Summary
The spread of misfolded proteins has been implicated in a wide variety of neurodegenerative diseases. Prions associated with spongiform encephalopathy are currently the only misfolded proteins in which high specific biological infectivity can be produced in vitro. Using a system that generates infectious prions de novo from purified recombinant PrP and conversion cofactors palmitoyl-oleoyl-phosphatidylglycerol (POPG) and RNA, we examined by deuterium exchange mass spectrometry (DXMS) the stepwise protein conformational changes that occur during prion formation. We found that initial incubation with POPG causes major structural changes in PrP involving all three α-helices and one β-strand, with subsequent RNA rendering the N-terminus highly exposed. Final conversion into the infectious PrPSc form was accompanied by globally decreased solvent exposure, with persistence of the major cofactor-induced conformational features. Thus, we report that cofactor molecules appear to induce major structural rearrangements during prion formation, initiating a dynamic sequence of conformational changes resulting in biologically active prions.
Keywords: deuterium exchange, protein misfolding, PrP, scrapie, cofactor
Introduction
The propagation of aberrant protein folding has emerged as a potentially important mechanism for many neurodegenerative diseases. Intracerebral inoculation of samples containing misfolded proteins can induce the formation and spread of neuropathological features associated with these diseases throughout the brain with varying degrees of efficiency (Castilla et al., 2005; Clavaguera et al., 2013; Deleault et al., 2007; Legname et al., 2004; Luk et al., 2012; Meyer-Luehmann et al., 2006; Stohr et al., 2012; Wang et al., 2010a). Of these diseases, spongiform encephalopathies such as Creutzfeldt-Jakob Disease and bovine spongiform encephalopathy are the best characterized for studying the phenomenon of transmissible protein misfolding (Prusiner, 1998), as well as the only ones in which proteins possessing significant levels of specific infectivity have been produced in vitro (Deleault et al., 2007; Deleault et al., 2012b; Shikiya and Bartz, 2011). Spongiform encephalopathies are caused by prions, contain PrPSc, a misfolded conformer of the normal cellular prion protein PrPC (Prusiner, 1998). The production of high-titer infectious prions in vitro requires the addition of non-protein cofactor molecules such as RNA and phospholipids (Deleault et al., 2007; Deleault et al., 2012b). Misfolded PrP prepared in the absence of cofactors is able to induce prion formation in vivo, but possesses ≥105-fold less specific infectivity (Colby et al., 2010; Deleault et al., 2012b; Kim et al., 2010; Legname et al., 2004; Makarava et al., 2010; Makarava et al., 2012). Cofactors maintain the infectious conformation of PrPSc in purified recombinant prions (Deleault et al., 2012b), though the mechanism of this is unclear.
Preparations of PrPSc made with purified recombinant PrP and the cofactors palmitoyl-oleoyl-phosphatidylglycerol (POPG) and RNA recapitulate the major features of prion disease, causing transmissible spongiform encephalopathy in wild-type animals with a rapid incubation time (Wang et al., 2010a). POPG, RNA, and the protein misfolding cyclic amplification (PMCA) conversion method are each necessary to form PrPSc and prion infectivity in this system (Deleault et al., 2012a; Wang et al., 2010a). Moreover, in this pathway, the sequential steps of cofactor addition result in stable insoluble intermediate species, enabling examination of the prion formation process in discrete steps. Taken together, the chemically defined components, stable intermediates, de novo prion formation, transmission efficiency, and rapid incubation in this system provide a unique opportunity for studying the formation of biologically active prions. Although it is not known how closely the POPG and RNA cofactor pathway resembles the process of prion formation in vivo, it provides a tractable model in which the generation of bona fide prion infectivity can be dissected at a structural level.
The atomic structure of PrPC contains three α-helices, two short β-strands, and a flexibly disordered N-terminus (Riek et al., 1996; Riek et al., 1997). PrPSc contains significantly more β-sheet content (Caughey et al., 1991; Moore et al., 2011) and is protease-resistant (Bolton et al., 1982), but its insolubility (McKinley et al., 1991) has hindered the standard structural techniques of X-ray crystallography and solution NMR. Hydrogen/deuterium exchange mass spectrometry (DXMS) has proven to be a powerful method for studying protein dynamics, domain structure, regional stability, and function (Chalmers et al., 2011; Dai et al., 2008; Engen, 2009; Englander, 2006; Hamuro et al., 2003; Konermann et al., 2011; Zhang et al., 2010). DXMS can reveal structural information about insoluble proteins, by measuring the solvent accessibility of peptide backbone amide hydrogens, reflective of the degree of steric blocking and hydrogen-bonded secondary structure at each position. Smirnovas et al. previously applied DXMS to the study of native PrPSc lacking most post-translational modifications (Smirnovas et al., 2011), finding a conformation with very low solvent access.
In this study, we apply the DXMS approach to the POPG and RNA cofactor system for prion generation, providing a unique opportunity to study the dynamics of protein structural rearrangement in prion formation. We report here a stepwise progression of conformational changes during prion protein conversion into the infectious form.
Results
In order to assess protein conformational changes during PrPSc formation, we examined the solvent accessibility of PrP at incremental steps of the conversion process by which PrPSc is formed using the cofactors POPG and RNA (Fig. 1A). We took advantage of specific properties of each PrP species that allowed us to isolate them prior to performing deuterium exchange analysis of solvent accessibility (Fig. S1a). Anionic phospholipids, such as POPG, bind to PrP and induce conformational change to an insoluble state (Wang et al., 2007), which can be washed and separated from other components of the initial mixture. We refer to this conformation as PrP conversion intermediate 1 (PrPInt1). Following incubation of PrPInt1 with RNA molecules, PrP remains insoluble (Fig. S1b), in a form we refer to as PrP conversion intermediate 2 (PrPInt2). PrP-POPG-RNA complexes (PrPInt2) can be used as a substrate for protein misfolding cyclic amplification (PMCA) to form PrPSc that is infectious to wild-type animals (Wang et al., 2010a). However, this PMCA reaction does not convert 100% of the PrP molecules into PrPSc. Therefore, to study PrPSc in isolation, we required a processing step to eliminate unconverted PrPInt2. PrPSc is resistant to limited proteolysis, with proteinase K treatment digesting only the N-terminus to residue ~89 (Prusiner, 1998). In order to preserve more of the N-terminus of PrPSc, we used trypsin, an endopeptidase which cleaves C-terminal to lysine and arginine residues (Olsen et al., 2004). Following treatment with 5 ng/µL trypsin, PrPInt2 is removed (Fig. S1c). The same trypsin treatment of the PMCA reaction product yields only the substrate molecules that were converted into PrPSc. Most of these PrPSc molecules are truncated to a protease-resistant core, while a small amount show the same electrophoretic mobility as full-length PrP.
Figure 1. Experimental paradigm for deuterium exchange mass spectrometry analysis of prion formation process.
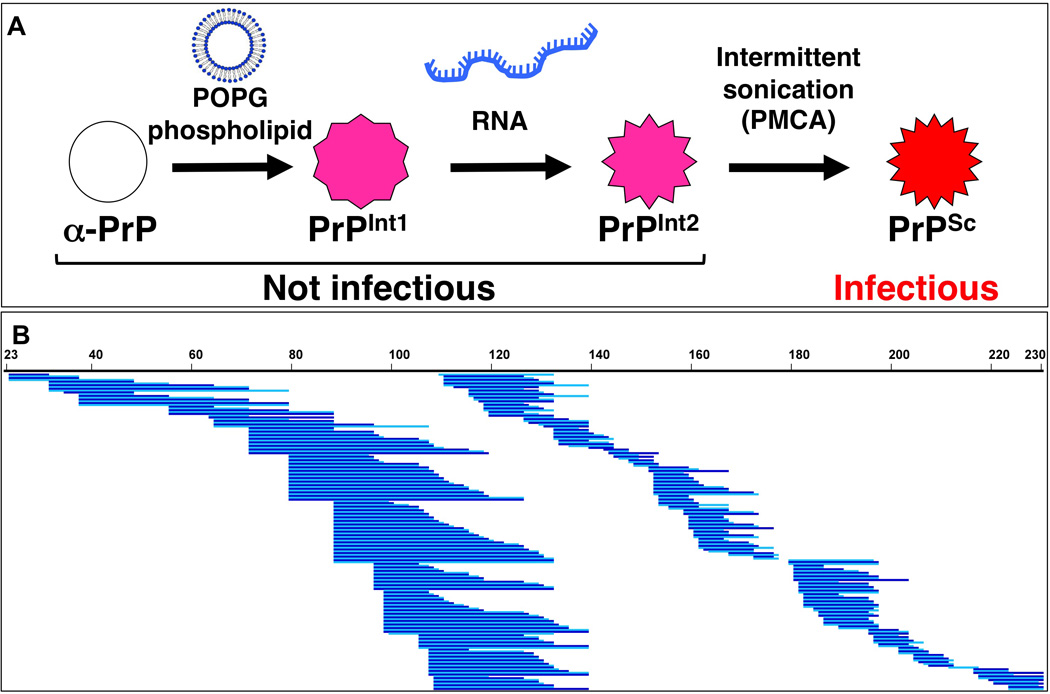
(A) Schematic diagram of PrP conformations during in vitro formation of PrPSc, which is infectious to wild-type animals (Wang et al., 2010a). (B) Coverage map of prion protein proteolytic peptides obtained by treatment of deuterium-exchanged and quenched samples with fungal protease XII and pepsin. Unique high-quality peptides (including multiple charge states of the same peptide) were identified by LC-MS and are shown here as blue lines. Two hues are used to visually distinguish adjacent peptides. Of 355 total peptides identified and shown here, an average of 160 were recovered in each experiment. Numbers refer to mouse PrP amino acid sequence. Differences between nested peptides permitted fine resolution of deuterium exchange levels.
Generation of overlapping peptides from prion protein
To apply deuterium exchange to examine the conformational changes in PrP during the prion formation process, we developed PrP fragmentation methods to enable resolution of solvent protection differences to specific sequence locations. Previous deuterium exchange studies of PrP have reported data from 28–44 peptic peptides, covering most of the C-terminus with minimal coverage of the N-terminus (Lu et al., 2007; Nazabal et al., 2009; Smirnovas et al., 2011; Smirnovas et al., 2009). Through a series of experiments, we identified conditions of reaction quenching and extended digestion with fungal protease XIII and pepsin that consistently yielded ~160 high-quality peptides (Supplemental Experimental Procedures and Table S1). This dense peptide coverage created extensive overlap in most areas of the PrP sequence (Fig. 1B), enabling fine resolution of local deuterium exchange differences, in some regions to the level of single amino acid residues.
Deuterium exchange of α-helical recombinant PrP
Using the optimized conditions for recovery of PrP-derived peptides, we performed DXMS experiments on soluble recombinant prion protein, α-PrP, which has the same structure as native PrPC (Hornemann et al., 2004). Based on the insolubility of the other analytes in this study and the previously reported low solvent accessibility of native PrPSc lacking a glycosylphosphatidylinositol anchor and most glycans (GPI− PrPSc) (Smirnovas et al., 2011), we focused this study on longer periods of deuterium incubation, between 30 min. and 24 hrs. However, to permit comparison with historical results, we also included a shorter time point for α-PrP (Fig. S2). Fig. S3 shows the mass spectra for a representative peptide, 99–110, to indicate the effect of deuterium exposure on the isotopic envelope. We found that the α-PrP N-terminus is highly exposed, rapidly showing full deuteration (Fig. 2A). NMR spectroscopy has determined that the structure of α-PrP has three α-helices and two small β-strands (Riek et al., 1996). As expected, these motifs showed protection from deuterium uptake, indicating lower solvent exposure than other regions of PrP. β-strand 1 (β1) was more exposed than strand 2 (β2), possibly due to the deeper position of β2 in the tertiary structure. These results are consistent with previously reported deuterium exchange data for recombinant α-PrP (Hosszu et al., 1999; Nazabal et al., 2009).
Figure 2. Regional solvent accessibility of prion protein conformations at distinct stages during PrPSc formation.
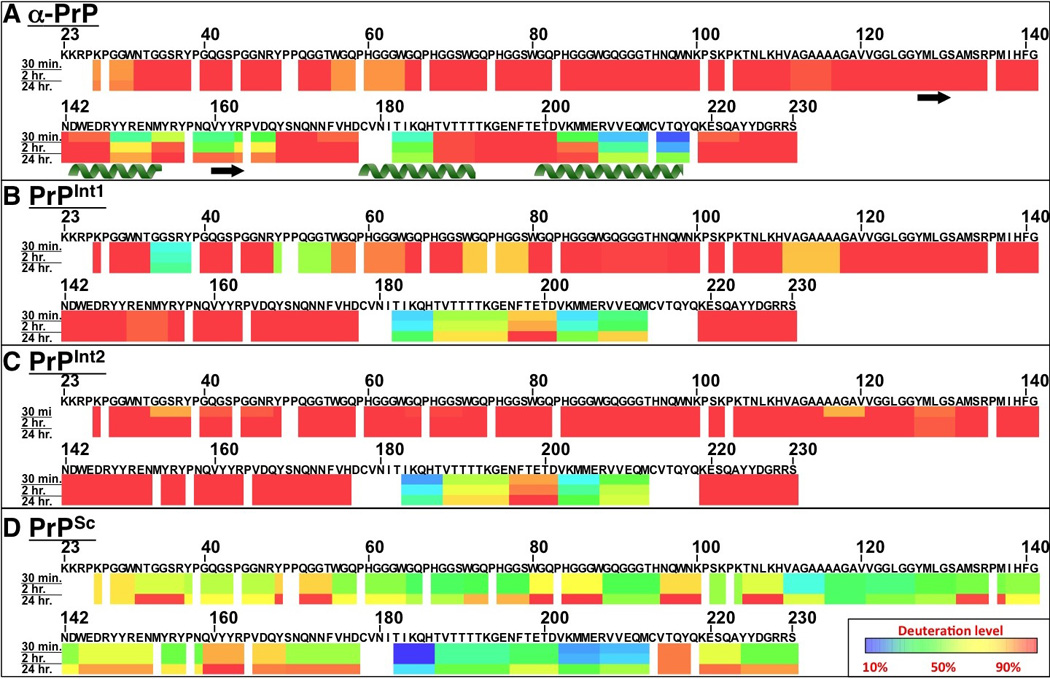
Prion protein conformations α-PrP (A), PrPInt1 (B), PrPInt2 (C), and PrPSc (D) were each purified and then incubated separately in deuterated water (D2O) for varying time durations in order to assess regional solvent exposure. Deuterium incorporation was quenched, followed by proteolytic fragmentation of prion proteins and liquid chromatography-mass spectrometry detection. Using centroid masses, deuterium incorporation for each peptide was determined relative to an equilibrium-deuterated sample, from which deuteration values for PrP regions were computed. For each PrP conformer and incubation time, deuterium incorporation is indicated by color, with areas of protein with low deuterium incorporation (highly protected) are indicated in blue, while areas with high deuterium exchange (highly exposed) are indicated in red. The α-helix and β-strand secondary structural motifs from α-PrP NMR (Riek et al., 1996) are indicated below the solvent accessibility map.
Deuterium exchange of PrP complexed to POPG
We next examined the conformation of PrP after incubation with POPG (PrPInt1). As PrPInt1 is formed during PrP conversion into PrPSc, it may be considered an intermediate in the process. PrPInt1 is insoluble (Wang et al., 2007) but lacks the infectivity of PrPSc (Wang et al., 2010a). Deuterium exchange of PrPInt1 shows an area of significant protection at 182–212 (Fig. 2B). The rest of the PrPInt1 structure is highly exposed, except for a few short protected areas in the N-terminus. When PrPInt1 is compared to α-PrP, significant changes are evident (Fig. 3A). A few areas of the PrPInt1 N-terminus, notably 34–38 and 50–55, show less deuteration than the highly exposed α-PrP N-terminus. This is consistent with the formation of small α-helices or β-strands. PrPInt1 deuterium exchange is reduced at 182–206, a region that in α-PrP encompasses distal α-helix 2 (α2), proximal α-helix 3 (α3), and the α2-α3 loop. This increased protection may reflect an increase in packing of the endogenous α-helical structure or a secondary structural change, such as to β-sheet. PrPInt1 also shows increased exchange at 143–167, suggesting the unfolding of helix 1 (α1) and β-strand 2 when POPG is added to PrP. To visualize the three-dimensional locations of these regions, we threaded the POPG-induced changes in deuterium uptake onto the 3-dimensional atomic structure of PrPC (Fig. 4). It is notable that the major areas of increased exposure (α1 and β2) and decreased exposure (α2 and α3) are in physical proximity, suggesting that the two changes may be related and occur as part of a major structural rearrangement in PrP.
Figure 3. Changes in prion protein solvent accessibility during each step of PrPSc formation.
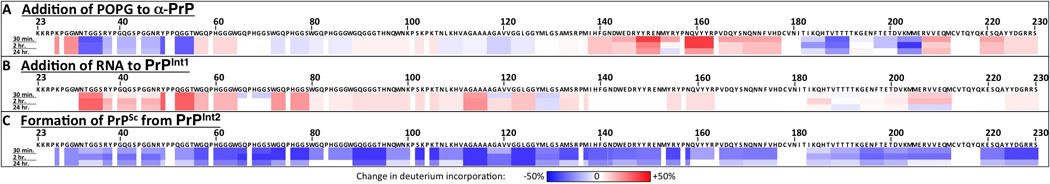
(A–C) Based on the deuterium exchange solvent accessibility data of each prion protein conformation, differences were calculated for each subsequent step in PrPSc formation. Increases in deuterium incorporation (indicating increases in solvent accessibility) are depicted in shades of red, while decreases in deuterium incorporation are depicted in shades of blue.
Figure 4. Atomic structural location of POPG-induced prion protein conformational changes.
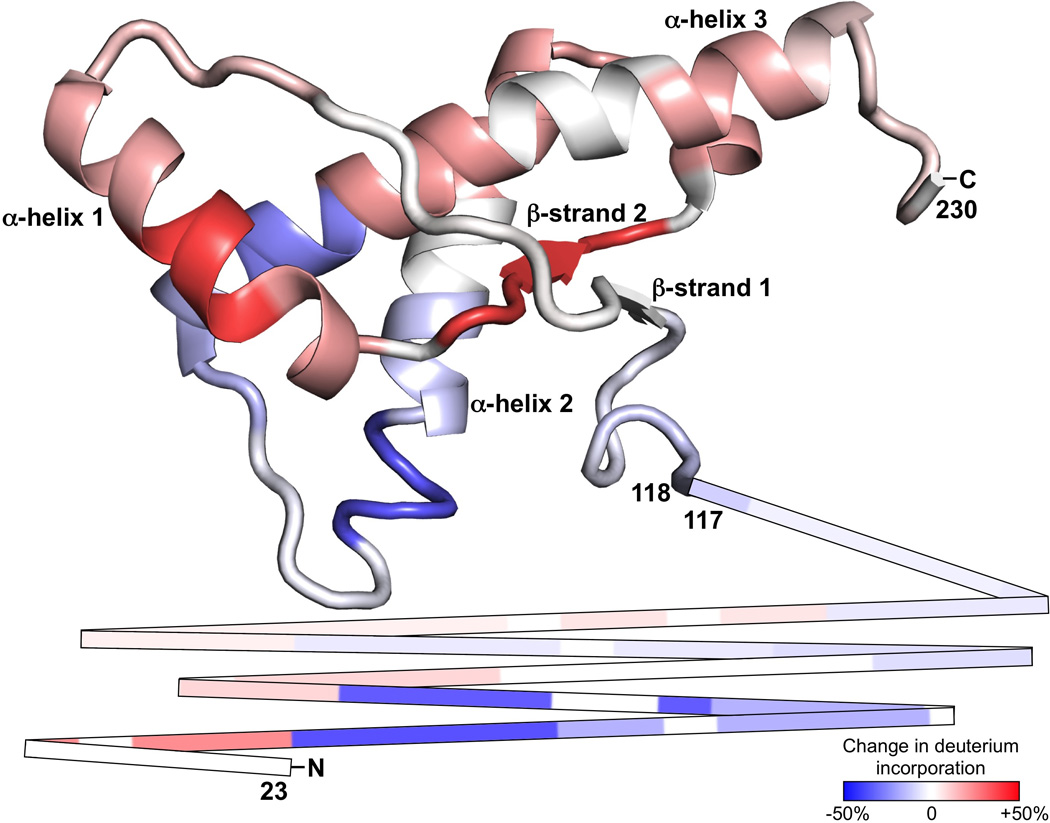
Changes in prion protein solvent accessibility upon conversion from α-PrP to PrPInt1, depicted by color shades, were threaded onto the normal prion protein NMR structure using PyMOL. The unstructured N-terminus is shown as lines for the sequence 23–117. The N-terminus, C-terminus, and major structural features of α-PrP are indicated.
Deuterium exchange of PrP following incubation with POPG and RNA
When PrP-POPG complexes (PrPInt1) are incubated with RNA, the resulting protein species (termed PrPInt2) remains insoluble. Deuterium exchange experiments indicate that PrPInt2 has a conformation very similar to PrPInt1 (Fig. 2C). Much of the C-terminus shows high solvent exposure, and the 183–212 region shows a significant degree of protection. The entire N-terminus shows a highly exposed structure, including exposure of the small areas of N-terminal protection in PrPInt1 (Fig. 3B).
Deuterium exchange of PrPSc
Following conversion of PrPInt2 into PrPSc by PMCA, we isolated PrPSc molecules using trypsin digestion and then performed deuterium exchange experiments (Fig. 2D). As with PrPInt1 and PrPInt2, the C-terminal region 182–212 is solvent protected, but to an even greater degree in PrPSc. For example, peptide 196–201 shows high exposure in α-PrP, some protection in PrPInt1 and PrPInt2, and is further protected in PrPSc (Fig. S4). The rest of PrPSc also shows significant solvent protection, with dramatically reduced exchange compared to the PrPInt2 predecessor (Fig. 3C). This protection is particularly notable in the hydrophobic domain (111–126) and in much of the octapeptide repeat region, including 65–79.
Discussion
While the precise natural pathway of prion formation is not known, PrP conversion in vitro using the cofactors POPG and RNA has been shown to generate infectious prions that cause spongiform encephalopathy in wild-type animals (Wang et al., 2010a). Formation of prions in vitro in this manner provided us the opportunity to examine the conformation of PrP during the conversion process, by isolating intermediate forms of PrP that occur after binding cofactor molecules but before conversion into infectious PrPSc. In this study, we used deuterium exchange mass spectrometry to identify the specific locations of PrP conformational changes that occur during prion formation. These data provide a dynamic map of specific folding events that indicates that conversion cofactor molecules are responsible for the major structural rearrangement that occurs during prion formation.
Incubation with the phospholipid POPG caused a major structural rearrangement in PrP involving all three α-helices and β-strand 2. Specifically, helix 1 and β-strand 2 appear to unfold, as they change from a rather protected to a highly exposed state. In contrast, there is a significant decrease in solvent exposure in the area surrounding the α2-α3 loop. This could be due to the transformation of this region into a β-sheet structure since circular dichroism and Fourier transform infrared spectroscopy experiments show that PrP binding to POPG is accompanied by an increase in β-sheet content (Kazlauskaite et al., 2003; Sanghera and Pinheiro, 2002). The two aforementioned areas that display the most significant change (i.e. increase or decrease) in solvent exposure are adjacent in PrPC, suggesting that their rearrangements may be related and occur as part of a broad structural transformation in PrP upon interaction with POPG. In addition to the C-terminal changes, POPG appears to induce some areas of the PrP N-terminus to form a structure with increased protection, consistent with the formation of new secondary structure such as small α-helices or β-strands. Furthermore, the increased protection seen at 111–117 may be a result of POPG binding to the hydrophobic domain (Wang et al., 2010b).
In contrast to the substantial conformational change that occurs with POPG, the subsequent addition of the polyanion RNA to PrPInt1 has a relatively small immediate effect on PrP structure. While the combination of POPG and RNA supports PrPSc propagation (Wang et al., 2010a), POPG alone is not sufficient (Deleault et al., 2012a), indicating an essential role for RNA in this prion formation pathway. Given its meager immediate structural effect on PrP, RNA may act subsequently during PMCA to induce conversion to PrPSc. Nonetheless, addition of RNA does have the immediate structural effect of increasing the solvent exposure of the N-terminus, possibly by destabilizing secondary structural elements formed in the presence of POPG. As such, the cumulative effect of POPG and RNA cofactors renders an exposed PrP structure from the N-terminus to position ~182. Interestingly, both the N-terminal (23–28) and central (100–109) polybasic domains remain highly exposed following incubation with POPG and RNA, consistent with evidence that they bind PrPSc during templated conversion (Miller et al., 2011).
The conversion of PrPInt2 into PrPSc is accompanied by a broad decrease in deuterium incorporation. This general increase in solvent protection contrasts with the domain-specific structural rearrangements in PrP that occurred with addition of POPG. In the global closure of PrPSc structure, one notable area of significant protection is the hydrophobic domain (111–126), thought to be a critical region in PrPSc (Holscher et al., 1998) for reasons that are not well understood. It has been postulated that this region serves as a transition point during PrPSc formation (Peretz et al., 1997). During PrPSc formation, there is further closure of the α2-α3 region, which first showed increased protection in the presence of POPG. Indeed, based on statistical coupling analysis, the α2-α3 region has also been hypothesized to initiate transition to PrPSc (Chen and Thirumalai, 2013). Our findings indicate that the α2-α3 region (along with α1-β2) participates in the initial PrP conformational change during cofactor incubation, while the hydrophobic domain is not involved until subsequent PrPSc formation.
Recently, Timmes et al. reported that sPMCA propagation in the presence of POPG and RNA could also stochastically produce a non-infectious PrPSc conformer (Timmes et al., 2013) similar to a self-propagating, non-infectious PrPSc conformer formed in the absence of cofactors (Deleault et al., 2012b). Therefore, it is important to note that for this study we used a PrPSc sample whose infectivity has been confirmed by bioassay in wild type mice. It is also worth considering the possibility that our preparations of PrPInt1 and PrPInt2 might be precursors of the non-infectious rather than the infectious PrPSc conformer. However, the data suggest that the intermediate preparations are on-pathway to the infectious PrPSc conformer because most of the cofactor-induced structural changes persist in the final, infectious PrPSc product (Fig. 2). Specifically, helix 1 of PrPC opens and helices 2 and 3 close upon cofactor introduction, and these relative exchange protection features are also present in PrPSc. The relatively low yield of PrPSc from PrPInt2 suggests that the infectious form may not be the only end point of the pathway, but PrPSc indeed appears to come from the cofactor-induced intermediate conformations.
Smirnovas et al. utilized the unique properties of GPI-anchorless PrP expressed by transgenic mice to perform DXMS. They found that GPI− PrPSc molecules show very low solvent accessibility in the sequence 83–223(Smirnovas et al., 2011). Nearly all peptides in this region from GPI− PrPSc of prion strains 22L, Chandler, and Me7 showed less than 40% deuterium incorporation after prolonged incubation. Our results for OSU-strain PrPSc produced in vitro with POPG and RNA cofactors also show a structure with low solvent accessibility. However, with longer incubation (24 hrs.), some areas of OSU POPG/RNA PrPSc show deuterium incorporation of >70%, though the C-terminal domain 182–212 and other regions remain highly protected even with prolonged D2O incubation. Indeed, various approaches have shown that there exist multiple PrPSc structures corresponding to distinct prion strains(Bessen and Marsh, 1994; Smirnovas et al., 2011). The deuterium exchange differences between our PrPSc results and those reported by Smirnovas et al. are likely related to prion strain differences. Furthermore, whereas the pathway studied here requires two different cofactors (POPG and RNA), a distinct strain of infectious prions is produced via an alternative pathway that uses only a single endogenous cofactor, phosphatidylethanolamine(Deleault et al., 2012a). RNA depletion studies also suggest the existence of different folding pathways employed by different prion strains, which can be distinguished by their polyanion requirement(Saa et al., 2012).
Forms of PrPSc have been reported that are sensitive to protease digestion (Colby et al., 2010; Kim et al., 2011; Pastrana et al., 2006). The PrPSc findings we report here were obtained from protease-resistant PrPSc, which has been more extensively characterized as a conformer associated with infectious prions. Furthermore, recent findings indicate that protease-sensitive and protease-resistant PrPSc have similar structures (Sajnani et al., 2012), suggesting that our results may reflect information about the full population of PrPSc formed by this pathway.
Studies of secondary structure indicate that PrPSc has significantly more overall β-sheet content than PrPC (Caughey et al., 1998; Pan et al., 1993; Smirnovas et al., 2011). Two theoretical structural models of PrPSc retain α-helices 2 and 3 from PrPC (DeMarco and Daggett, 2004; Govaerts et al., 2004). Our solvent accessibility data for OSU PrPSc formed with POPG and RNA cofactors show significant exposure changes in the region of PrPC α-helices 2 and 3, as does the data for GPI− PrPSc (Smirnovas et al., 2011). This argues against PrPSc containing these structures, though we cannot exclude the possibility that the helices are retained but with greater packing or another cause of increased solvent protection.
By studying the step-by-step conformational changes in PrP during prion formation, we have found that conversion cofactor molecules drive structural rearrangements that initiate and persist throughout the process of infectious PrPSc formation. These results provide evidence that cofactors strongly influence PrPSc structure, complementing previous observation of such cofactor influence on PrPSc behavior (Deleault et al., 2012b). Taken together, these findings illuminate an essential mechanistic role for non-protein cofactors in driving dynamic structural changes during protein misfolding.
Experimental Procedures
Preparation of α-PrP
Mouse prion protein (MoPrP-A 23–230) was expressed in E. coli and purified as described previously, by nickel (Ni) affinity and ion exchange chromatography (IEC) (Wang et al., 2010a), or by nickel affinity, size exclusion chromatography, and high performance liquid chromatography (HPLC) (Makarava and Baskakov, 2008). Recombinant α-helical prion protein (α-PrP) was used to represent cellular PrP (PrPC), which has the same structure, with the exception of mammalian post-translational modifications of glycosylation and glycophosphatidylinositol (GPI) anchor on native PrPC. Initial peptide recovery optimization experiments (using 1 or 4 µg PrP) used 0.5 µg/mL HPLC α-PrP preparation in water. For subsequent peptide recovery optimization experiments, HPLC α-PrP was concentrated to 5 µg/mL by dialyzing against absorbent resin in 10 mM acetate, 0.01 mM EDTA, pH 4.5 (Riek et al., 1997). Following peptide recovery optimization, all deuterium exchange experiments used α-PrP prepared by the Ni/IEC purification method.
Preparation of PrP-cofactor intermediate species
Mixtures of prion protein, 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (POPG, Avanti Polar Lipids, Alabaster, AL), and mouse liver total RNA (extracted with Trizol, Invitrogen, Carlsbad, CA) were prepared as described (Wang et al., 2010a). Briefly, POPG was incubated at 0.2 mg/mL with 0.25 mg/mL recombinant α-PrP for 10 min. at room temperature. This mixture was diluted 9-fold into 0.25% Triton X-100, 150mM NaCl, 10mM tris pH 7.5, generating PrP conversion intermediate 1 (PrPInt1). Following 5 min. incubation, RNA was added to 0.15 mg/mL concentration, generating PrP conversion intermediate 2 (PrPInt2). To prepare for deuterium exchange and remove excess phospholipid, each intermediate species was washed twice with 1% n-octyl-β-D-glucopyranoside (nOG) (Anatrace Affymetrix, Maumee, OH), 150 mM NaCl, 8.3 mM tris pH 7.2. Each wash was performed by centrifugation at 100,000×g for 30 min. at 4°C, removal of supernatant, and resuspension in wash buffer by vortexing for 10 sec. and sonicating at 85% power (Misonix 4000 with Microplate Horn, Qsonica, Newtown, CT) for 1 min.
Preparation of PrPSc by Protein Misfolding Cyclic Amplification
The recombinant prion strain designated OSU was used in this study. OSU is the recPrPSc sample originally produced de novo by F.W. as previously described, and which is infectious to wild type mice as determined by bioassay (Wang et al., 2010a). Sufficient quantities of PrPSc for this study were generated by propagating OSU PrPSc in the same manner using protein misfolding cyclic amplification (PMCA). Mixtures of PrP, POPG, and mouse liver RNA were prepared as described above. Each reaction mixture was then seeded with 10% volume PMCA product OSU PrPSc, followed by incubation at 37°C with intermittent sonication (30 sec. per 30 min.) at 75% power for 24 hours per round of PMCA.
Analytical Methods provided in Supplemental Information
Supplementary Material
Highlights.
Non-protein cofactors induce a major structural rearrangement in prion protein
Specific cofactor-induced changes in PrP persist through infectious PrPSc formation
Infectious PrPSc formation also involves global closure in PrP conformation
PrP rearrangement suggests key cofactor role in specifying prion structure
Acknowledgements
This manuscript is dedicated to Virgil Woods, whose expertise, enthusiasm, and mentorship was invaluable to the development of the project. Virgil’s fearless vision and detailed guidance for those under his leadership continue to inspire and encourage others following his footsteps. His unfortunate passing prevented him from seeing the full fruits of his labor in the completion of this work.
We thank Dan Walsh for providing HPLC-purified recombinant prion protein that was used in peptide recovery optimization experiments. We thank David Lee and Danny Tran for their assistance with peptide identification. MBM thanks Jon Engen and Thomas Wales for assistance in teaching the hydrogen-exchange mass spectrometry technique. MBM was supported by an NIH MD/PhD NRSA (F30 NS646373) and by the Burroughs Wellcome Fund Travel Award (1009071). This work was funded by NIH research grants to SS (2R01 NS046478), to JM (R01 NS071035, R01 NS060729), and a NIH High End Instrumentation grant to VLW, Jr. (S10 RR029388).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem. 1998;273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert review of proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Thirumalai D. Helices 2 and 3 are the initiation sites in the PrP(C) -->PrP(SC) transition. Biochemistry. 2013;52:310–319. doi: 10.1021/bi3005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB. Protease-sensitive synthetic prions. PLoS Pathog. 2010;6:e1000736. doi: 10.1371/journal.ppat.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SY, Chalmers MJ, Bruning J, Bramlett KS, Osborne HE, Montrose-Rafizadeh C, Barr RJ, Wang Y, Wang M, Burris TP, et al. Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc Natl Acad Sci U S A. 2008;105:7171–7176. doi: 10.1073/pnas.0710802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal componenets in vitro. Proc Natl Acad Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A. 2012a;109:8546–8551. doi: 10.1073/pnas.1204498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A. 2012b;109:E1938–E1946. doi: 10.1073/pnas.1206999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco ML, Daggett V. From conversion to aggregation: protofibril formation of the prion protein. Proc Natl Acad Sci U S A. 2004;101:2293–2298. doi: 10.1073/pnas.0307178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci U S A. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J Biomol Tech. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- Holscher C, Delius H, Burkle A. Overexpression of nonconvertible PrPc delta114–121 in scrapie-infected mouse neuroblastoma cells leads to trans-dominant inhibition of wild-type PrP(Sc) accumulation. J Virol. 1998;72:1153–1159. doi: 10.1128/jvi.72.2.1153-1159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann S, Schorn C, Wuthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu LL, Baxter NJ, Jackson GS, Power A, Clarke AR, Waltho JP, Craven CJ, Collinge J. Structural mobility of the human prion protein probed by backbone hydrogen exchange. Nat Struct Biol. 1999;6:740–743. doi: 10.1038/11507. [DOI] [PubMed] [Google Scholar]
- Kazlauskaite J, Sanghera N, Sylvester I, Venien-Bryan C, Pinheiro TJ. Structural changes of the prion protein in lipid membranes leading to aggregation and fibrillization. Biochemistry. 2003;42:3295–3304. doi: 10.1021/bi026872q. [DOI] [PubMed] [Google Scholar]
- Kim C, Haldiman T, Cohen Y, Chen W, Blevins J, Sy MS, Cohen M, Safar JG. Protease-sensitive conformers in broad spectrum of distinct PrPSc structures in sporadic Creutzfeldt-Jakob disease are indicator of progression rate. PLoS Pathog. 2011;7:e1002242. doi: 10.1371/journal.ppat.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, Surewicz WK. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285:14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- Lu X, Wintrode PL, Surewicz WK. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci U S A. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O'Brien P, Trojanowski JQ, Lee VM. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Baskakov IV. Expression and purification of full-length recombinant PrP of high purity. Methods Mol Biol. 2008;459:131–143. doi: 10.1007/978-1-59745-234-2_10. [DOI] [PubMed] [Google Scholar]
- Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Kovacs GG, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Stabilization of a prion strain of synthetic origin requires multiple serial passages. J Biol Chem. 2012;287:30205–30214. doi: 10.1074/jbc.M112.392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner SB. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- Miller MB, Geoghegan JC, Supattapone S. Dissociation of infectivity from seeding ability in prions with alternate docking mechanism. PLoS Pathog. 2011;7:e1002128. doi: 10.1371/journal.ppat.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Timmes AG, Wilmarth PA, Safronetz D, Priola SA. Identification and removal of proteins that co-purify with infectious prion protein improves the analysis of its secondary structure. Proteomics. 2011;11:3853–3865. doi: 10.1002/pmic.201100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazabal A, Hornemann S, Aguzzi A, Zenobi R. Hydrogen/deuterium exchange mass spectrometry identifies two highly protected regions in recombinant full-length prion protein amyloid fibrils. J Mass Spectrom. 2009;44:965–977. doi: 10.1002/jms.1572. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR. Isolation and Characterization of a Proteinase K-Sensitive PrP(Sc) Fraction. Biochemistry. 2006;45:15710–15717. doi: 10.1021/bi0615442. [DOI] [PubMed] [Google Scholar]
- Peretz D, Williamson RA, Matsunaga Y, Serban H, Pinilla C, Bastidas RB, Rozenshteyn R, James TL, Houghten RA, Cohen FE, et al. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–321) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- Saa P, Sferrazza GF, Ottenberg G, Oelschlegel AM, Dorsey K, Lasmezas CI. Strain-specific role of RNAs in prion replication. J Virol. 2012;86:10494–10504. doi: 10.1128/JVI.01286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajnani G, Silva CJ, Ramos A, Pastrana MA, Onisko BC, Erickson ML, Antaki EM, Dynin I, Vazquez-Fernandez E, Sigurdson CJ, et al. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog. 2012;8:e1002547. doi: 10.1371/journal.ppat.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera N, Pinheiro TJ. Binding of prion protein to lipid membranes and implications for prion conversion. J Mol Biol. 2002;315:1241–1256. doi: 10.1006/jmbi.2001.5322. [DOI] [PubMed] [Google Scholar]
- Shikiya RA, Bartz JC. In vitro generation of high-titer prions. J Virol. 2011;85:13439–13442. doi: 10.1128/JVI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2011;18:504–506. doi: 10.1038/nsmb.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnovas V, Kim JI, Lu X, Atarashi R, Caughey B, Surewicz WK. Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by hydrogen/deuterium exchange. J Biol Chem. 2009;284:24233–24241. doi: 10.1074/jbc.M109.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr J, Watts JC, Mensinger ZL, Oehler A, Grillo SK, DeArmond SJ, Prusiner SB, Giles K. Purified and synthetic Alzheimer's amyloid beta (Abeta) prions. Proc Natl Acad Sci U S A. 2012;109:11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. Generating a Prion with Bacterially Expressed Recombinant Prion Protein. Science. 2010a;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang F, Hu Y, Wang X, Wang X, Jin C, Ma J. Lipid Interaction Converts Prion Protein to a PrP(Sc)-like Proteinase K-Resistant Conformation under Physiological Conditions. Biochemistry. 2007;46:7045–7053. doi: 10.1021/bi700299h. [DOI] [PubMed] [Google Scholar]
- Wang F, Yin S, Wang X, Zha L, Sy MS, Ma J. Role of the highly conserved middle region of prion protein (PrP) in PrP-lipid interaction. Biochemistry. 2010b;49:8169–8176. doi: 10.1021/bi101146v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, McLoughlin SM, Frausto SD, Tang H, Emmett MR, Marshall AG. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal Chem. 2010;82:1450–1454. doi: 10.1021/ac902550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


