Abstract
The activity of natural killer (NK) cells is controlled by a balance of signals derived from inhibitory and activating receptors. TIGIT is a novel inhibitory receptor, recently shown in humans to interact with two ligands: PVR and Nectin2 and to inhibit human NK-cell cytotoxicity. Whether mouse TIGIT (mTIGIT) inhibits mouse NK-cell cytotoxicity is unknown. Here we show that mTIGIT is expressed by mouse NK cells and interacts with mouse PVR (mPVR). Using mouse and human Ig fusion proteins we show that while the human TIGIT (hTIGIT) cross-reacts with mPVR, the binding of mTIGIT is restricted to mPVR. We further demonstrate using surface plasmon resonance (SPR) and staining with Ig fusion proteins that mTIGIT binds to mPVR with higher affinity than the co-stimulatory PVR-binding receptor mDNAM1. Functionally, we show that triggering of mTIGIT leads to the inhibition of NK-cell cytotoxicity, that IFN-γ secretion is enhanced when mTIGIT is blocked and that the TIGIT-mediated inhibition is dominant over the signals delivered by the PVR-binding co-stimulatory receptors. Additionally, we identify the inhibitory motif responsible for mTIGIT inhibition. In conclusion we show that TIGIT is a powerful inhibitory receptor for mouse NK cells.
Keywords: NK cells, PVR, TIGIT
Introduction
NK cells, which belong to the innate immune system, are programmed to distinguish between healthy cells and virally-infected, stressed or malignant cells [1]. This NK-cell property is made possible by an array of activating and inhibitory receptors that regulate NK-cell activity. The ligands recognized by the NK activating receptors are mostly pathogen-derived, stress-induced, tumor-specific molecules or sometimes even self ligands (for example CD48 and AICL that serve as ligands for 2B4 and NKp80 respectively) [1]. In contrast, the inhibitory receptors mostly recognize self molecules, in particular they interact with MHC class I proteins [1, 2]. Several families of MHC class I-binding inhibitory receptors exist in humans which include the KIR (killer cell immunoglobulin-like receptors) and LIR (leukocyte immunoglobulin-like receptors) families [1], the Ly49 family in mice [1] and the CD94/NKG2 heterodimers in both species [1]. These receptors protect target cells from NK-cell-mediated killing upon interaction with classical and non-classical MHC class I proteins [1].
In addition, several inhibitory NK receptors interacting with non-MHC class I molecules also exists [3]. Among them is CEACAM1 which interacts homophilically with CEACAM1 [4]; CD300a which binds phosphatydylserine and phosphatidylethanolamine [5, 6]; Mafa (KLRG1) which binds cadherins [7–9]; NKR-P1A which interacts with LLT1 [10, 11]; LAIR1 which binds collagens [12, 13]; SIGLEC7 which binds α−2,8 disialic acid [14, 15]; and most importantly in regard to this manuscript TIGIT (also known as WUCAM and VSTM3) which interacts with PVR and PVR-related proteins [16–20].
All of the above mentioned inhibitory receptors deliver their inhibitory signals through 1–4 motifs present in their cytoplasmic tails, named immune-receptor tyrosine-based inhibitory motifs (ITIM) [2].
We have previously identified human TIGIT as a new receptor expressed on human NK cells, and demonstrated that it inhibits NK-cell cytotoxicity upon interaction with PVR and Nectin2 [17]. Human TIGIT possesses some unique characteristics: it is expressed on the entire peripheral blood NK-cell population [17], it contains one ITIM [16, 17, 19] and it binds PVR and Nectin2 with high affinity [16, 17, 19, 20]. It is also expressed by immune cells other than NK cells such as activated, regulatory and memory human T cells [16–19].
Initially, it was demonstrated that hTIGIT inhibits T cells indirectly by modulating cytokine production by dendritic cells [16]. Later we demonstrated that hTIGIT directly inhibits NK-cell-mediated cytotoxicity and others have shown that even in regard to T cells, hTIGIT is a direct inhibitory receptor as T-cell activity was inhibited by hTIGIT in the absence of APCs [18, 21, 22].
Human TIGIT interacts with PVR (CD155) and with Nectin2 (CD112) [16–20]. It was suggested that Nectin3 is also recognized by hTIGIT [16], however our data did not support this conclusion [17]. Interestingly, PVR and Nectin2 serve as ligands for the co-stimulatory receptor DNAM1 (CD226) [23–26], and PVR is also a ligand for another co-stimulatory receptor CD96 (Tactile) [27, 28].
Far less is known regarding TIGIT in mice. It was recently shown that the TIGIT murine orthologue (mTIGIT) binds mPVR and that it can inhibit the function of T cells [18, 21]. Whether TIGIT inhibits mouse NK-cell activity (cytotoxicity and cytokine secretion) is unknown.
Here we studied in detail the function of mTIGIT on mouse NK cells. We show that mTIGIT is expressed by mouse NK cells, that it interacts specifically with mPVR and possibly with an additional unknown ligand found on PBMCs and that these interactions lead to the inhibition of NK-cell activities. We also identify the mTIGIT motif that transduces its inhibitory signal and demonstrate that the mTIGIT-mediated inhibition is dominant over the signals mediated by the PVR-binding co-stimulatory receptors.
Results
Human and mouse TIGIT recognition of PVR
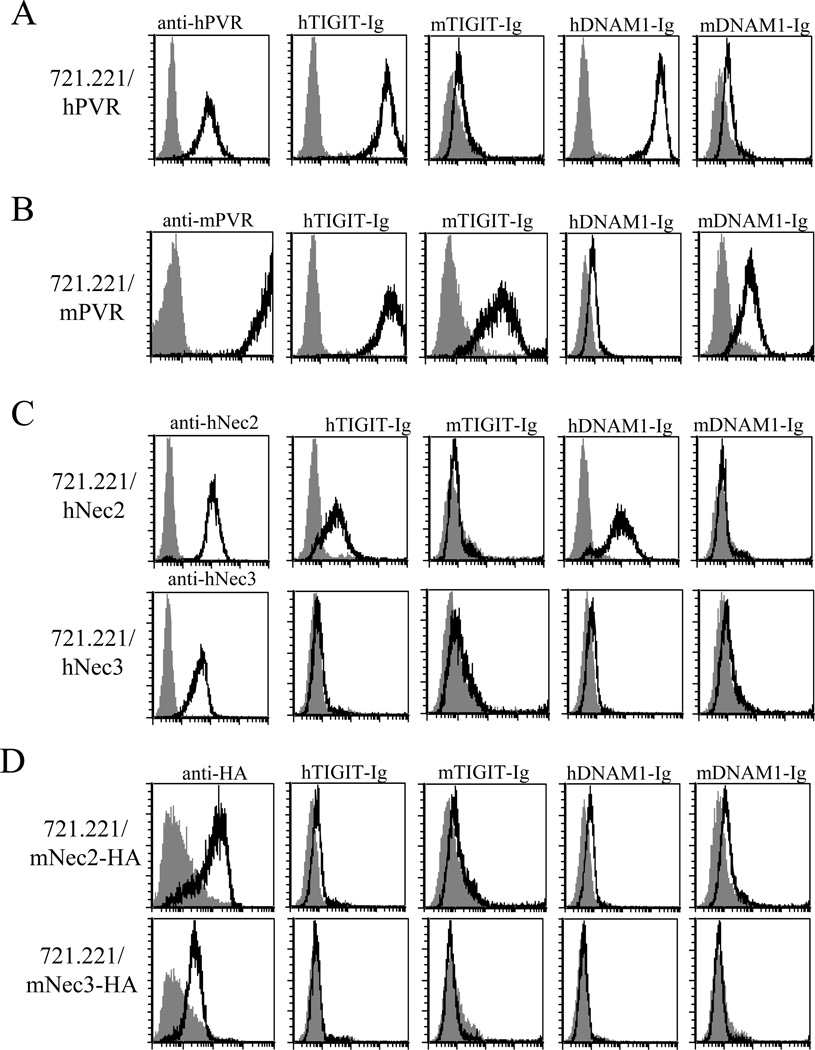
We have previously demonstrated that hTIGIT binds PVR and Nectin2, but not Nectin3 [17]. To test whether mTIGIT also recognizes mPVR, mNectin2 and possibly even mNectin3, we generated 721.221 transfectants expressing hPVR and mPVR (Fig. 1A and B respectively), hNectin2 and hNectin3 (Fig. 1C) as well as mNectin2 and mNectin3 (Fig. 1D). Because we initially lacked antibodies directed against mNectin2 and mNectin3, we tagged both proteins with HA to detect their expression on the cell surface. In addition, we generated 4 fusion proteins that compose the extracellular domains of the human and mouse TIGIT and DNAM1 fused to the Fc portion of human IgG1 (Fig. 1) and used these proteins to stain the various 721.221 transfectants. All fusion proteins used in this study were highly purified (purity was around 95%, data not shown). As demonstrated before [16–20], hPVR was recognized by hTIGIT-Ig and by hDNAM1-Ig fusion proteins, whereas little or no recognition of hPVR was observed when the mouse fusion receptors were used (Fig. 1A). In contrast, mPVR was recognized by all human and mouse receptors expect for hDNAM1-Ig (Fig. 1B). Human Nectin2 was recognized by the human receptors only and hNectin3 was not recognized by any of the receptors tested (neither human nor mouse, Fig. 1C). Mouse Nectin2 and Nectin3 proteins were not recognized by any of the receptors tested (Fig. 1D). We were surprised to observe that mNectin2 was not recognized by any of the receptors tested, as hTIGIT does interact with hNectin2 and it was shown that mNectin2 interacts with mDNAM1 [26]. Thus, we wondered whether the presence of the HA tag on mNectin2 prevents its recognition. To test this we generated 721.221 transfectants that express mNectin2 without an HA tag. The expression levels of both mNectin2-HA and mNectin2 were detected with a specific mAb and both transfectants were also stained with anti-HA mAb (Supporting Information Fig. 1, upper and lower histograms respectively). As can be seen, mNectin2 was not recognized by any of the receptors even in the absence of the HA tag (Supporting Information Fig. 1). Thus, we demonstrate that while hTIGIT binds hNectin2 and hPVR, and also cross-reacts with mPVR, the binding of mTIGIT is more restricted and this protein binds mPVR only. Similarly, we show that while hDNAM1 interacts with hPVR and hNectin2, mDNAM1 in our experimental system interacts with mPVR only.
Figure 1. Cross recognition of various human and mouse PVR binding receptors.
Flow cytometric analysis of 721.221 transfectants expressing (A) human PVR (hPVR), (B) mouse PVR (mPVR), (C) human Nectin2 (hNec2) and human Nectin3 (hNec3) and (D) mouse Nectin2 (mNec2) and mouse Nectin3 (mNec3) is shown. The transfectants in (D) are fused to HA tag. Cells were stained with specific antibodies (black lines, left histograms, indicated above the histograms) or with mouse and human PVR-binding receptors fused to Ig (black lines, indicated above the histograms). The gray-filled histograms are the basal staining of the parental 721.221 cell line. Data shown are from one representative experiment out of three performed.
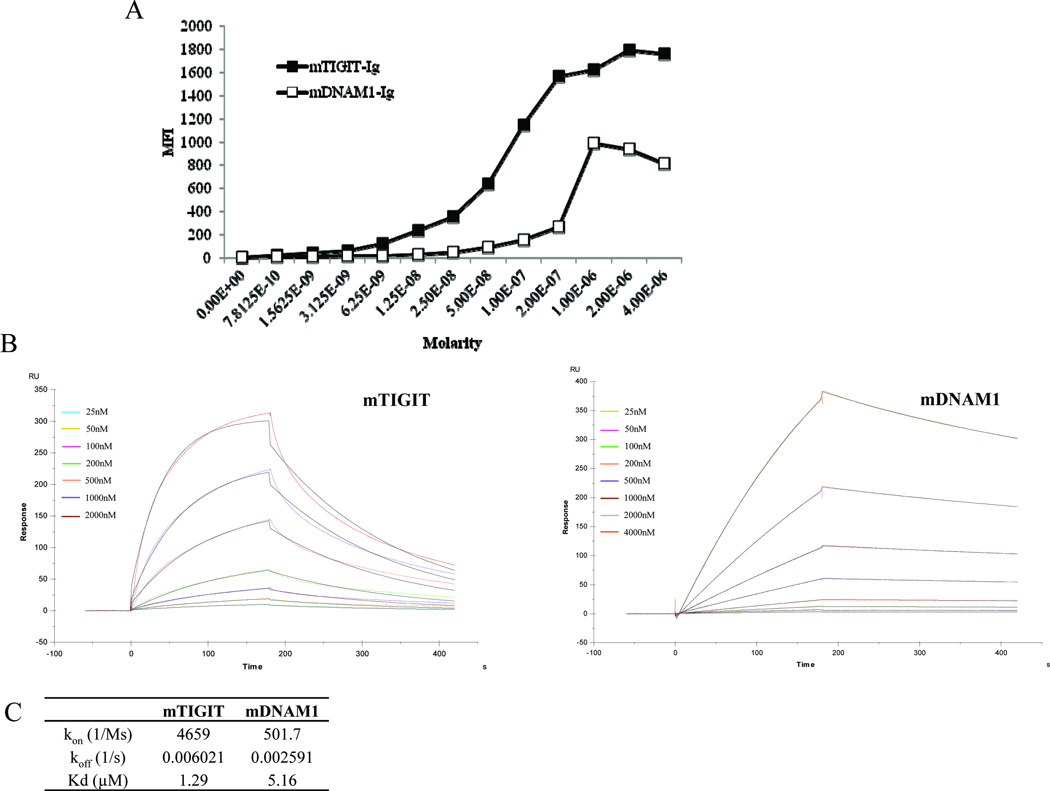
Because mPVR was the only ligand recognized by both mTIGIT and mDNAM1, we next compared the binding efficiencies of these two proteins to mPVR by using 721.221 transfectants expressing mPVR (the parental cell line does not express any other ligand for TIGIT or DNAM1, as depicted in figure 1) and increasing molar concentrations of mTIGIT-Ig and mDNAM1-Ig fusion proteins. As can be seen in figure 2A which depicts the median fluorescence intensity (MFI) of the binding of the two fusion proteins, a dose-dependent binding was observed and thus the binding of mTIGIT-Ig seemed to be of higher affinity as compared with that of mDNAM1-Ig (similar results were observed when the mean binding was calculated, data not shown). To directly compare the binding affinities mTIGIT and mDNAM1 to mPVR we performed SPR experiments using the mPVR-Ig fusion protein immobilized on a CM5 sensor chip and the two Ig fusion proteins: mTIGIT-Ig and mDNAM1-Ig. As can be seen in Figure 2B and C, mTIGIT-Ig indeed binds mPVR-Ig in a higher affinity as compared to mDNAM1-Ig (Kd values of 1.29 µM and 5.16 µM respectively) and while the koff for mTIGIT is almost 3 times faster than mDNAM (0.006021/s and 0.002591/s respectively), the kon of mTIGIT was of an order of magnitude faster (4659/Ms and 501.7/Ms respectively, Fig. 2B–C).
Figure 2. mTIGIT recognizes mPVR with higher affinity as compared with mDNAM1.
(A) The median fluorescent intensity (MFI) values from flow cytometric analysis of 721.221 transfectants expressing mPVR stained with mTIGIT-Ig and mDNAM1-Ig fusion proteins in the indicated molar concentrations are shown. Binding of the Ig fusion proteins was detected by phycoerythrin-conjugated anti-human IgG mAb. One representative experiment out of three performed is shown. (B) SPR analysis of mPVR-Ig interactions with mTIGIT-Ig (left) versus mDNAM1-Ig (right) using the bivalent analyte model. The molar concentration tested for mTIGIT-Ig and mDNAM1-Ig are indicated. (C) Binding parameters calculated based on the SPR analysis in (B).
hTIGIT and mTIGIT inhibit YTS cytotoxicity when interacting with mPVR
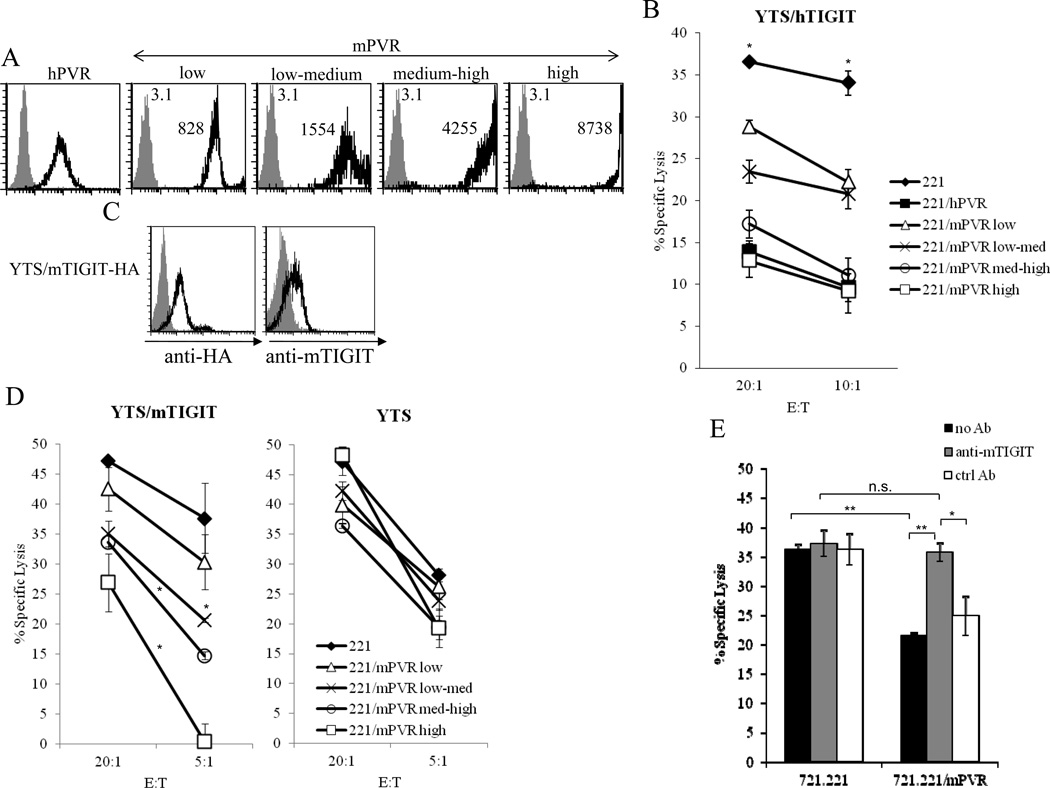
Because we observed (Fig. 1) that hTIGIT recognizes mPVR, we next tested whether this interaction can lead to the inhibition of NK cytotoxicity. To this end we conducted killing assays using the human NK cell line YTS, which do not express TIGIT and CD96 endogenously but do express DNAM1 (which is unable, due to an unknown reason, to interact with PVR ([17] and Supporting Information Fig. 2). YTS cells were transfected to express hTIGIT and assayed against target 721.221 cells expressing either hPVR, or 721.221 transfectants expressing mPVR at various levels (low, low-medium, medium-high and high, Fig. 3A). In agreement with the binding data presented in figure 1, we observed that the killing of the various 721.221 transfectants by YTS cells expressing hTIGIT was inhibited in a direct correlation with the expression levels of mPVR (Fig. 3B).
Figure 3. Human and mouse TIGIT inhibit YTS cytotoxicity upon interaction with mouse PVR.
(A) Flow cytometric analysis of 721.221 transfectants expressing either human PVR (left histogram) or mouse PVR (four right histograms) stained with anti-human PVR antibody or with rat anti-mPVR antibody respectively. Gray-filled histograms are the basal staining of the parental 721.221 cell line. The MFI values of mPVR expression are indicated. (B) Killing of the indicated 721.221 transfectants by YTS cells expressing human TIGIT. The effector-to-target (E:T) ratios are indicated on the x axis. The killing of each of the transfectants compared with that of the parental 721.221 cells showed a significant difference. *p<0.005, Student’s t test. Data are shown as mean + SD of three replicates and one representative experiment out of three performed is shown. (C) Flow cytometric analysis of YTS cells transfected to express mouse TIGIT attached to HA tag (YTS/mTIGIT-HA). The cells were stained with anti-HA mAb (left histogram, black line) or with rat anti-mTIGIT serum (right histogram, black line). The gray-filled histograms are the basal staining of the parental untransfected YTS cells. (D) Killing of the indicated 721.221 transfectans by YTS cells expressing mouse TIGIT (left) and the parental YTS cell line (right). The E:T ratios are indicated on the x axis. *p<0.05, Student’s t test, relative to the parental 721.221 cells. Data are shown as mean + SD of three replicates and one representative experiment out of three performed is shown. (E) Blocking of mTIGIT-PVR interactions. 721.221 cells or 721.221 cells expressing mPVR were incubated with YTS/mTIGIT-HA cells that were pre-incubated either with rat anti-mTIGIT sera, or with rat control sera. The E:T ratio was 10:1. *p<0.05, **p<0.005, n.s. non-significant, Student’s t test. Data are shown as mean + SD of three replicates and one representative experiment out of three performed is shown.
To demonstrate that mTIGIT can also inhibit NK cell cytotoxicity we used the YTS cell line once again and transfected it to express mTIGIT. When we started this project antibodies against mTIGIT were not available and thus we tagged the N terminus of mTIGIT with HA and expressed it in the YTS NK tumor line (Fig. 3C, left). Then, we proceeded to generate specific anti-mTIGIT antibodies by immunizing rats with mTIGIT-Ig and used these antibodies to demonstrate that they recognize the YTS/mTIGIT transfectants (Fig. 3C, right). Next we tested whether the various 721.221/mPVR transfectants (displayed in Fig. 3A) can inhibit YTS/mTIGIT cytotoxicity. As can be seen in figure 3D left, the interaction between mTIGIT and mPVR is indeed functional and the killing by YTS/mTIGIT was inhibited in a dose-dependent manner. No inhibition was observed when the parental YTS cells were used (Fig. 3D, right), or when 721.221 expressing hPVR were used as targets (in agreement with the lack of binding of mTIGIT to hPVR, Fig. 1) (data not shown). Finally, to directly demonstrate that the inhibition observed is due to mTIGIT interaction with mPVR, we blocked this interaction by using the anti-mTIGIT polyclonal antibodies we generated and, as can be seen in figure 3E, the YTS/mTIGIT inhibition was abolished.
Mechanism of mTIGIT inhibition
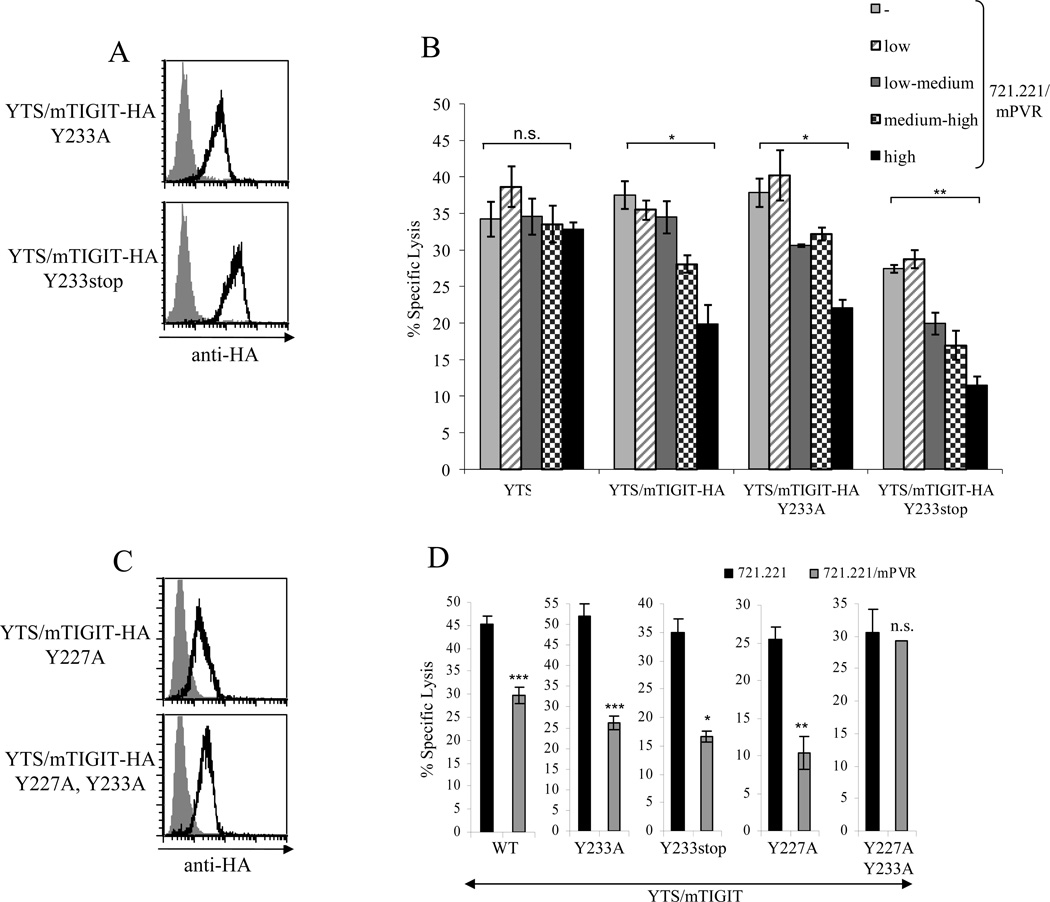
We have previously shown that the ITIM found in hTIGIT is involved in the transduction of its inhibitory signal [17]. The human and mouse TIGIT proteins share 58% identity in their amino acid sequences and importantly, the sequence in the cytoplasmic tail of hTIGIT (which contains the ITIM) is identical to that of mTIGIT (Fig. 4). To test whether the putative ITIM of mTIGIT is involved in the transduction of its inhibitory signal we generated two new mTIGIT constructs; one in which the tyrosine residue of the ITIM, located at position 233, was replaced with alanine (Y233A) and a second construct in which a stop codon was inserted instead of the tyrosine at position 233 (Y233stop). We tagged both mTIGIT mutants with HA and expressed them in YTS cells (Fig. 5A). The different YTS transfectants and the parental YTS cells were tested for the killing of 721.221 cells expressing various levels of mPVR (Fig. 3A). Since different YTS clones were generated they differ in the basal killing of 721.221 cells (Fig. 5B). For example, the killing of the untransfected 721.221 cells by YTS/mTIGIT-HA Y233stop was significantly lower as compared to the other YTS transfectants. Therefore, the inhibition mediated by the various TIGIT transfectants should be assayed per each clone individually and not compared between various YTS transfectants. As can be seen in figure 5B, inhibition of YTS cytotoxicity was observed in all cases, regardless of whether mTIGIT was mutated or not.
Figure 4. Alignment of the sequences of mouse and human TIGIT.
Alignment of the amino acid sequences of mouse and human TIGIT. The residues that are involved in the generation of hTIGIT homodimers are indicated in blue boxes. The amino acids implicated in hTIGIT binding to hPVR are indicated in green boxes which are numbered 1–3. The filled green residues mark the three contact amino acids that are different between human and mouse and human TIGIT. The conserved sequence in the TIGIT cytoplasmic tail which contains a classic ITIM (on the right side of the dashed red line) and a partial ITIM (on the left side of the dashed red line) is marked is by a red box. The tyrosine residues responsible for transducing mTIGIT’s inhibitory signal at positions 227 and 233 are highlighted in gray (the numbers refer to the mouse sequence only).
Figure 5. Identification of the inhibitory motif of mouse TIGIT.
(A) Flow cytometric analysis of YTS cell lines transfected to express mutated mTIGIT that contains HA tag and either a point mutation in the tyrosine residue located at position 233 (Y233A, upper histogram), or a stop codon at position 233 (Y233stop, bottom histogram). The various YTS cells were stained with anti-HA mAb. The gray-filled histograms are the basal staining of the parental YTS cells. (B) Killing of 721.221 cells, and 721.221 transfectants expressing different levels of mPVR, by the various YTS transfectants (indicated on the x axis). The E:T ratio was 40:1. Data are shown as mean + SD of three replicates and are from one experiment representative of three performed. *p<0.005, **p<0.001, Student’s t test. (C) Flow cytometric analysis of YTS transfectants expressing mTIGIT fused to HA tag, containing either point mutation at position 227 (YTS/mTIGIT-HA Y227A) or double mutations in positions 227 and 233 (YTS/mTIGIT-HA Y227A, Y233A). Transfectants were stained with anti-HA mAb (black lines). Gray-filled histograms are the basal staining of the parental YTS cell line. (D) Killing of 721.221 expressing mPVR or 721.221 parental cell line by the various YTS transfectants (indicated in the x axis). The E:T ratio was 40:1. Data are shown as mean + SD of three replicates and are from one experiment representative of three performed. *p<0.05, **p<0.005, ***p<0.001, Student’s t test.
An additional tyrosine residue is located near tyrosine 233, at position 227, in a sequence that partially resembles an ITIM (Fig. 4). To test whether this residue in mTIGIT is involved in transducing its inhibitory signal we mutated the tyrosine 227 residue into alanine (Y227A) and in addition generated a double-mutated mTIGIT in which both tyrosine residues were replaced with alanine (Y227A, Y233A). Both mTIGIT mutants were tagged with HA and expressed in YTS cells (Fig. 5C). Importantly, as shown in figure 5D, only when both tyrosine residues were mutated the TIGIT-mediated inhibition was abolished, indicating that each of these tyrosine residues is sufficient for the transduction of the mTIGIT inhibitory signal.
Mouse TIGIT inhibition of NK-cell cytotoxicity
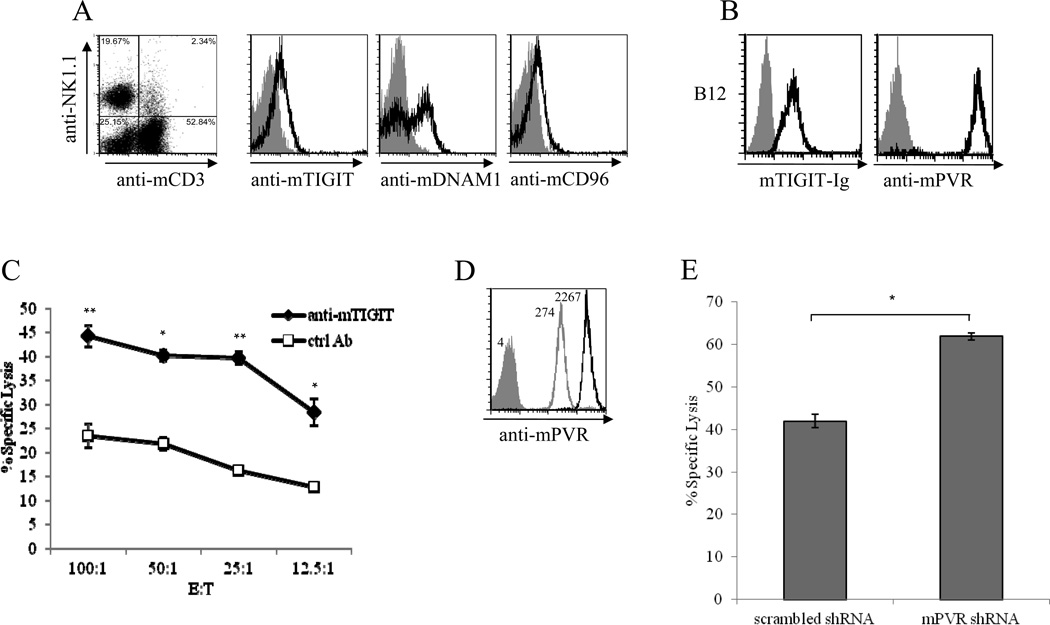
Our next aim was to demonstrate that mTIGIT inhibits the cytotoxicity of primary mouse NK cells. For this purpose we analyzed the expression of mTIGIT and the other mPVR-binding receptors, mDNAM1 and mCD96, on the surface of NK cells derived from C57BL mice. We observed that while the entire population of mouse NK cells expresses mTIGIT and mCD96, only a fraction of the NK cells tested express mDNAM1 (Fig. 6A).
Figure 6. Mouse TIGIT is an inhibitory receptor expressed by mouse NK cells.
(A) Flow cytometric analysis of mouse PBMCs isolated from C57BL mice 18 hours after poly(I:C) injection. The analysis was performed on the gated NK1.1-positive CD3-negative cells (NK cells, left dot plot). Cells were stained with the antibodies indicated in the x axis (black line histogram) following blocking of Fc-receptors with anti-mouse CD16/32 mAb. The gray-filled histograms represent staining with the appropriate isotype controls. (B) Flow cytometric analysis of B12 mouse fibroblasts cell line, stained with mTIGIT-Ig (left) or with rat anti-mPVR antibody (right). The gray-filled histograms are the background staining of the secondary antibodies only. (C) Blocking of TIGIT activity enhances the killing of B12 cells. C57BL mice were injected with poly(I:C) and 18 hours later PBMCs were isolated and pre-incubated with commercial sheep anti-mouse TIGIT polyclonal antibodies or with control sheep antibodies. These PBMCs were then incubated with B12 cells in the indicated E:T ratios and specific lysis was determined 5 hours later. Data are shown as mean + SD of three replicates and are from one experiment representative of three performed. *p<0.01, **p<0.001, Student’s t test. (D) Flow cytometric analysis of B12 cells transduced with shRNA vector against mPVR (gray-line histogram) or scrambled shRNA vector (black-line histogram), stained with rat anti-mPVR antibody. The gray-filled histogram is the background staining with the secondary antibody only. Numbers indicate MFI. (E) C57BL mice were injected with poly(I:C). 18 hours later PBMCs were isolated and incubated with the various B12 cells presented in (D) in E:T ratio of 60:1. Specific lysis was determined 5 hours later. Data are shown as mean + SD of three replicates and are from one experiment representative of three performed. *p<0.0005, Student’s t test).
To test whether mTIGIT can inhibit NK cell cytotoxicity, we used the murine B12 cell line which expresses mPVR and is recognized by mTIGIT-Ig (Fig. 6B). Importantly, as can be seen in figure 6C, blocking of mTIGIT using sheep anti-mTIGIT polyclonal antibodies significantly increased the killing of B12 cells in all effector to target (E:T) ratios tested.
In the next set of experiments we investigated whether the activity of mTIGIT is dominant over that of the PVR-binding co-stimulatory receptors, DNAM1 and CD96. Because we observed that mTIGIT binds mPVR with higher affinity compared to mDNAM1 (Fig. 2), since mDNAM1 is not expressed on the entire NK cell population (Fig. 6A) and since the function of mCD96 is not yet clear, we hypothesized that mTIGIT binding to mPVR will be dominant over mDNAM1 and mCD96. We assumed that if the co-stimulation signals dominate, we should see less killing in cells expressing reduced levels of PVR (as compared to the same cells expressing high levels of PVR) and if inhibition is dominant we should see increased killing after lowering the mPVR levels. To investigate this we used the murine B12 cell line, which endogenously expresses mPVR (Fig. 6B), and knocked down mPVR by using an shRNA construct (a scrambled shRNA was used as control). As can be seen, B12 cells transduced with a specific anti-mPVR shRNA expressed much lower levels of mPVR as compared to B12 cells transduced with a scrambled shRNA (Fig. 6D, gray and black empty histograms respectively). The transduced B12 cells were incubated with poly(I:C)-activated mouse PBMCs and elevated killing was observed in the B12 cells in which mPVR was knocked down (Fig. 6E). These results suggest that inhibition is the dominant signal mediated by the PVR-binding receptors.
The activity of mTIGIT is dominant over mCD96 and mDNAM1
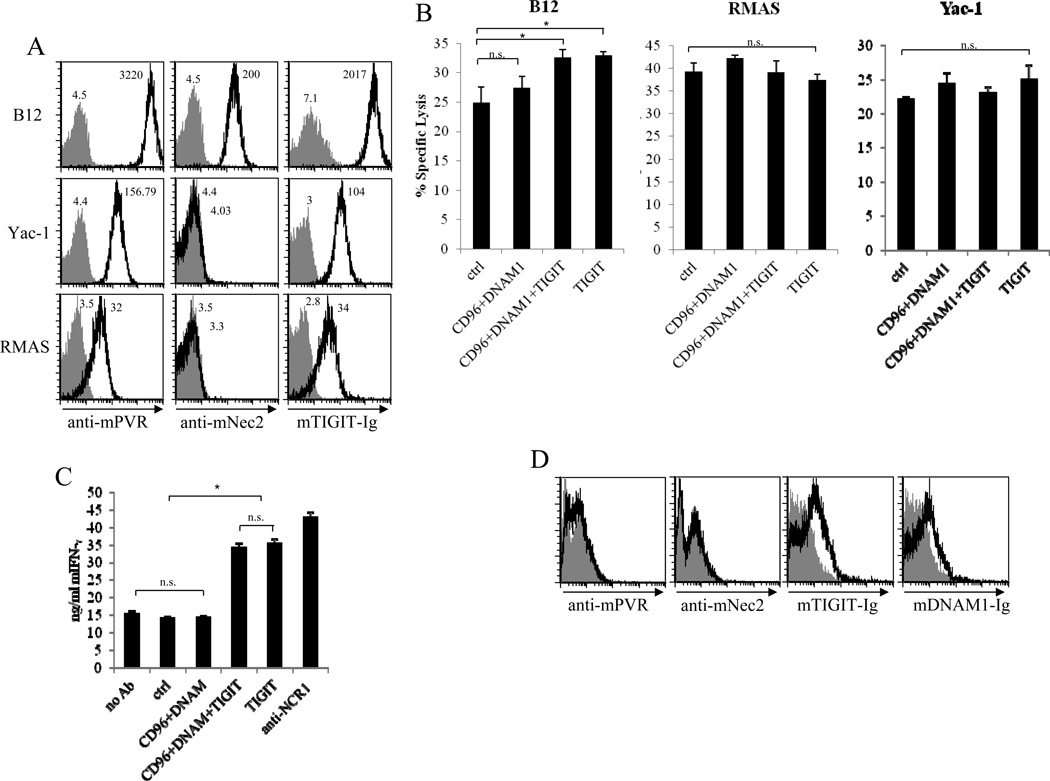
Next, we were interested to study the TIGIT activity against additional classic NK cell targets such as Yac-1 and RMAS. For that, we first examined whether RMAS and Yac-1 express mPVR and mNectin2 and observed that the expression of mPVR on both Yac-1 and RMAS cells was significantly lower than that of B12 and even lower than the transfectant of 721.221/mPVR expressing mPVR at the lowest levels (compare Figs. 7A and 3A). However, despite this lower expression mTIGIT-Ig binding was still detected on both RMAS and Yac-1 cell lines, albeit in much lower levels as compared to the B12 recognition (Fig. 7A).
Figure 7. Mouse TIGIT plays a dominant role in the inhibition of cytotoxicity and IFN-γ secretion.
(A) Flow cytometric analysis of B12, Yac-1 and RMAS cell lines, stained with anti-mPVR, anti-mNectin2 and mTIGIT-Ig (indicated in the x axis, black lines). The gray-filled histograms represent the staining with either isotype control or with secondary antibody only. (B, C) C57BL mice were injected with poly(I:C) and 18 hours later PBMCs were isolated and pre-incubated with anti-mouse CD16/32 to block the Fc receptors. Next, the cells were incubated with the various combinations of antibodies (indicated in the x axis) or with their appropriate isotype controls (ctrl) and were assayed either (B) for killing against B12, Yac-1 or RMAS cells, or (C) for IFN-γ secretion. Anti-NCR1 was bound to the plate and used to stimulate IFN-γ production by NK cells. Both killing and ELISA assays were performed for 5 hours. Data are shown as mean + SD of three replicates and are from one experiment representative of three performed. (B) *p<0.05, (C) *p<0.001, Student’s t test. (D) Flow cytometric analysis of PBMCs isolated from C57BL mice following poly(I:C) stimulation. Cells were first incubated with anti-mouse CD16/32 to block the Fc receptors and then stained with anti-mPVR and anti-mNectin2 antibodies and with mTIGIT-Ig and mDNAM1-Ig (black lines). The gray-filled histograms represent either isotype control or are the background staining of the secondary antibody only. Data shown are from one experiment representative of two performed.
To further demonstrate that mTIGIT inhibition is dominant over the co-activation signals of mCD96 and mDNAM1 we used all 3 cell lines (B12, RMAS and Yac-1) in NK cytotoxicity assays in the presence or in the absence of either anti-mTIGIT antibodies, with a combination of anti-mDNAM1 and mCD96 antibodies (we used antibodies that were previously shown to block the activity of both receptors [26, 28]) and with a triple combination of anti-mTIGIT, mDNAM1 and mCD96 antibodies. As shown in figure 7B, the addition of each of the antibodies did not affect the killing of either Yac-1 or RMAS cells, probably because of the low expression levels of PVR (the PVR expression levels on both cells were lower than the low PVR expression on 721.221 transfectants, compare Fig. 7A and 3A, and such low expression was not sufficient to significantly inhibit YTS/mTIGIT cytotoxicity, Fig. 3D). In contrast, in B12 cells mTIGIT had a dominant inhibitory role because when mCD96 and mDNAM1 were blocked no change in B12 cytotoxicity was observed and increased killing was noticed only when mTIGIT was blocked, either alone, or together with mCD96 and mDNAM1 (Fig. 7B).
In our final set of experiments we wanted to test whether blocking of mTIGIT will also affect cytokine secretion. When we started calibrating these experiments we were surprised to observed that increased IFN-γ secretion was observed even in the absence of targets, either when we incubated the PBMCs with antibodies against mTIGIT alone, or with antibodies against mTIGIT, mDNAM1 and mCD96 (Fig. 7C). In contrast, no change in IFN-γ secretion was observed when PBMCs were incubated either with control antibodies or with the combination of anti-mCD96 and mDNAM1 antibodies (Fig. 7C). The elevation in IFN-γ secretion, following the blocking of mTIGIT, was very pronounced and was equivalent to the IFN-γ secretion observed following triggering of NCR1 with anti-NCR1 antibodies (Fig. 7C). No change in the secretion of TNF-α was observed (data not shown).
We speculated that the increased IFN-γ secretion observed following the inclusion of anti-mTIGIT antibodies might result from expression of TIGIT ligands on the PBMCs themselves. To test this, we stained PBMCs derived from C57BL mice activated with poly(I:C) (as these were used in the cytotoxicity and cytokines experiments) for the expression of mPVR and mNectin2. In addition, we stained these PBMCs with mTIGIT-Ig and mDNAM1-Ig (Fig. 7D). Unexpectedly, we observed that although no mPVR nor mNectin2 expression could be detected, the PBMCs were recognized by mTIGIT-Ig and by mDNAM1-Ig (Fig. 7D), suggesting that they express a yet unknown ligand/s for these two receptors.
Discussion
We demonstrate here that mTIGIT inhibits mouse NK-cell cytotoxicity. Our data are in agreement with previous reports in which the physiological significance of mTIGIT was demonstrated through the generation of TIGIT knockout mice [18, 21]. It was demonstrated that mTIGIT is an inhibitory receptor for mouse T cells and that it interacts with mPVR [18]. However, it remained unknown whether mTIGIT can inhibit mouse NK-cell activity; whether it can bind ligands other than mPVR; and whether its activity is dominant over the co-stimulatory receptors that also interact with PVR.
The TIGIT knockout mice were not available to us and we have therefore used several fusion proteins, cell transfectants, SPR experiments and functional experiments to provide answers to all of the above questions. We showed that mPVR is recognized by mTIGIT while mNectin2 and mNectin3 are not. These results are in partial agreement with our previous observations regarding hTIGIT [17] as we have shown that hNectin3 is not recognized by hTIGIT. In addition, we showed that transfectants expressing mNectin2, which was previously shown to be mDNAM1 ligand [26], were not recognized neither by mTIGIT-Ig nor by mDNAM1-Ig fusion proteins. One of the possible explanations for these discrepancies might be that the expression levels of mNectin2 and mNectin3 on the surface of 721.221 cells are low and therefore they were not detected by the Ig fusion proteins. Indeed, the staining levels of the mouse Nectins were lower than that of mPVR, which may indicate lower expression levels (this comparison is a bit problematic since different antibodies are used to stain the various cells and they might differ in their binding affinities). Transfectants expressing higher levels of mNectin2 and mNectin3 could not be obtained. Another explanation might be the set of conditions used, i.e. the cells on which mNectin2 was overexpressed (RMA in [26] as compared with 721.221 (this study)). Whether mNectin2 is recognized by mTIGIT or by mDNAM1 will be investigated in the future using SPR.
Another difference in the binding properties of hTIGIT and mTIGIT was the cross-species specificity of these proteins. While hTIGIT interacts with both human and mouse PVR, mTIGIT interacts only with mPVR. The crystal structure of hTIGIT with hPVR was recently solved [20] and revealed that hTIGIT can form TIGIT/TIGIT homodimer that interacts with hPVR [20]. The core contact residues forming this homodimer were identified (blue boxes in Fig. 4) as well as the hTIGIT contact residues that interact with hPVR (green boxes 1–3 in Fig. 4). Alignment of the mouse and human amino acid sequences demonstrates that of all the hTIGIT contact residues (those required for the homodimer formation and those required for the binding to hPVR), only the three residues located at positions 69–71 differ between the mouse and the human proteins (included in green box number 2 and marked in light green, Fig. 4). Thus, these residues might contribute to the differences observed in the binding properties of human and mouse TIGIT. Further support of this assumption are mutational analysis experiments which demonstrated that mutating one of these residues, N70, in hTIGIT disrupted the binding to hPVR [20]. Whether these three residues are responsible for the differences in the cross-species specificity of the human and mouse TIGIT will be investigated in the future.
The observations that mPVR is recognized by mTIGIT, mDNAM and mCD96 (here and [18, 26, 28]) place mTIGIT in the pair-wise receptors category, in which the same ligand is recognized by both inhibitory and activating (co-stimulatory) receptors. The pair-wise receptors family includes pairs of proteins such as the KIR and the KAR [29], CTLA4 and CD28 [30], CD300a and possibly CD300c [31] and TIGIT together with DNAM1 and CD96 in mice (here) and in humans [17, 32]. One of the main characteristics of the pair-wise receptor family is that in all cases the binding of the inhibitory receptors is stronger than that of the corresponding co-stimulatory/activating receptors. For example, it was shown that the binding of CTLA4 to its ligands B7.1/2 is 20 times higher than that of its co-stimulatory counterpart, CD28 [33]. The binding of the KIR inhibitory receptors to MHC class I is much more efficient as compared with the activating KAR receptors[29, 32]. Consistent with this, it was previously shown that the binding of hTIGIT to PVR is stronger than that of human DNAM1 [16].
Using SPR experiments we observed that the binding of mTIGIT to mPVR is of higher affinity and faster kinetics as compared to mDNAM1. The fast association and dissociation rates of the inhibitory mTIGIT resembles those of other inhibitory receptors and this correlates with the need for inhibition to occur very rapidly, but only transiently [34]. The fact that mTIGIT binds PVR with high affinity has functional consequences since, as we show here, functionally TIGIT inhibition is dominant.
Interestingly, we observed that blocking of mCD96 and mDNAM1 activity against B12, RMAS and Yac-1 cells had no effect on NK-cell cytotoxicity. Because mDNAM1 and mCD96 are co-stimulatory receptors, the killing mediated by these two receptors is heavily dependent on the expression of other ligands expressed by target cells. It will be interesting to test in the future under what conditions the expression and function of all PVR-binding receptors is differentially regulated and whether in other targets the function of mDNAM1 and mCD96 will be more significant.
These observations might shed new light on previous data regarding the in vivo function of the co-stimulatory receptors for PVR. For example, it was shown that DNAM1-deficient mice manifest enhanced tumor spread, suggesting that DNAM1 might contribute to tumor surveillance [35, 36]. However, it should be taken into account that without the activation signals of DNAM1, the inhibition of immune cells mediated by TIGIT might be even more pronounced and therefore it is possible that much of the effect observed is not due to the lack of DNAM1 but rather to the increased TIGIT-mediated inhibition.
We found that mTIGIT inhibits cytotoxicity and also affect IFN-γ secretion even in the absence of targets and that poly(I:C)-activated PBMCs express an unknown ligand/s for mTIGIT and mDNAM1. This new ligand/s might be responsible for the protection of immune cells from an autoreactive response of NK cells.
Most of the NK inhibitory receptors transmit their signal through 2–4 ITIMs found in their cytoplasmic tail. 26 amino acids usually space between two ITIMs [37] and following the engagement of a particular inhibitory receptor, primarily SHP1 is recruited [37]. TIGIT, both in humans and in mice is quite unusual in this regard as it contains only one classical ITIM, which is preceded by a partial ITIM, found only 5 amino acids away (Fig. 4). We have previously shown in humans that the classical ITIM is responsible (at least in part) for delivering the inhibitory signal [17]. However, since we could not precipitate SHP1 with hTIGIT we suggested that the inhibitory mechanism of hTIGIT is different from that of the classical NK inhibitory receptors [17]. Indeed a recent publication identified this supposedly ‘partial ITIM’ as an ITT-like motif and showed it has a dominant role in transferring the inhibitory signal of human TIGIT, involving Grb2 and SHIP1 recruitment [38]. Here we demonstrate that the ITIM of mTIGIT together with the ITT-like motif are involved in the inhibition mediated by this novel and powerful inhibitory receptor.
Materials and Methods
Generation of Fusion Proteins
The generation of hTIGIT-Ig and hDNAM1-Ig was previously described [17]. For the production of mTIGIT-Ig fusion protein, the sequence encoding the extracellular portion of mTIGIT was amplified by PCR using the 5’ primer CCGAATTCGCCGCCACCATGCATGGCTGGCTGCTCCT (including EcoRI restriction site) and the 3’ primer CGGATCCGGGGCAGTCTGGAACTGAG (including BamHI restriction site). For the production of mDNAM1-Ig fusion protein, the sequence encoding the extracellular portion of mDNAM1 was amplified by PCR using the 5’ primer CCGATATCGCCGCCACCATGGCTTATGTTACTTGGCTTTTG (including EcoRV restriction site) and the 3’ primer CGGATCCGGAAGGATAAAATGTTTATTGGTTC (including BamHI restriction site). For the production of mPVR-Ig fusion protein, the sequence encoding the extracellular portion of mPVR was amplified by PCR using the 5’ primer CCGAATTCGCCGCCACCATGGCTCAACTCGCCCGAG (including EcoRI restriction site) and the 3’ primer CCAGATCTAATCTTGTATTTTGCTGCATATTC (including BglII restriction site). These PCR-generated fragments were cloned into the expression vector containing the mutated Fc portion of human IgG1 (Fc mut pIRESpuro3) which was previously described [17]. SDS/PAGE analysis revealed that the Ig fusion proteins were approximately 95% pure.
Generation of transfectants
Generation of 721.221 cells expressing hPVR, hNectin2 and hNectin3 was previously described [17]. For the generation of 721.221 cells expressing mPVR, the cDNA was amplified by PCR using the 5’ primer CCGAATTCGCCGCCACCATGGCTCAACTCGCCCGAG (including EcoRI restriction site) and the 3’ primer GGGCTCGAGTCACCTTGTGCTGTTTGGCTC (including XhoI restriction site). For the generation of 721.221 cells expressing mNectin2 fused to HA tag the signal peptide of mNectin2 was amplified by PCR using the 5’ primer CCCAAGCTTGCCGCCACCATGGCCCGGGCCGCAGTC (including HindIII restriction site) and the 3’ primer AGCGTAATCTGGAACATCGTATGGGTAATCTTGGGCTCCTGTTTCCTG (including the twenty seven nucleotides encoding the HA tag). mNectin2 was amplified by PCR using the 5’ primer TACCCATACGATGTTCCAGATTACGCTGTGCGGGTACGAGTGCTTCC (including the 27 nucleotides encoding the HA tag) and the 3’ primer GGTCTAGATCACACATACATGGCCCGTGA (including XbaI restriction site).
For the generation of 721.221 cells expressing mNectin3 fused to HA tag the signal peptide of mNectin3 was amplified by PCR using the 5’ primer CCCAAGCTTGCCGCCACCATGGCGCGGACCCCGGGC (including HindIII restriction site) and the 3’ primer AGCGTAATCTGGAACATCGTATGGGTATGATCCAGCTAAGGCACCACA (including the twenty seven nucleotides encoding the HA tag). mNectin3 was amplified by PCR using the 5’ primer TACCCATACGATGTTCCAGATTACGCTATTATTGTGGAGCCACATGTCAC (including the 27 nucleotides encoding the HA tag) and the 3’ primer GGTCTAGATTAGACATACCACTCCCTCCTG (including XbaI restriction site). The mPVR, mNectin2-HA and mNectin3-HA constructs were cloned into pcDNA3 vector (Invitrogen Life Technologies) and were stably expressed in 721.221 cells.
For the generation of 721.221 cells expressing mNectin2 (without HA tag), mNectin2 cDNA was amplified by PCR using the 5’ primer CGGATCCGCCGCCACCATGGCCCGGGCCGCAGTC (including BamHI restriction site) and the 3’ primer GGAATTCTCACACATACATGGCCCGTGA (including EcoRI restriction site). The mNectin2 was then cloned into the lentiviral vector SIN18-pRLL-hEFI_p-EGFPWRPE [39], instead of the GFP coding sequence. Lentiviral viruses were produced by transient three-plasmid transfection as described [40].
The generation of YTS NK tumor line expressing hTIGIT fused to HA tag was previously described [17]. For the generation of YTS cells expressing mTIGIT fused to HA tag the signal peptide of mTIGIT was amplified by PCR using the 5’ primer CGGATCCGCCGCCACCATGCATGGCTGGCTGCTCCT (including BamHI restriction site) and the 3’ primer AGCGTAATCTGGAACATCGTATGGGTAGAAGGCAGCCTGTATCAGCC (including the twenty seven nucleotides encoding the HA tag). mTIGIT was amplified by PCR using the 5’ primer TACCCATACGATGTTCCAGATTACGCTCTCGCTACAGGAGCCACAGC (including the 27 nucleotides encoding the HA tag) and the 3’ primer CCTGTACATTAGCCAGTCTTCGATACAGCA (including Bsp1407 restriction site).
For generation of the mTIGIT protein mutated in Y233A, we used the mTIGIT-HA construct as a template and amplified by PCR the 3’ end of the gene by using an internal 5’ primer GTCCTGAGTGCCAGAAGCCT bearing the mutation and the gene-specific 3’-edge primer (including the Bsp1407 restriction site). The PCR product was used as a 3’ primer together with the gene-specific 5’-edge primer (including the BamHI restriction site) for the generation of the mutated full-gene cDNA. For generation of the mTIGIT protein mutated in Y227A, we used the mTIGIT-HA construct as a template and amplified by PCR the 3’ end of the gene by using an internal 5’ primer CCACAGGAAGCCTTTAATGTCC bearing the mutation and the gene-specific 3’-edge primer (including the Bsp1407 restriction site). The PCR product was used as a 3’ primer together with the gene-specific 5’-edge primer (including the BamHI restriction site) for the generation of the mutated full-gene cDNA.
For generation of the mTIGIT protein double mutated in Y233A and Y227A, we used the mTIGIT-HA Y233A construct as a template for the PCRs generating Y227A, as described above.
For generation of the truncated mTIGIT protein Y233stop, we used the mTIGIT-HA construct as a template and amplified mTIGIT by PCR using the gene-specific 5’-edge primer (including the BamHI restriction site) and the 3’ primer CCTGTACACTAGCTCAGGACATTAAAGTATTCC (including a stop codon and Bsp1407 restriction site). The mTIGIT-HA and the four mutated constructs were then cloned into the lentiviral vector SIN18-pRLL-hEFI_p-EGFPWRPE [39], instead of the GFP coding sequence. Lentiviral viruses were produced by transient three-plasmid transfection as described [40].
Antibodies
The Abs used in this work were anti-human CD155/PVR clone 300907 (R&D Systems), anti-mouse PVR clone 3F1 (Hycult biotech), anti-human PVRL2/CD112 clone TX31 (BioLegend), anti-human PVRL3/CD113 clone N3.12.4 (Santa Cruz Biotechnology), anti-mouse and human PVRL2/CD112 clone 502-57 (abcam), 12CA5 directed against HA. For detection of mouse NK cells PE-conjugated anti-mouse NK1.1 clone PK136 (PharMingen) and FITC-conjugated anti-mouse CD3 clone 17A2 (BioLegend) were used. For mTIGIT staining and blocking allophycocyanin-conjugated and purified sheep polyclonal anti-mouse TIGIT were used respectively (R&D Systems). Additional antibodies used were allophycocyanin-conjugated and purified sheep IgG control antibodies (R&D Systems), anti-mouse CD96 clone 6A6 (Hycult biotech, used in the blocking experiments), anti-mouse DNAM1 clone TX42.1 (BioLegend, used in the blocking experiments), Rat IgG2a Isotype control clone RTK2758 (BioLegend), allophycocyanin-conjugated anti-mouse CD96 clone 3.3 (BioLegend), allophycocyanin-conjugated Rat IgG1 Isotype control clone RTK2071 (BioLegend), Alexa Fluor 647-conjugated anti-mouse DNAM1 clone TX42.1 (BioLegend) and Alexa Fluor 647-conjugated Rat IgG2a Isotype control clone RTK2758 (BioLegend). The anti-mouse CD16/32 clone 93 (eBioscience) mAb was used to block Fc receptors. Polyclonal antibodies to mouse TIGIT were generated in rat, immunized with mTIGIT-Ig fusion protein. The rat serum was tested for specific recognition of mTIGIT-Ig fusion protein by WB and of YTS/mTIGIT transfectants by flow cytometry.
Mouse IFN-γ secretion
C57BL mice were injected with poly(I:C) and 18 hours later PBMCs were isolated and pre-incubated with anti-mouse CD16/32 to block the Fc receptors. Next, 500,000 PBMCs per well were incubated for 5 hours with various combinations of blocking antibodies or with their appropriate isotype controls. Anti-NCR1 was bound to the plate and used to stimulate IFN-γ production by NK cells. Secreted Mouse IFN-γ was measured by ELISA using purified anti-mouse IFN-γ clone R4-6A2 (BioLegend) as capture antibody and biotin-conjugated anti-mouse IFN-γ clone XMG1.2 (BioLegend) as detection antibody, followed by peroxidase-conjugated streptavidin (Jackson ImmunoResearch). The concentrations of mouse IFN-γ were estimated by comparing the absorbance at 650 nm, after subtraction of the blank control absorbance values, with a standard curve made by recombinant mouse IFN-γ standard (PROSPEC). All samples were run in triplicates.
Cytotoxicity Assay
The cytotoxic activity of primary mouse PBMCs and YTS cells against the mouse B12, Yac-1 and RMAS cell lines and against 721.221cells was assessed in 5-h 35S release assays as previously described [41].
Surface plasmon resonance analysis
A Biacore™ T100 (GE Healthcare) Biosensor was used to assay the interaction of mPVR-Ig with mTIGIT-Ig and mDNAM1-Ig. mPVR-Ig was immobilized to CM5 sensor chip via amine coupling kit by activating both the active and reference surfaces with 1:1 EDC:NHS mixture for 7 minutes at a flow rate of 10 mL/min. Then, solution of 50nM of mPVR-Ig in immobilization buffer was passed over the active surface only for 6 minutes at 10 mL/min. Blocking of both active and reference channels was accomplished by flowing 1M Ethanolamine blocking solution for 7 minutes at 10 mL/min. mPVR was immobilized at 4875RU. Ligands (mTIGIT-Ig and mDNAM1-Ig) were prepared at a range of concentrations from 25nM to 4000nM diluted in running buffer and were injected onto both active and reference surfaces for 180sec at 50 mL/min. Dissociation was monitored for 180 sec. Double referencing was accomplished by subtracting signal from both reference channel and blank injection with running buffer. Surface was regenerated between injection cycles with regeneration solution for 60 sec at 10 mL/min. BiaEvaluation software was used to perform kinetic analysis fit in the Bivalent analyte analysis.
Supplementary Material
Acknowledgements
This study was supported by grants from the Israeli Science Foundation, The Croatia Israel Research Grant, by the Rosetrees trust, by the ICRF by the Israeli ICORE, by the Association for International Cancer Research (AICR), and by the ERC advanced grant all to OM and by NIH grant 1R01AI083201-01 to S. Jonjic. O.M is a Crown professor of Molecular Immunology.
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar V, McNerney ME. A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat Rev Immunol. 2005;5:363–374. doi: 10.1038/nri1603. [DOI] [PubMed] [Google Scholar]
- 4.Markel G, Lieberman N, Katz G, Arnon TI, Lotem M, Drize O, Blumberg RS, Bar-Haim E, et al. CD66a interactions between human melanoma and NK cells: a novel class I MHC-independent inhibitory mechanism of cytotoxicity. J Immunol. 2002;168:2803–2810. doi: 10.4049/jimmunol.168.6.2803. [DOI] [PubMed] [Google Scholar]
- 5.Nakahashi-Oda C, Tahara-Hanaoka S, Honda S, Shibuya K, Shibuya A. Identification of phosphatidylserine as a ligand for the CD300a immunoreceptor. Biochem Biophys Res Commun. 2012;417:646–650. doi: 10.1016/j.bbrc.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Simhadri VR, Andersen JF, Calvo E, Choi SC, Coligan JE, Borrego F. Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood. 2012;119:2799–2809. doi: 10.1182/blood-2011-08-372425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundemann C, Bauer M, Schweier O, von Oppen N, Lassing U, Saudan P, Becker KF, Karp K, et al. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J Immunol. 2006;176:1311–1315. doi: 10.4049/jimmunol.176.3.1311. [DOI] [PubMed] [Google Scholar]
- 8.Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med. 2006;203:289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzkopff S, Grundemann C, Schweier O, Rosshart S, Karjalainen KE, Becker KF, Pircher H. Tumor-associated E-cadherin mutations affect binding to the killer cell lectin-like receptor G1 in humans. J Immunol. 2007;179:1022–1029. doi: 10.4049/jimmunol.179.2.1022. [DOI] [PubMed] [Google Scholar]
- 10.Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175:7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 11.Aldemir H, Prod'homme V, Dumaurier MJ, Retiere C, Poupon G, Cazareth J, Bihl F, Braud VM. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 12.Lebbink RJ, de Ruiter T, Kaptijn GJ, Bihan DG, Jansen CA, Lenting PJ, Meyaard L. Mouse leukocyte-associated Ig-like receptor-1 (mLAIR-1) functions as an inhibitory collagen-binding receptor on immune cells. Int Immunol. 2007;19:1011–1019. doi: 10.1093/intimm/dxm071. [DOI] [PubMed] [Google Scholar]
- 13.Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, Farndale RW, Lisman T, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falco M, Biassoni R, Bottino C, Vitale M, Sivori S, Augugliaro R, Moretta L, Moretta A. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med. 1999;190:793–802. doi: 10.1084/jem.190.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker PR, Varki A. Siglecs, sialic acids and innate immunity. Trends Immunol. 2001;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 17.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin SD, Taft DW, Brandt CS, Bucher C, Howard ED, Chadwick EM, Johnston J, Hammond A, et al. Vstm3 is a member of the CD28 family and an important modulator of T-cell function. Eur J Immunol. 2011;41:902–915. doi: 10.1002/eji.201041136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boles KS, Vermi W, Facchetti F, Fuchs A, Wilson TJ, Diacovo TG, Cella M, Colonna M. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur J Immunol. 2009;39:695–703. doi: 10.1002/eji.200839116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stengel KF, Harden-Bowles K, Yu X, Rouge L, Yin J, Comps-Agrar L, Wiesmann C, Bazan JF, et al. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc Natl Acad Sci U S A. 2012;109:5399–5404. doi: 10.1073/pnas.1120606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, Sharpe AH, Kuchroo VK. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J Immunol. 2011;186:1338–1342. doi: 10.4049/jimmunol.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 axis regulates human T cell function. J Immunol. 2012;188:3869–3875. doi: 10.4049/jimmunol.1103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pende D, Bottino C, Castriconi R, Cantoni C, Marcenaro S, Rivera P, Spaggiari GM, Dondero A, et al. PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: involvement in tumor cell lysis. Mol Immunol. 2005;42:463–469. doi: 10.1016/j.molimm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 25.Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, et al. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112) Int Immunol. 2004;16:533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 26.Tahara-Hanaoka S, Miyamoto A, Hara A, Honda S, Shibuya K, Shibuya A. Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem Biophys Res Commun. 2005;329:996–1000. doi: 10.1016/j.bbrc.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 28.Seth S, Maier MK, Qiu Q, Ravens I, Kremmer E, Forster R, Bernhardt G. The murine pan T cell marker CD96 is an adhesion receptor for CD155 and nectin-1. Biochem Biophys Res Commun. 2007;364:959–965. doi: 10.1016/j.bbrc.2007.10.102. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Botet M, Bellon T, Llano M, Navarro F, Garcia P, de Miguel M. Paired inhibitory and triggering NK cell receptors for HLA class I molecules. Hum Immunol. 2000;61:7–17. doi: 10.1016/s0198-8859(99)00161-5. [DOI] [PubMed] [Google Scholar]
- 30.Oosterwegel MA, Greenwald RJ, Mandelbrot DA, Lorsbach RB, Sharpe AH. CTLA-4 and T cell activation. Curr Opin Immunol. 1999;11:294–300. doi: 10.1016/s0952-7915(99)80047-8. [DOI] [PubMed] [Google Scholar]
- 31.Lankry D, Simic H, Klieger Y, Levi-Schaffer F, Jonjic S, Mandelboim O. Expression and function of CD300 in NK cells. J Immunol. 2010;185:2877–2886. doi: 10.4049/jimmunol.0903347. [DOI] [PubMed] [Google Scholar]
- 32.Stanietsky N, Mandelboim O. Paired NK cell receptors controlling NK cytotoxicity. FEBS Lett. 2010;584:4895–4900. doi: 10.1016/j.febslet.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 33.McAdam AJ, Schweitzer AN, Sharpe AH. The role of B7 co-stimulation in activation and differentiation of CD4+ and CD8+ T cells. Immunol Rev. 1998;165:231–247. doi: 10.1111/j.1600-065x.1998.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 34.Vales-Gomez M, Reyburn HT, Mandelboim M, Strominger JL. Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors. Immunity. 1998;9:337–344. doi: 10.1016/s1074-7613(00)80616-0. [DOI] [PubMed] [Google Scholar]
- 35.Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, Andrews DM, Smyth MJ, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med. 2008;205:2965–2973. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, Yasui T, Kikutani H, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205:2959–2964. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Zhang H, Li M, Hu D, Li C, Ge B, Jin B, Fan Z. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013;20:456–464. doi: 10.1038/cdd.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu K, Ma H, McCown TJ, Verma IM, Kafri T. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- 40.Blomer U, Naldini L, Kafri T, Trono D, Verma IM, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.