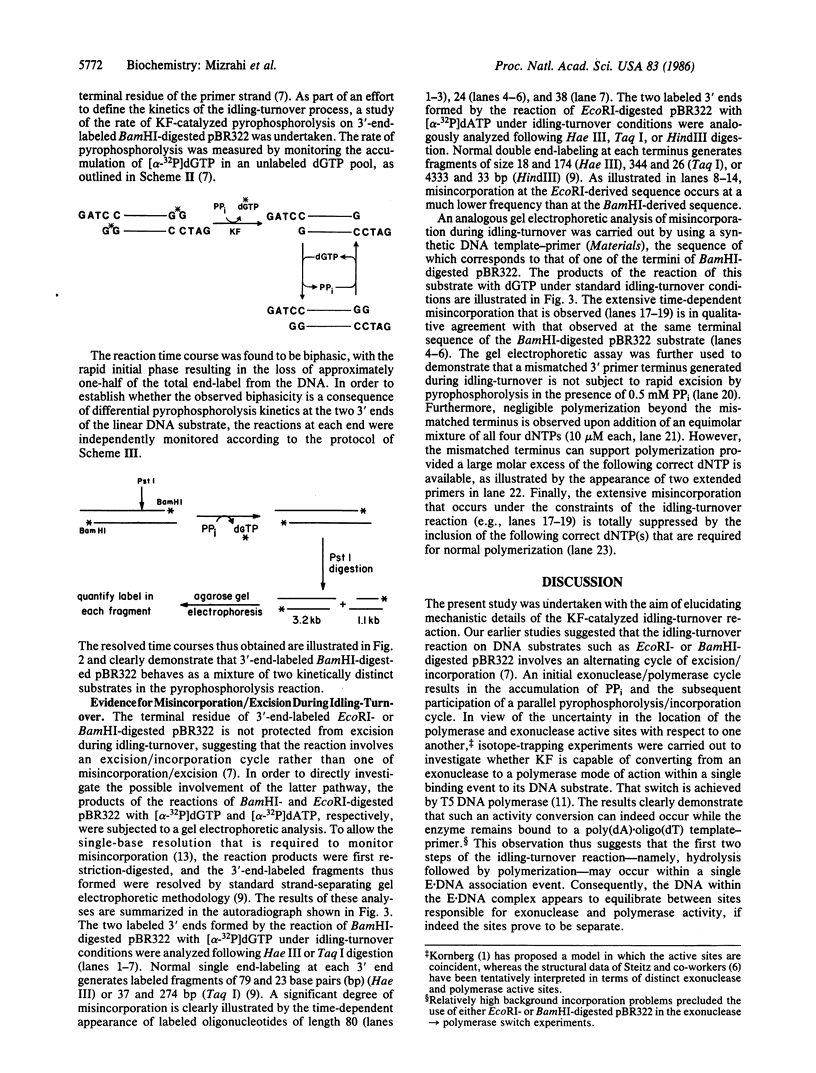
Abstract
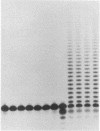
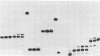
Mechanistic features of several processes involved in the idling-turnover reaction catalyzed by the large (Klenow) fragment of Escherichia coli DNA polymerase I have been established. The exonuclease----polymerase activity switch involved in the excision/incorporation mode of idling-turnover occurs without an intervening dissociation of the enzyme from its DNA substrate. Comparative studies on the pyrophosphorolysis kinetics of related DNA substrates indicate a significant dependence of the reaction rate upon the DNA sequence within the duplex region upstream of the primer-template junction. Finally, a gel electrophoretic analysis of the products of the idling-turnover reaction has provided direct evidence for an alternative DNA sequence-dependent misincorporation/excision pathway.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Bryant F. R., Johnson K. A., Benkovic S. J. Elementary steps in the DNA polymerase I reaction pathway. Biochemistry. 1983 Jul 19;22(15):3537–3546. doi: 10.1021/bi00284a001. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Eckstein F. A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate. J Biol Chem. 1979 Aug 10;254(15):6889–6893. [PubMed] [Google Scholar]
- Das S. K., Fujimura R. K. Mechanism of primer-template-dependent conversion of dNTP leads to dNMP by T5 DNA polymerase. J Biol Chem. 1980 Aug 10;255(15):7149–7154. [PubMed] [Google Scholar]
- Deutscher M. P., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 28. The pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):3019–3028. [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Tsui W. C. Kinetics of base misinsertion by DNA polymerase I of Escherichia coli. J Mol Biol. 1983 Apr 25;165(4):655–667. doi: 10.1016/s0022-2836(83)80272-1. [DOI] [PubMed] [Google Scholar]
- Gupta A. P., Benkovic S. J. Stereochemical course of the 3'----5'-exonuclease activity of DNA polymerase I. Biochemistry. 1984 Nov 20;23(24):5874–5881. doi: 10.1021/bi00319a029. [DOI] [PubMed] [Google Scholar]
- Hillebrand G. G., McCluskey A. H., Abbott K. A., Revich G. G., Beattie K. L. Misincorporation during DNA synthesis, analyzed by gel electrophoresis. Nucleic Acids Res. 1984 Apr 11;12(7):3155–3171. doi: 10.1093/nar/12.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Mizrahi V., Benkovic P. A., Benkovic S. J. Mechanism of the idling-turnover reaction of the large (Klenow) fragment of Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1986 Jan;83(2):231–235. doi: 10.1073/pnas.83.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985 Jul 16;24(15):4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]