
0.35 ± 0.07 |
0.63 ± 0.07 |
0.02 ± 0.02 |
| Native American |
23 |
0.53 ± 0.29 |
0.47 ± 0.29 |
0.00 ± 0.00 |
| African American |
25 |
0.03 ± 0.04 |
0.24 ± 0.20 |
0.73 ± 0.19 |
| Parental populations |
|
|
|
|
| European |
144 |
0.03 ± 0.02 |
0.97 ± 0.02 |
0.00 ± 0.00 |
| African |
153 |
0.00 ± 0.00 |
0.01 ± 0.01 |
0.97 ± 0.02 |
| Native American | 64 | 0.93 ± 0.09 | 0.07 ± 0.09 | 0.00 ± 0.00 |
Definition of abbreviations: LSC = Lovelace Smokers cohort; NAA = Native American ancestry; NHW = non-Hispanic whites.
Statistical Analysis
The association analyses between ethnicity (Hispanic vs. NHW) and risk for gene methylation were restricted to 1,199 NHWs and 341 Hispanics in LSC with complete data for methylation, ethnicity, and covariates (Table 2). The association was assessed using generalized estimating equations (GEE) in gee package (25) with a vector of the methylation status of 12 genes (1 for methylated status and 0 for unmethylated status for each individual gene) for each individual smoker as the outcome. GEE modeling was conducted under the assumption that the methylation status for each individual gene in the outcome vector was binomially distributed and a logit link function was used. Furthermore, GEEs incorporate the correlation matrix among the 12 genes; the fixed covariance structure was derived from the combined sample with the assumption of no ethnic differences under the null hypothesis. Odds ratios (OR) and 95% confidence intervals (CI) were calculated to quantify the magnitude of the associations. This modeling approach is superior to others that usually use a composite methylation index as the end point because it considers the correlation structure among the 12-gene panel and increases the precision of the estimates. Several clinical variables, including age, sex, smoking history (smoking status and pack-years), overweight or obesity (body mass index ≥25), and chronic mucous hypersecretion status were selected a priori and included in the final GEE models for covariate adjustment. Social economic indicators (income and education) were not included in the final models because of lack of association with risk for gene methylation and having missing values (not shown, Table 2). The protection of NAA against acquiring gene methylation was assessed in Hispanics using the GEE models with adjustment for the covariates and European and African ancestry. Because of the low prevalence of RASSF1A methylation (<0.7%), the association analyses introduced above were also conducted based on an 11-gene panel. Results similar to the analyses based on the 12-gene panel were obtained (not shown). The association of individual gene methylation with ethnicity or NAA was also explored using logistic regression models, but viewed as secondary analyses to reduce the concern of multiple comparisons.
TABLE 2.
CHARACTERISTICS OF LSC SUBJECTS BY ETHNICITY
| Variable | Self-identified Ethnicity |
P Value | |
|---|---|---|---|
| Hispanic | NHW | ||
| n |
341 |
1,199 |
|
| Age, yr, mean ± SD |
54.4 ± 9.0 |
56.9 ± 9.5 |
1.4 × 10−5* |
| Sex, male, % |
27.8 |
20.5 |
0.004† |
| Current smokers, % |
73.0 |
54.2 |
5.1 × 10−10† |
| Pack-years, mean ± SD |
34.3 ± 16.1 |
41.1 ± 20.9 |
<0.0001‡ |
| 10–26, % |
34.0 |
22.8 |
2.2 × 10−6† |
| 26–37, % |
33.4 |
30.6 |
|
| 37–145, % |
32.6 |
46.5 |
|
| Age started smoking, yr, mean ± SD |
16.3 ± 3.7 |
16.7 ± 3.6 |
0.09* |
| Duration of smoking, yr, mean ± SD |
34.0 ± 8.8 |
33.6 ± 9.7 |
0.41‡ |
| Age quit smoking, yr, mean ± SD |
50.0 ± 9.7 |
48.5 ± 10.1 |
0.17* |
| Body mass index, kg/m2, mean ± SD |
28.8 ± 5.4 |
28.1 ± 5.7 |
0.048* |
| 16–25, % |
24.9 |
33.4 |
0.01† |
| 25–30, % |
39.9 |
36.0 |
|
| 30–61, % |
35.2 |
30.5 |
|
| Annual household income, %§ |
|
|
|
| Under $20,000 |
34.3 |
24.9 |
0.03† |
| $20,000–40,000 |
29.6 |
32.4 |
|
| Over $40,000 |
36.1 |
42.7 |
|
| Education level, %|| |
|
|
|
| Less than college |
55.6 |
24.5 |
<1 × 10−18† |
| Some college or above |
44.4 |
75.5 |
|
| Ancestry component, mean ± SD |
|
|
|
| European ancestry |
0.62 ± 0.07 |
0.97 ± 0.03 |
NC¶ |
| Native American ancestry |
0.36 ± 0.07 |
0.03 ± 0.03 |
NC¶ |
| African ancestry |
0.02 ± 0.02 |
0.00 ± 0.02 |
NC¶ |
| Chronic obstructive pulmonary disease, yes, %** |
17.1 |
39.2 |
1.5 × 10−14† |
| Chronic mucous hypersecretion, yes, %†† | 35.9 | 30.5 | 0.06† |
Definition of abbreviations: LSC = Lovelace Smokers cohort; NHW = non-Hispanic whites.
Student t test.
Chi-square test.
Wilcoxon rank sum test.
Data missing for 340 subjects.
Data missing for 45 subjects.
Not calculated.
Data missing for 14 subjects. Chronic obstructive pulmonary disease was defined according to the Global Initiative for Chronic Obstructive Lung Disease criteria (FEV1/FVC < 0.70) (12).
Participants with self-identified cough productive of phlegm for at least 3 months per year for at least 2 consecutive years were considered to have chronic mucous hypersecretion (12).
Differences in consumption of three dietary factors (folate, leafy green vegetables, and multivitamins) were assessed between 214 Hispanics and 1,047 NHWs with both gene methylation and dietary data available. The effect of inclusion of dietary factors on the association between ethnicity and risk for gene methylation in LSC subjects (n = 1261) was assessed using GEE models as well.
NSCLC cases and control subjects enrolled in the New Mexico lung cancer case-control study were unmatched for ethnicity and pack-years, allowing assessment of the interaction between these two variables. Because a direct comparison of ethnicity distribution between cases and control subjects is sensitive to selection bias in a population-based case-control design, we conducted an ethnicity-stratified analysis to estimate the coefficients for pack-years using a logistic regression model with adjustment for covariates. We also assessed the interaction between pack-years and ethnicity for the risk for NSCLC using the likelihood ratio test in an unstratified analysis. The Hosmer-Lemeshow test was used to assess the goodness of fit for logistic regression models. No evidence for a lack of fit of the models in the overall analysis and ethnicity-stratified analyses was identified (P > 0.25). All statistical analyses were conducted in SAS 9.2 (SAS Institute Inc., Cary, NC) and R 2.14 (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org/).
Results
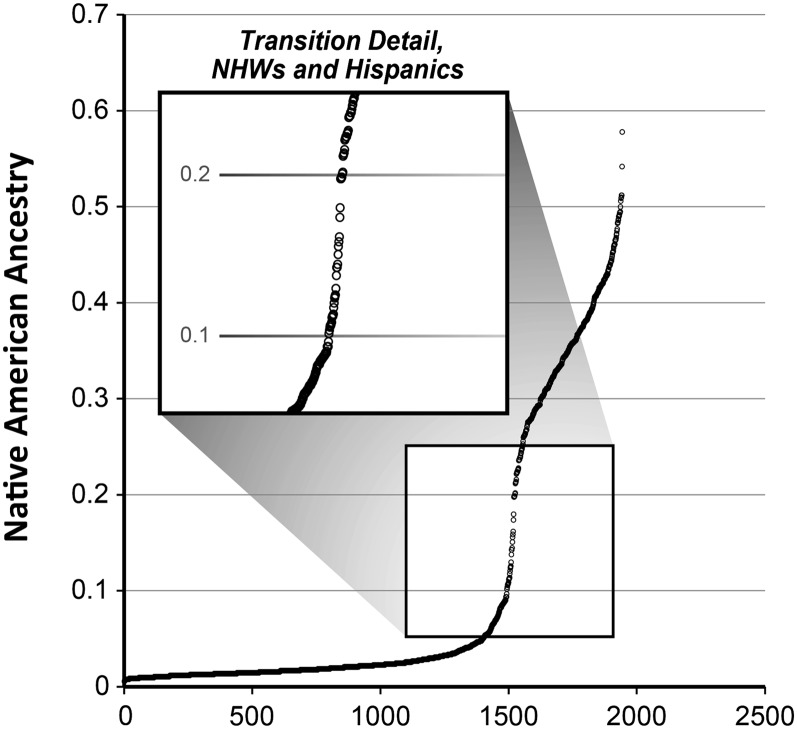
Genetic ancestry estimates in the three reference populations correlated well with expected values (Table 1). Significant admixture was identified in LSC subjects with self-identified Hispanic, Native American, and African American ethnicity. The distribution of NAA in NHWs and Hispanics is depicted in Figure 1 (n = 1,944). NAA of 418 Hispanic smokers ranged from 0.198 to 0.578 with an average of 0.35. In addition, only 5 of 1,526 NHW smokers had NAA greater than 0.198. This suggests a strong corroboration of self-identified ethnicity by genetic ancestry analysis.
Figure 1.
The distribution of Native American ancestry (NAA) in the Lovelace Smokers cohort subjects with self-identified non-Hispanic whites (NHW) (n = 1,526) and Hispanic ethnicity (n = 418). The smaller figure is the zoom-in of the NAA distribution in Lovelace Smokers cohort subjects with NAA between 0.05 and 0.25. The two populations were separated at NAA of 0.198. All Hispanic smokers had NAA greater than 0.198. In contrast, only 5 of 1,526 NHW smokers had NAA greater than 0.198.
Compared with NHWs, Hispanics were more likely to be younger (54.4 vs. 56.9 yr); male (27.8% vs. 20.5%); current smokers (73.0% vs. 54.2%); have lower pack-years (34.3 vs. 41.1 packs per year); and have body mass index greater than or equal to 25 (75.1% vs. 66.5%). Socioeconomic status as reflected by annual household income and college education was significantly lower in Hispanics than in NHWs. Interestingly, Hispanics had lower prevalence of COPD (17.1% vs. 39.2%) but higher prevalence of chronic mucous hypersecretion (35.9% vs. 30.5%) (Table 2).
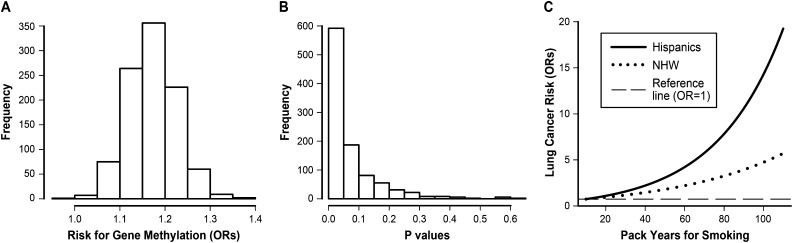
Methylation prevalence of the 12 genes ranged from 0.7% to 37.4% (Table 3). In 30 of 66 pairwise correlation tests, low to moderate correlations with Pearson correlation coefficients greater than 0.10 (P values < 0.0001) were identified for the methylation status between genes (see Table E3). Hispanics and NHWs had similar correlation matrices of the 12 genes (P = 0.65, not shown). GEE modeling identified that Hispanics had significantly increased risk for gene methylation compared with NHWs with adjustment for covariates (OR, 1.18; 95% CI, 1.05–1.32; P = 0.007) (Table 4). In addition, older age, male sex, and former smoker were also identified as three major clinical variables significantly (P values between 10−3 and 10−8) associated with increased risk for gene methylation (Table 4). Because Hispanics were more likely to be younger, male, and current smoker (Table 2), additional analyses were conducted to assess whether the effect of ethnicity on the risk for gene methylation was confounded by these three risk factors. Hispanics and NHWs were frequency-matched on age group (by 5-yr intervals), sex (male vs. female), and smoking status (current vs. former) using a random sampling approach. This approach resulted in Hispanic and NHW samples of the same size and with the same distribution of age, sex, and smoking status. Random sampling was repeated 1,000 times to avoid bias and for each random sampling the data was modeled using GEE to assess the association between self-identified ethnicity and risk for gene methylation. On average, Hispanics had 17.3% increased risk for gene methylation compared with NHWs (Figure 2A). The median significance level obtained in analyses matched for age, sex, and current smoking status was P = 0.03 with approximately 80% of all P values falling below the 0.10 over 1,000 random samplings (Figure 2B). These analyses suggest that Hispanics have increased risk for gene methylation that is independent of the effect of age, sex, and current smoking status.
TABLE 3.
METHYLATION PREVALENCE OF INDIVIDUAL GENES IN ALL LSC SUBJECTS, HISPANICS, AND NHWS
| Variable | Prevalence of Gene Methylation (%) |
P Value* | ||
|---|---|---|---|---|
| All Subjects (n = 1,540) | Hispanics (n = 341) | NHWs (n = 1,199) | ||
| P16 |
19.3 |
20.2 |
19.1 |
0.65 |
| MGMT |
26.2 |
26.3 |
26.1 |
0.94 |
| RASSF1A |
0.7 |
0.3 |
0.8 |
0.47† |
| DAPK |
16.9 |
20.2 |
15.9 |
0.06 |
| GATA4 |
37.4 |
43.9 |
35.5 |
0.005 |
| GATA5 |
15.4 |
20.2 |
14.1 |
0.006 |
| PAX5α |
14.5 |
13.2 |
14.9 |
0.43 |
| PAX5β |
8.4 |
8.8 |
8.3 |
0.80 |
| SULF2 |
35.8 |
44.2 |
33.4 |
0.0002 |
| PCDH20 |
36.9 |
44.2 |
34.9 |
0.0016 |
| DAL1 |
7.1 |
7.9 |
6.8 |
0.50 |
| JPH3 | 22.4 | 20.5 | 22.9 | 0.33 |
Definition of abbreviations: LSC = Lovelace Smokers cohort; NHWs = non-Hispanic whites.
Chi-square test compared the prevalence of gene methylation between 341 Hispanics and 1,199 NHWs.
Fisher exact test.
TABLE 4.
MULTIVARIATE ANALYSIS IDENTIFIED HISPANIC ETHNICITY AS A RISK FACTOR FOR GENE METHYLATION*
| Variable | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|
| Age, 10 yr |
1.09 (1.03–1.16) |
0.0053 |
| Male sex |
1.38 (1.23–1.55) |
3.8 × 10−8 |
| Current smoker |
0.81 (0.72–0.90) |
1.7 × 10−4 |
| Pack-years, 10 |
1.01 (0.98–1.04) |
0.48 |
| Chronic mucous hypersecretion |
1.11 (0.99–1.24) |
0.082 |
| Body mass index ≥ 25 |
1.01 (0.90–1.13) |
0.90 |
| Hispanic ethnicity† | 1.18 (1.05–1.32) | 0.007 |
n = 1,540.
Compared with non-Hispanic whites.
Figure 2.
(A) The distribution of odds ratios (ORs) for the 1,000 times random sampling for association of ethnicity with risk for gene methylation. The average OR was 1.17 with 25th and 75th percentiles as 1.14 and 1.21. (B) The distribution of P values for the 1,000 times random sampling for association of ethnicity with risk for gene methylation. The average P value was 0.03 with 80% of P values less than 0.10. (C) Predicted risk for non–small cell lung cancer using the New Mexico lung cancer study stratified by ethnicity. The range of pack-years shown was from 10 to 108 pack-years, which were within the 5th–95th percentiles of pack-years in cases. The ORs were generated with smokers with 10 pack-years smoking history as the reference. The P value for the interaction between pack-years and ethnicity was 0.058. NHW = non-Hispanic whites.
The Hispanics (n = 341) included in this study had a wide range of NAA (0.198–0.578) (Figure 1), providing the opportunity to study the effect of NAA on their risk for gene methylation. GEE modeling assessed the influence of NAA as a continuous variable for modulating the risk for gene methylation in Hispanics with adjustment for the effects of African and European ancestry and covariates. The difference between the 5th and 95th percentiles of NAA (0.24 vs. 0.48) in Hispanics was used as the unit of change in GEE models to estimate the ORs and 95% CIs. A difference of 0.24 in NAA was associated with a 32% reduced risk for gene methylation in Hispanics (OR, 0.68; P = 0.032) (Table 5). A secondary analysis was conducted to associate dichotomized NAA by its median (0.35) in Hispanics with the risk for gene methylation and found that Hispanics with greater than 0.35 NAA had 30% reduced risk (P = 0.03) for gene methylation compared with those with less than 0.35 NAA (not shown). Consistent associations seen between these two analyses suggest that higher NAA is protective against acquiring gene methylation in the lungs of Hispanic smokers.
TABLE 5.
MULTIVARIATE ANALYSIS IDENTIFIED THAT NAA IN HISPANICS IS PROTECTIVE AGAINST GENE METHYLATION*
| Variable | OR (95% Confidence Interval) | P Value |
|---|---|---|
| Age, 10 yr |
1.05 (0.92–1.19) |
0.48 |
| Male sex |
1.11 (0.89–1.39) |
0.33 |
| Current smoker |
0.81 (0.64–1.03) |
0.086 |
| Pack-years, 10 |
1.04 (0.98–1.10) |
0.24 |
| Chronic mucous hypersecretion |
1.17 (0.95–1.45) |
0.14 |
| Body mass index ≥25 |
1.22 (0.97–1.54) |
0.093 |
| NAA, 0.24† |
0.68 (0.48–0. 97) |
0.032 |
| African ancestry, 0.04‡ | 1.26 (0.97–1.62) | 0.079 |
Definition of abbreviations: NAA = Native American ancestry; OR = odds ratio.
n = 341.
The 5th and 95th percentiles of NAA in Hispanics were 0.24 and 0.48. Thus, 0.24 was selected as a unit change of NAA in Hispanics to calculate the OR.
The 5th and 95th percentiles of African ancestry in Hispanics were 0.008 and 0.048. Thus, 0.04 was selected as a unit change of African ancestry in Hispanics to calculate the OR.
Analyses of individual gene methylation identified multiple genes whose methylation was associated with self-identified ethnicity or NAA (see Table E4). Five of 12 genes had 4.3–10.8% increased prevalence for methylation in Hispanics compared with NHWs (P < 0.04). Furthermore, 3 of 11 genes had more than 48% reduced risk for methylation associated with an increase of 0.24 in NAA in Hispanics (P < 0.10). These results exclude the possibility that the global association between ethnicity or NAA and risk for gene methylation described above was driven by a strong signal associated with methylation of one particular gene.
Our previous studies identified that dietary intake of folate, leafy green vegetables, and multivitamins protected against gene methylation in the aerodigestive tract of smokers using an eight-gene panel (4). These associations were confirmed using GEE models in a larger sample set (214 Hispanics and 1,047 NHWs) with methylation data of a 12-gene panel including the original eight genes (see Table E5). In addition, significantly lower levels of folate intake (median [Q1–Q3], 665 [358–1,197] vs. 1,031 [474–1,304]; P = 0.00038) and lower prevalence of current multivitamin use (47.9% vs. 66.8%; P = 3.5 × 10−6) was identified in Hispanics compared with NHWs with adjustment for demographic variables and total caloric intake. No difference in consumption of leafy green vegetables was identified between ethnicities (P = 0.22). We used GEE models to determine whether the higher risk for gene methylation among Hispanics may be in part caused by their lower dietary intake of folate and lower prevalence of multivitamin use. Inclusion of food folate and multivitamin use in the GEE models resulted in a less significant P value for the association between self-identified Hispanic ethnicity and risk for gene methylation (Table 6). The effect of dietary factors on the association between NAA and risk for gene methylation in Hispanics only was not conducted because of the limited sample size in this ethnic group with available dietary information (n = 214).
TABLE 6.
MULTIVARIATE ANALYSIS FOR THE EFFECT OF FOOD FOLATE AND MULTIVITAMIN SUPPLEMENTATION ON THE ASSOCIATION BETWEEN ETHNICITY AND RISK FOR GENE METHYLATION IN LSC SUBJECTS*
| Variable | Model 1 |
Model 2 |
||
|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | |
| Age, 10 yr |
1.07 (1.00–1.15) |
0.041 |
1.10 (1.02–1.18) |
0.011 |
| Male sex |
1.45 (1.26–1.66) |
1.6 × 10−7 |
1.44 (1.65–1.26) |
2.1 × 10−7 |
| Current smoker |
0.81 (0.71–0.92) |
0.0011 |
0.79 (0.70–0.90) |
3.5 × 10−4 |
| Pack-years, 10 |
1.01 (0.98–1.04) |
0.52 |
1.00 (0.97–1.04) |
0.77 |
| Chronic mucous hypersecretion |
1.10 (0.96–1.25) |
0.16 |
1.10 (0.97–1.26) |
0.13 |
| Body mass index ≥ 25 |
0.99 (0.87–1.12) |
0.82 |
0.98 (0.86–1.11) |
0.73 |
| Hispanic ethnicity |
1.18 (1.02–1.37) |
0.025 |
1.15 (0.99–1.34) |
0.06 |
| Total calorie intake, 1 kcal |
1.03 (0.94–1.13) |
0.59 |
1.06 (0.96–1.17) |
0.28 |
| Food folate, 750 μg† |
NC‡ |
|
0.91 (0.83–1.01) |
0.078 |
| Multivitamin use | NC‡ | 0.87 (0.77–0.98) | 0.018 | |
Definition of abbreviations: CI = confidence interval; LSC = Lovelace Smokers cohort; OR = odds ratio.
n = 1,261.
Food folate was equal to total folate minus folate contained in the multivitamin, which was calculated by linear interpolation.
Not calculated.
The increased risk for gene methylation in Hispanic smokers suggests that this ethnic group may be more susceptible to cigarette smoking–induced lung cancer compared with NHW. This hypothesis was assessed using the New Mexico lung cancer study. Using a generalized linear model with adjustment for age, sex, smoking duration, and current smoking status, we found that Hispanic NSCLC cases had significantly lower pack-years (51.4 vs. 59.1; P = 0.007) and cigarette consumption per day (23.2 vs. 29.1; P = 0.006) compared with NHW cases (see Table E1). Hispanic cases were older and smoked for more years than NHW cases, although the difference is not statistically significant (P > 0.09). More importantly, an interaction (P = 0.058) between pack-years and Hispanic ethnicity was identified for the risk for NSCLC (Figure 2C). Ethnicity-stratified analyses identified that with smokers having 10 pack-years as the reference, the ORs associated with smokers with 50 pack-years is 3.2 and 2.0 for Hispanic and NHW smokers, respectively (Figure 2C). A sensitivity analysis was conducted to truncate the pack-years at 110 and a similar interaction between pack-years and ethnicity for lung cancer risk was observed (not shown).
Discussion
A cross-sectional study was conducted to assess the genetic ancestry of smokers enrolled in the LSC and its association with the risk for gene methylation detected in the exfoliated lung cells collected in the sputum samples. New Mexican Hispanics on average had 35% NAA and 63% European ancestry. In Hispanic smokers, higher NAA was associated with a 32% reduced risk for gene methylation, suggesting that higher NAA in Hispanics may be protective against smoking-induced chronic lung damage. Our previous study identified that New Mexican Hispanic smokers with higher NAA were more resistant to acquiring COPD (12). A similar association was also identified in a Hispanic smoker population (n = 579) from Costa Rica that contains 28% NAA and 70% European ancestry (16). In that study, a 0.1 increase in NAA was associated with 40% reduction in the prevalence of COPD and a 97-ml increase in FEV1 (P = 0.005 for both). Thus, higher NAA in Hispanics is protective against smoking-induced chronic lung damage and pulmonary diseases, although the genetic or environmental-lifestyle factors that are associated with NAA and causal for this association remain to be elucidated.
Although NAA is protective against gene methylation, Hispanics on average still had an 18% increased risk for gene methylation compared with NHW smokers even after strictly controlling for the confounding effects from age, sex, and current smoking status using a random sampling approach. This indicates that gene methylation in sputum, already known as a multifactorial phenotype may be affected by undisclosed risk factors that are related to Hispanic culture. Our previous studies identified that increased dietary intake of folate, leafy green vegetables, and multivitamins were associated with reduced risk for gene methylation (4). Compared with NHWs, Hispanics had significantly lower dietary intake of folate and multivitamins. Inclusion of food folate and multivitamin use in the GEE models reduced the significance for the association between ethnicity and risk for gene methylation. This suggests that the higher risk for gene methylation in Hispanics may be partially explained, although to a small extent, by the insufficient intake of protective factors in the diet. In addition, although Hispanic smokers have significantly lower social economic indicators than NHWs, inclusion of education level in the model only showed a very minimal effect on the association between ethnicity and risk for gene methylation (not shown). In addition, only minimal correlations were identified between dietary factors and social economic indicators (see Table E6). These findings suggest that the lower social economic indicator associated with Hispanic ethnicity cannot explain the increased risk for gene methylation in this ethnic group.
Hispanics on average have lower age-standardized incidence rates for lung cancer compared with NHWs in New Mexico and nationwide (8, 13). A population-based lung cancer case-control study previously conducted in New Mexico suggests that this ethnic difference in lung cancer incidence may be largely explained by the different patterns of cigarette smoking of these two groups (14, 26). However, we report for the first time that Hispanic NSCLC cases have significantly lower (by 8 pack-years) cumulative exposure to cigarette smoke than NHW counterparts and this difference is mainly caused by lower smoking intensity in Hispanic smokers. This suggests that Hispanic smokers may be more susceptible to cigarette smoking–induced lung cancer. Support for this premise is seen within the New Mexico lung cancer study where Hispanic smokers had a more rapidly increased risk for lung cancer as a function of cumulative exposure to cigarettes compared with NHWs (Figure 2C). In addition, a careful reevaluation of the lung cancer study conducted by Humble and coworkers (14, 26) identified that the coefficient for cigarettes per day in a logistic regression model was 34% greater in Hispanics than NHWs. Furthermore, a subgroup analysis was conducted using 11 lung cancer cases and 27 control subjects who are Hispanic smokers from our previous study (3). We identified that the composite methylation index of the 12 genes (same 12 genes as measured in the current study) was significantly associated with the risk for lung cancer (OR, 1.4; 95% CI, 1.0–2.0 per gene methylation; P = 0.05) in Hispanic smokers. This increased risk is not different from what we observed in NHWs in the same prior study (24 cases and 56 control subjects; OR, 1.34; 95% CI, 1.07–1.67; P = 0.01). This suggests that methylation based on this 12-gene panel in sputum could predict risk for lung cancer in both NHW and Hispanic smokers. Thus, these findings, combined with the increased risk for gene methylation in Hispanic smokers, support a greater susceptibility to NSCLC for New Mexico Hispanics smokers than NHWs. However, caution needs to be taken when extrapolating these findings to other Hispanic populations because of the heterogeneity in the acculturation status, environmental and dietary exposure, the level of genetic admixture, and the source of NAA associated with Hispanic populations living in different geographic areas.
Acknowledgments
Acknowledgment
The authors thank Drs Elizabeth A. Burki at Lovelace Respiratory Research Institute (LRRI) and Richard E. Crowell at University of New Mexico for enrolling lung cancer patients; Kieu C. Do and Randall P. Willink at LRRI for conducting nested MSP assays; Dr. Nicholas J. Schork at The Scripps Research Institute La Jolla CA for critical advice on admixture analyses; and Thomas J. Gagliano at LRRI for scientific editing of the figures.
Footnotes
Supported by National Cancer Institute grant R01 CA097356 and the State of New Mexico as a direct appropriation from the Tobacco Settlement Fund (to S.A.B.). The National Institutes of Health and the State of New Mexico had no influence in study design; collection, analysis, and interpretation of data; writing the report; or the decision to submit the report for publication.
Author Contributions: S.L., C.A.S., and S.A.B. conceived of and designed the study. S.L., C.L.T., and D.V.D.B. performed the genotyping. S.L., Y.L., M.A.P., X.Z., and C.A.S. conducted the data analyses and tabulated the results. W.J.G. commented critically on generalized estimating equation modeling and interpretation. S.E.B. commented critically on admixture analyses. S.L. and Y.L. interpreted the results and drafted the manuscript. S.L., S.E.B., K.G.F., C.A.S., F.D.G., and S.A.B. critically edited the manuscript. All authors have read the manuscript and approved its submission.
Originally Published in Press as DOI: 10.1164/rccm.201305-0925OC on September 13, 2013
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 2.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 3.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, Feser WJ, Baron AE, Franklin WA, Brock MV, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res. 2012;18:3387–3395. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stidley CA, Picchi MA, Leng S, Willink R, Crowell RE, Flores KG, Kang H, Byers T, Gilliland FD, Belinsky SA. Multivitamins, folate, and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer Res. 2010;70:568–574. doi: 10.1158/0008-5472.CAN-09-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leng S, Stidley CA, Willink R, Bernauer A, Do K, Picchi MA, Sheng X, Frasco MA, Van Den Berg D, Gilliland FD, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer Res. 2008;68:3049–3056. doi: 10.1158/0008-5472.CAN-07-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores KG, Stidley CA, Mackey AJ, Picchi MA, Stabler SP, Siegfried JM, Byers T, Berwick M, Belinsky SA, Leng S. Sex-specific association of sequence variants in CBS and MTRR with risk for promoter hypermethylation in the lung epithelium of smokers. Carcinogenesis. 2012;33:1542–1547. doi: 10.1093/carcin/bgs194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng S, Stidley CA, Liu Y, Edlund CK, Willink RP, Han Y, Landi MT, Thun M, Picchi MA, Bruse SE, et al. Genetic determinants for promoter hypermethylation in the lungs of smokers: a candidate gene-based study. Cancer Res. 2012;72:707–715. doi: 10.1158/0008-5472.CAN-11-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin. 2012;62:283–298. doi: 10.3322/caac.21153. [DOI] [PubMed] [Google Scholar]
- 9.Pew Hispanic Center Demographic profile of Hispanics in New Mexico, 2010. [accessed 2013 Mar 21]. Available from: http://www.pewhispanic.org/states/state/nm/
- 10.Bertoni B, Budowle B, Sans M, Barton SA, Chakraborty R. Admixture in Hispanics: distribution of ancestral population contributions in the Continental United States. Hum Biol. 2003;75:1–11. doi: 10.1353/hub.2003.0016. [DOI] [PubMed] [Google Scholar]
- 11.Arias E. United States life tables by Hispanic origin. Vital Health Stat 2. 2010;152:1–33. [PubMed] [Google Scholar]
- 12.Bruse S, Sood A, Petersen H, Liu Y, Leng S, Celedón JC, Gilliland F, Celli B, Belinsky SA, Tesfaigzi Y. New Mexican Hispanic smokers have lower odds of chronic obstructive pulmonary disease and less decline in lung function than non-Hispanic whites. Am J Respir Crit Care Med. 2011;184:1254–1260. doi: 10.1164/rccm.201103-0568OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.New Mexico Cancer Council New Mexico cancer facts and figures, 2007. [accessed 2013 Mar 21]. Available from: http://www.cancernm.org/cancercouncil
- 14.Humble CG, Samet JM, Pathak DR, Skipper BJ. Cigarette smoking and lung cancer in Hispanic whites and other whites in New Mexico. Am J Public Health. 1985;75:145–148. doi: 10.2105/ajph.75.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, Le Marchand L. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Brehm JM, Boutaoui N, Soto-Quiros M, Avila L, Celli BR, Tesfaigzi Y, Bruse S, Celedon JC. Native American ancestry is associated with reduced risk of COPD and higher Fev1 in Costa Ricans. Am J Respir Crit Care Med. 2012;185:A6882. [Google Scholar]
- 17.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 18.Leng S, Bernauer AM, Hong C, Do KC, Yingling CM, Flores KG, Tessema M, Tellez CS, Willink RP, Burki EA, et al. The A/G allele of rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinomas. Clin Cancer Res. 2011;17:2014–2023. doi: 10.1158/1078-0432.CCR-10-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, Gamble MV, Susser E. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–2310. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6:e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng S, Bernauer AM, Zhai R, Tellez CS, Su L, Burki EA, Picchi MA, Stidley CA, Crowell RE, Christiani DC, et al. Discovery of common SNPs in the miR-205/200 family-regulated epithelial to mesenchymal transition pathway and their association with risk for non-small cell lung cancer. Int J Mol Epidemiol Genet. 2011;2:145–155. [PMC free article] [PubMed] [Google Scholar]
- 22.Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, Bergen AW, Swan GE, Tyndale RF, Benowitz NL, et al. Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SPSmart [accessed 2013 Mar 21]. Available from: http://spsmart.cesga.es/
- 25.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 26.Humble CG, Samet JM. Smoking and lung cancer in New Mexico. Am J Public Health. 1986;76:1361. doi: 10.2105/ajph.76.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]