Abstract
A drawback of gene therapy using adeno-associated virus (AAV) is the DNA packaging restriction of the viral capsid (<4.7 kb). Recent observations demonstrate oversized AAV genome transduction through an unknown mechanism. Herein, AAV production using an oversized reporter (6.2 kb) resulted in chloroform and DNase-resistant particles harboring distinct “fragment” AAV (fAAV) genomes (5.0, 2.4, and 1.6 kb). Fractionation experiments determined that only the larger “fragments” mediated transduction in vitro, and relatively efficient transduction was also demonstrated in the muscle, the eye, and the liver. In contrast with concatemerization-dependent large-gene delivery by split AAV, fAAV transduction is independent of the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) in vitro and in vivo while disproportionately reliant on the DNA strand–annealing protein Rad51C. Importantly, fAAV's unique dependence on DNA repair proteins, compared with intact AAV, strongly suggests that the majority of oversized AAV transduction is mediated by fragmented genomes. Although fAAV transduction is less efficient than intact AAV, it is enhanced fourfold in muscle and sevenfold in the retina compared with split AAV transduction. Furthermore, fAAV carrying codon-optimized therapeutic dysferlin cDNA in a 7.5 kb expression cassette restored dysferlin levels in a dystrophic model. Collectively, oversized AAV genome transduction requires unique DNA repair pathways and offers an alternative, more efficient strategy for large-gene therapy.
Introduction
Adeno-associated virus (AAV) is a small (25 nm) single-stranded DNA (ssDNA, 4.7 kb) virus routinely used as a viral vector for human gene therapy. Many genetic variants of AAV exist, which allow transduction of a wide range of species and specific types of human tissues. AAV vectors (rAAV) carry only the 145-nucleotide inverted terminal repeats (ITRs) and have contributed significantly to the field of gene transfer, with >70 clinical trials initiated for a spectrum of genetic diseases.1,2 Among the many favorable attributes of rAAV, the fact that the AAV capsid is among the smallest described, imposes a DNA-packaging limitation (<5 kb) that has restricted its widespread application. This size constraint has traditionally rendered rAAV deficient for gene addition approaches requiring large transgenic cDNA, such as in hemophilia A, cystic fibrosis, and several types of muscular dystrophies, including dysferlinopathy.
rAAV cellular uptake relies on receptor-mediated endocytosis and culminates in the presence of ssDNA within the host nucleus.3,4 There, the palindromic T-shaped ITR is extended by host-catalyzed second-strand synthesis in a manner independent of the cell cycle phase or cell division. The now-linear duplex DNA molecule is capable of transcription/transgene production and primarily persists as a double-strand episome. Previous work using circularization-dependent single-stranded rAAV demonstrated that intramolecular circularization of the linear duplexed transgenic DNA occurs rapidly and is, initially, the primary persistent form of rAAV transgenic DNA.5 To a lesser extent, intermolecular linkages of rAAV genomes occur and increase in prevalence over time to form larger concatemers. Both of these processes, rAAV circularization and concatemer formation, are carried out by host DNA repair proteins representative of the major canonical pathways, homologous recombination (HR), and nonhomologous end joining (NHEJ).5,6,7,8,9 Previous reports have demonstrated that the ITRs are “recombinogenic” and play a role in circularization/concatemerization because they are bound and processed by effectors of both HR and NHEJ.6,10 Work from the Nakai lab has implicated two proteins involved in NHEJ, DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Artemis, for ITR processing, which were found necessary for concatemer formation in vivo.11 In another study using dividing cells, ITR-induced HR was demonstrated by the ability to tether distinct rAAV genomes using a DNA oligonucleotide, which also acted as a repair template to induce intermolecular recombination.10 Additionally, the ITRs have been implicated in p53 activation, subsequent cell cycle arrest, and apoptosis in cells deficient for an “intact” DNA damage response.12,13
To overcome the DNA-packaging limitation of AAV vectors, creative strategies based on the mechanistic understanding of genome processing have demonstrated large-gene transduction. Perhaps the most efficient approach is the use of split AAV (also called trans-splicing) vectors, which rely on cotransduction of distinct (and defined) AAV genomes in which vector “A” contains a promoter, coding sequence, and splice donor site, whereas vector “B” has a splice acceptor site and the remaining coding sequence followed by a poly-A tail.14,15,16 Cotransduction by these genomes will sometimes result in a desired concatemerization event, creating a functional cassette that is larger than the AAV capsid's packaging capacity. This split vector context has been reported to occur in vitro and in vivo; however, the success of this approach is limited by the orientation of intermolecular concatemerization and the preferred tendency for AAV genomes to form circular monomers.10,14,15
Even though rAAV transduction was initially thought to be limited by its small packaging capacity (thus creating the need for split AAV vectors), several reports have recently described aspects of “oversized” gene transduction.17,18,19,20,21,22,23,24 This collective work suggests that viral genome (vg) packaging initiates from the 3′ end of ssDNA molecules of both polarities until the capsid reaches its capacity.18 At that point, the external 5′ end of the large genome is truncated by an unknown process.18,19,20,21 To explain this phenomenon, the current theory states that 5′-end single-strand annealing of opposite polarity genome “fragments” is followed by 3′ palindrome-mediated extension to construct transgenic DNA that is larger than the capacity of a single particle.19 This “fragment AAV” (fAAV) genome reassembly model is primarily based on the absence of observable large genomes using Southern blotting.18,19,20,23,24 These reports contrast with an earlier observation, also relying on Southern blotting, which maintains that the AAV5 capsid can package genomes up to 8.9 kb.17 Adding to the controversy, a recent report again failed to observe large AAV genomes (>5 kb) and concluded that fAAV transduction in muscle is mediated by HR.24 However, in that report, no direct evidence demonstrating a role for HR was provided. Furthermore, that hypothesis remains at odds with the canonical understanding that HR is restricted to dividing cells and the earlier report demonstrating packaged large AAV genomes (≈9 kb).17,24 Although the transduction mechanism of oversized AAV genomes remains uncertain, this approach has proven therapeutic in (i) mouse models of Stargardt syndrome and dysferlinopathy and (ii) a canine model of hemophilia A.17,23,24 Nonetheless, the void of molecular understanding describing the mechanism of AAV large-gene reconstruction limits the possibility of these novel vectors contributing to the clinical arena due to potential safety concerns (i.e., fragmented vectors yielding fragmented proteins).
In this work, we characterized fAAV vectors at several levels: (i) the packaged genome; (ii) the integrity of the “full” rAAV capsids; (iii) the transduction efficiency of different-sized fragments; (iv) the relative transduction efficiency of intact AAV in multiple tissues; (v) a quantitative comparison of fAAV and split AAV in cultured cells, skeletal muscle, and the retina; and (vi) the extension of this technology to an optimized dysferlin cDNA transgene cassette in a dystrophic mouse model. In addition, we demonstrate that fAAV transduction relies on a unique DNA repair mechanism compared with either intact or split AAV vectors. Our genetic analyses demonstrate the following: (i) intact vector transduction relies on Rad51C and XRCC3 while being independent of DNA-PKcs; (ii) fAAV vector transduction disproportionately requires Rad51C and XRCC3 to a relatively lesser extent while being DNA-PKcs independent; and (iii) split AAV requires Rad51C, XRCC3, and DNA-PKcs. The collective genetic analyses offer a characterized panel of repair-dependent transducing reporters, allowing investigations of the host DNA damage response in vitro and in vivo. Furthermore, the results demonstrate that the fAAV vectors are more efficient for large-gene delivery than traditional split AAV in cell culture, skeletal muscle, and retina. This work suggests that >90% of fAAV transduction is mediated by fragmented AAV genomes and not by unobserved packaged large genomes, which has been debated in the literature.17,18,19,20 Collectively, fAAV represents an emerging strategy for the treatment of genetic diseases requiring large-gene delivery.
Results
Characterization of 6.2-kb fragment AAV preparations
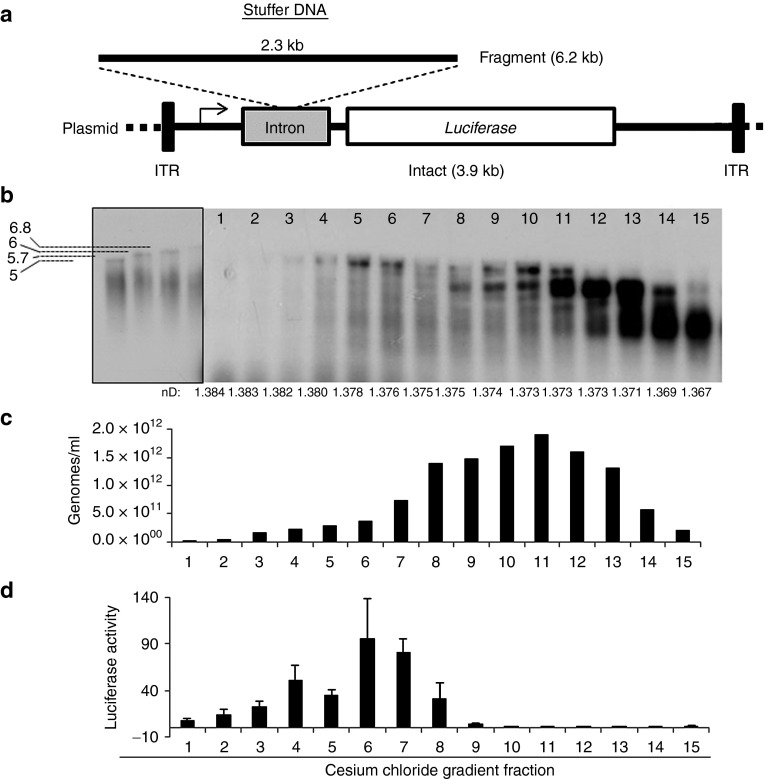
Previous work has demonstrated the ability of rAAV vectors to mediate large-gene (>5 kb) transduction using a novel vector format, where in the capsid is packaged with a 5′-truncated ssDNA genome (termed fragment AAV or fAAV).18,19,20,23,24 To characterize fAAV at a variety of levels, including its relative transduction efficiency compared with other AAV vector formats, we attempted to generate an rAAV vector packaged with a 6.2-kb genome. First, a plasmid vector was generated by inserting a 2.3-kb region of “stuffer” DNA into an intronic region of our standard ITR-flanked chick beta actin (CBA)-luciferase cassette (Figure 1a). Transfection experiments in human 293 cells demonstrated that this stuffer DNA did not affect luciferase activity when compared with the parental intact plasmid control (data not shown; (DNS)). Next, we generated a viral preparation of rAAV2-6.2 kb using a standard triple transfection protocol followed by cesium chloride gradient purification (twenty 0.5-ml fractions were obtained).25 Samples of fractions 1–15 (refractive index: 1.385–1.367) were dialyzed and treated with DNase to remove any “unprotected” DNA, followed by the addition of ethylenediamine tetraacetic acid to terminate the reaction. Then, the capsids were degraded, and the packaged genomes were denatured and electrophoresed on an alkaline gel. Southern blot detection demonstrated packaging of a single DNA species at ≈5 kb with less dense gradient fractions containing two additional smaller genomes at ≈2.4 and ≈1.6 kb (Figure 1b). In contrast, intact (3.9 kb) AAV vector analysis by Southern blotting of successive cesium chloride gradient fractions demonstrated the packaging of a single 3.9-kb genomic species (Supplementary Figure S1, online). Notably, the different genome species observed with the larger AAV (6.2 kb) preparation did not vary when harvested from AAV2, AAV6, and AAV8 serotype capsids (Figure 1b, DNS). To determine whether the larger and smaller species were competent for transduction, we first investigated viral titer by quantitative polymerase chain reaction (qPCR; Figure 1c). Then, fractions 1–15 were normalized for titer and used for 293 cell infection at a ratio of 10,000 vg/cell and luciferase activity was assayed 2 days posttransduction (Figure 1d). Consistent with the hypothesis that only the larger DNA species would be able to reconstruct the 6.2-kb reporter, fractions 1–9 mediated transduction, whereas the remaining fractions containing smaller species showed only slightly enhanced levels relative to the background (Figure 1d). Gradient fractions having refractive indexes of 1.381–1.376 g/cm3 or 1.381–1.371 g/cm3 (for in vitro or in vivo experiments, respectively) were then pooled to ensure a working titer, dialyzed, and used in the remainder of transduction experiments. The final titer of the in vivo fAAV preparation (1.381–1.371 g/cm3), as measured by quantitative Southern dot blot, was 1 × 1011 vg/15-cm plate, decreased ≈fivefold compared with the “intact” 3.9-kb control vector (Supplementary Figure S2a, online). However, this titer is within the standard range of AAV intact preparations, depending on the genome size and sequence packaged (DNS).
Figure 1.
Production and characterization of fragment AAV. (a) Illustration depicting the AAV plasmids that were used to generate “intact” and “fragment” AAV vectors. (b) Denaturing Southern blot of capsid-protected fragment AAV genomes from different cesium chloride gradient fractions following dialysis in 1× phoshate-buffered saline. A CBA-luciferase-radiolabeled probe (6.2 kb) corresponding to the region between the ITRs was used for detection. The boxed region represents DNA fragments of the indicated sizes. The refractive index (nD) of each fraction is depicted. (c) The quantity of viral genomes per milliliter was determined by quantitative polymerase chain reaction for the indicated cesium chloride gradient fractions. (d) Transduction analysis of the indicated genome species derived from the indicated gradient fractions. This experiment was performed using a viral genome to cell ratio of 10,000 for transduction of 293 cells. Luciferase activity was determined 48 hours posttransduction and normalized to the total protein recovered.
Fragment AAV transduction in vitro
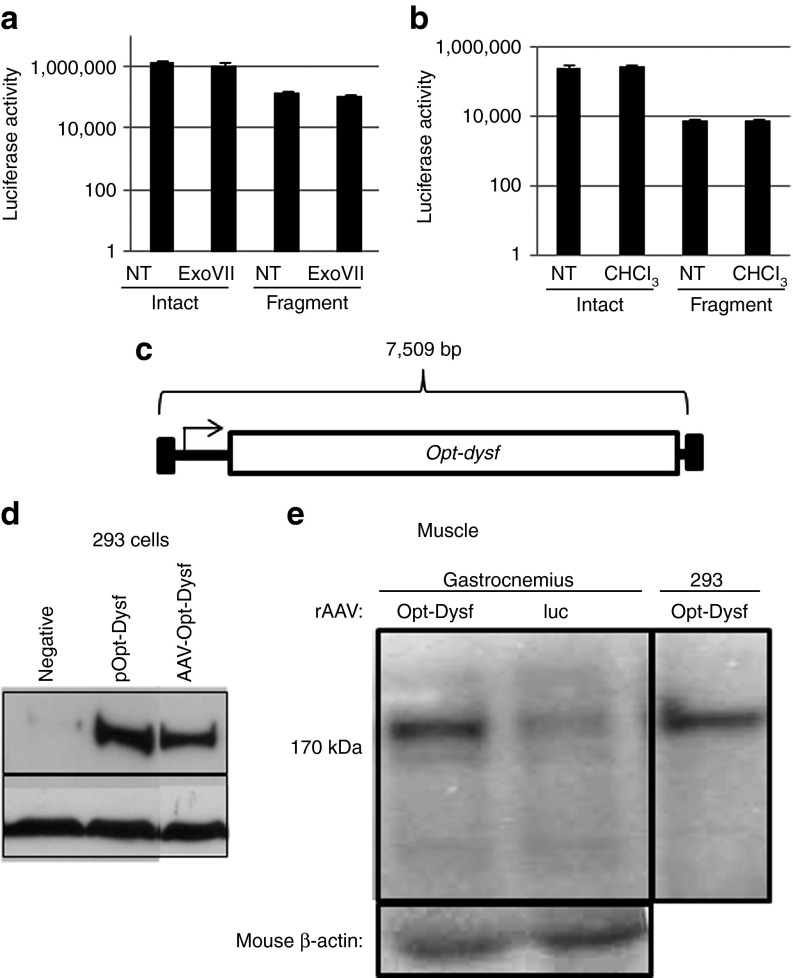
Next, fAAV transduction, using the viral preparation described above, was characterized under several conditions in human 293 cells. We performed a single-strand DNase treatment of intact and fragment vectors before transduction to eliminate the possibility that the full-length 6.2-kb genome was externally associated with the AAV capsid, yet below the limit of Southern blot detection. First, we verified that ExoVII, a bidirectional single-stranded nuclease, degrades ssDNA as reported by the manufacturer (DNS). Then, treatment of vector preparations using ExoVII resulted in a modest, yet statistically insignificant, decrease in the transduction efficiency of both vector types (Figure 2a). In additional experiments, the integrity of the AAV capsid was challenged by chloroform treatment, a condition known to dissociate improperly assembled capsids.26 Chloroform preincubation of both intact and fAAV vector types did not alter the transduction efficiency as measured by luciferase activity (Figure 2b). Collectively, these results suggest that fragment genomes of ≈5 kb are completely packaged in intact (chloroform-resistant) capsids and mediate AAV large-gene transduction of a 6.2-kb transgenic cassette (Figures 1b and 2a,b). Next, the relative transduction efficiency of the fAAV–luciferase reporter preparation, compared with the intact 3.9-kb control, was investigated at different multiplicities of infection (MOIs; vg/cell) in a kinetic manner. At an MOI of 5,000, fAAV transduction was decreased ≈25-fold for luciferase activity compared with that at similar MOI of intact AAV in 293 cells (Supplementary Figure S2b, online). The kinetics of fAAV expression were nearly identical to those of intact AAV as this transduction deficit was observed as early as 10 hour following virus administration and was maintained during the following 70-hour period (Supplementary Figure S2b, online). In addition, fAAV transduction demonstrated a standard saturation curve following transduction of 293 cells at escalating MOIs (Supplementary Figure S2c, online).
Figure 2.
Characterization of fragment AAV transduction in vitro. Fragment and intact AAV vectors were used for transduction following enzymatic or chemical treatment at 5e2 vector genomes per cell. The intact (3.9 kb) and fragment (6.2 kb) vectors were pretreated with either exonuclease VII (ExoVII) or chloroform (CHCl3) for 1 hour before 293 cell administration, respectively (a,b). Luciferase activity was determined 24 hours posttransduction. (c) A schematic illustration depicting the AAV plasmid used to generate fragment AAV–Opt–dysferlin. (d) Fragment AAV2–Opt–dysferlin transduction of 293 cells, along with cells treated with a plasmid containing a CMV–Opt–Dysf expression cassette, analyzed by western blot demonstrating the 237-kDa Dysferlin protein. (e) Fragment AAV–Opt–dysferlin was injected in to the hindlimb of the BlaJ dysferlin–deficient mouse model (N = 3). One month after injection of 5e10 viral genomes, total protein was extracted and analyzed for Dysferlin protein by western blotting. AAV2–Opt–dysferlin transduction of 293 cells was used as a positive control, whereas BlaJ muscle injected with fAAV–luciferase (6.2 kb) was used as the negative control (on the same blot).
These experiments using the 6.2-kb luciferase reporter demonstrate the ability of rAAV to mediate large-gene transduction. To specifically eliminate DNA sequence and size concerns that could be unique to the fAAV–luciferase reporter (Figure 1), fAAV transduction was also attempted using a larger gene expression cassette under the control of the cytomegalovirus (CMV) promoter. First, the 6.24-kb dysferlin coding sequence was optimized (Opt) for mouse/human codon usage and to eliminate the majority of alternative open reading frames (GenScript; Figure 2c). Next, dysferlin abundance from wild type (WT) or Opt-dysf cDNA was evaluated by western blot following transfection in vitro (Supplementary Figure S3a, online), which was then normalized to plasmid copy number abundance (Supplementary Figure S3b, online). The data demonstrate that the amount of dysferlin produced from either the WT or Opt-dysf cDNA was not significantly different in 293 cells or in mouse skeletal muscle (Supplementary Figure S3, online, DNS). However, because Opt-dysf contains less alternative open reading frames, which could result in unwanted immunological complications,27 we generated fAAV CMV-Opt-dysf (7.5 kb) in capsids 2 and 6 for in vitro and skeletal muscle transduction, respectively (Figure 2c). Again, no DNA species >5 kb were observed by Southern blot analysis in either preparation (DNS). Then, the ability of fAAV (7.5 kb) to mediate Opt-dysf transduction in 293 cells was demonstrated by western blotting (MOI of 10,000; Figure 2d). In addition, fAAV6 Opt-dysf was administered by intramuscular injection (5e10 vg) to the gastrocnemius muscle (N = 3) of the BlaJ Dysferlin-deficient mouse model (Figure 2e).28 Two weeks postinjection, total protein was recovered from the injected muscle and the dysferlin protein level was restored in tissue given AAV6 Opt-dysf but not in the control muscle injected with fAAV6–luciferase (Figure 2e). These collective experiments confirm the ability of fAAV to mediate the transduction using genomes of different sizes and sequences in vitro and in vivo (Figure 2).
Fragment AAV transduction in vivo
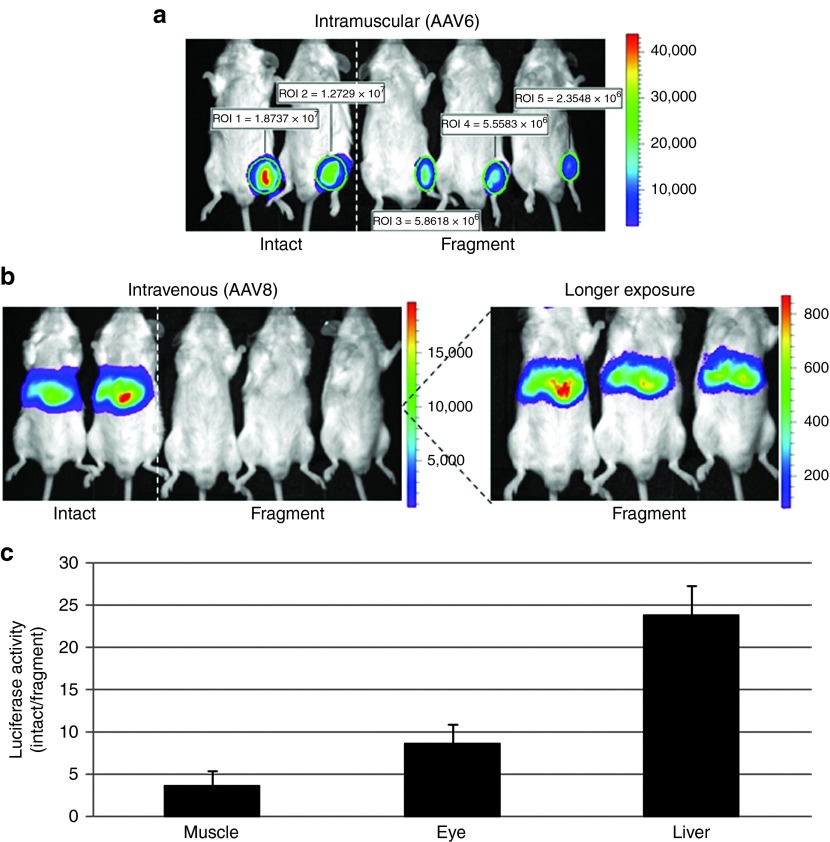
In previous work, the ability to use oversized vectors for gene delivery to the mouse eye, liver, and muscle was demonstrated (Figure 2e).17,23,24 However, the relative transduction efficiency in comparison with intact AAV (3.9 kb) was never evaluated. To answer this question in WT muscle, 5e10 vg of either intact or fAAV–luciferase (Figure 1a,b, Supplementary Figure S1, online) vectors were injected into the mouse gastrocnemius muscle (N = 4). For these experiments, the two vector types were produced using the AAV6 capsid, and Southern blot analysis of packaged DNA species demonstrated a similar profile to that observed with fAAV2–luciferase preparations (Figure 1b, DNS). As early as 3 days postinjection, luciferase activity could be observed by live imaging in both treatment groups. At day 10 postinjection, the luciferase activity was quantitated using Igor Pro software and the results demonstrate that at this MOI, a 3.5-fold deficit for fAAV transduction, compared with the value determined for intact vector, existed (Figure 3a,c). The relative luciferase values for both vectors were maintained for >6 months in WT mouse muscle (DNS).
Figure 3.
Characterization of fragment AAV transduction in vivo. (a). Either intact or fragment AAV6–CBA–luciferase vectors (5e10 viral genomes) were injected into wild-type mouse gastrocnemius skeletal muscle at the indicated dose (N = 4/treatment). The depicted image was taken 10 days postinjection. (b) The indicated AAV8 vectors were injected intravenously into wild-type mice (N = 3) at a dose of 1 × 1011 viral genomes. The depicted image was taken 12 days postinjection. The expanded inset labeled “longer exposure” refers to an increased integration time allowing visualization in the picture. (c) Quantitation of the luciferase values determined in the distinct data sets in a and b presented as fold change derived by the average value determined for the intact vector divided by the average value determined for the fragment vector. In addition, the fold change for values determined following mouse subretinal injections (N = 3) of either vector are presented (described in text).
Our previous report demonstrated that liver transduction using fAAV–factorVIII (5.8 kb) corrected the bleeding phenotype of the hemophilia A dog model.23 However, the relative transduction efficiency in the liver, compared with intact AAV, was not investigated, which is important for translation of fAAV into the clinic, especially considering the lower production titers (Supplementary Figure S1, online). Therefore, fAAV–luciferase or the intact control, packaged in AAV8 capsid, was administered intravenously to 8-week-old WT mice (N = 3). Twelve days following administration of 1 × 1011 vg, luciferase activity in the mouse liver was quantitated. Under these conditions, fAAV was clearly capable of liver transduction, albeit at a level 25-fold less than that of intact AAV8 vector (Figure 3b,c). As quantitative live imaging of the liver following systemic vector injection may also tally expression from neighboring tissues, an in vitro analysis of luciferase activity was performed. Mice of a separate cohort (N = 3) were injected intravenously with 1 × 1011 vg of intact or fragment AAV8–luciferase vectors and equivalent liver samples were harvested 15 days postinjection. After tissue lysis, luciferase activity was measured in vitro and normalized to the amount of recovered protein. Similar to the value obtained by live imaging, a 37-fold decrease in transduction efficiency was noted for fAAV compared with intact vectors (Supplementary Figure S4, online).
In addition to the liver and skeletal muscle, the transduction efficiency of fAAV was also directly investigated in the eye following subretinal injections (N = 3). Again, intact AAV–luciferase, which does not require intracellular gene reconstruction, served as a relative control. For these experiments, AAV2 was used at a dose of 5e8 vg for both vector types. Twelve days postinjection, luciferase activity from the total eye was quantitated, and the results demonstrate that fAAV transduction decreased about eightfold compared with intact AAV in this isolated tissue compartment (Figure 3c).
Comparison of AAV large-gene transduction strategies
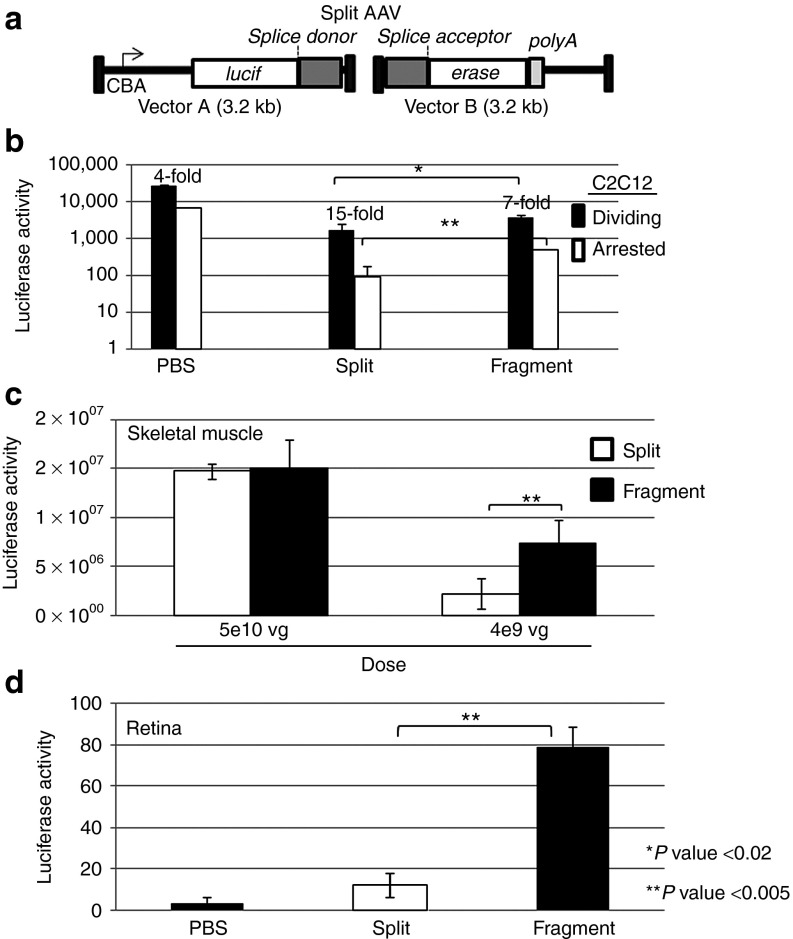
The finding that fAAV is capable of large-gene (6.2 and 7.5 kb) transduction in several mouse tissues expands the utility of AAV vectors for therapeutic applications requiring transgenic DNA >5 kb. However, additional AAV vector strategies for large-gene delivery have been described and of these vector formats, split AAV (also termed trans-splicing) has been deemed the most efficient.14 To determine the relative transduction efficiency of fAAV compared with that of split AAV, we generated split luciferase vectors that are genetically similar (Figure 4a). The split AAV approach relies on the cotransduction of two distinct vectors: (i) vector A contains the CBA promoter, followed by a partial luciferase coding sequence and a splice donor site; and (ii) vector B has a splice acceptor signal, the remaining luciferase coding sequence, and a poly-A tail (Figure 4a). Transduction by vector A or B alone did not result in luciferase activity (DNS); however, a particular intermolecular linkage event of the genomes of vectors A and B following cotransduction resulted in a functional luciferase cassette (Figure 4). AAV large-gene transduction was initially compared in vitro using fAAV2 and split AAV2 vectors. The results demonstrated that at the same final MOI (fAAV or split vectors A and B), fAAV is significantly enhanced for transduction, compared with split AAV, in dividing C2C12 myoblasts and myotubules (Figure 4b). Transduction of three different types of cells (hamster, mouse, and human) demonstrated that this enhancement was conserved across multiple species in cell culture, suggesting there is no cell type–specific discrepancy in the measured activities (Figure 4b, Supplementary Figure S5, online). We also compared the efficiency of large-gene transduction by split AAV and fAAV in vivo following intramuscular injections (N = 3). Similar luciferase activities were noted for both split and fAAV transduction when administered a high dose of vector, possibly representing saturation of our upper limit of detection (Figure 4c). However, when the administered dose was decreased tenfold, a significant increase in luciferase activity (fourfold) was noted in muscle-given fAAV relative to the value determined for split AAV (Figure 4c).
Figure 4.
Efficiencies of split and fragment AAV vector transduction. (a) An illustration depicting our split AAV reporter, which consists of two distinct vectors: (i) vector A contains a promoter, followed by partial luciferase coding sequence and a splice donor sequence and (ii) vector B has a splice acceptor sequence, the remaining luciferase coding sequence, and a polyA tail (intron derived from the β-globin intron II sequence). (b) Transduction of dividing and nondividing mouse C2C12 myoblasts and myotubules by the indicated type of AAV2 vector system at a total dose of 1 × 105 viral genomes per cell. Luciferase activity was quantitated 72 hour following vector administration, and the fold change between the values derived from dividing and arrested cells is presented. (c) The indicated rAAV6 vector type was injected through the intramuscular route at the indicated viral genome (vg) dose (N = 3). Luciferase activity was quantitated 20 days postinjection. (d) The indicated rAAV2 vector type was injected into the subretinal space (N = 3) and luciferase activity was determined as described in the text. For split AAV vectors, the final dosing refers to split vector A + split vector B. In all cases, an unpaired t test was used to determine statistical significance.
In addition, the transduction efficiencies of split and fAAV2 was evaluated in the eye following subretinal injections (5e8 vg; N = 3). Although live imaging of luciferase activity is possible following intact AAV vector transduction at this dose, the sensitivity of detection using the repair-dependent vectors was not (DNS). Therefore, 1 month postinjection, total eye lysate was prepared and luciferase activity was determined in vitro and normalized to total recovered protein. From this analysis, fAAV demonstrated a sevenfold enhancement in luciferase activity compared with that of split AAV (Figure 4d).
Mechanistic evaluation of AAV large-gene transduction strategies in vitro
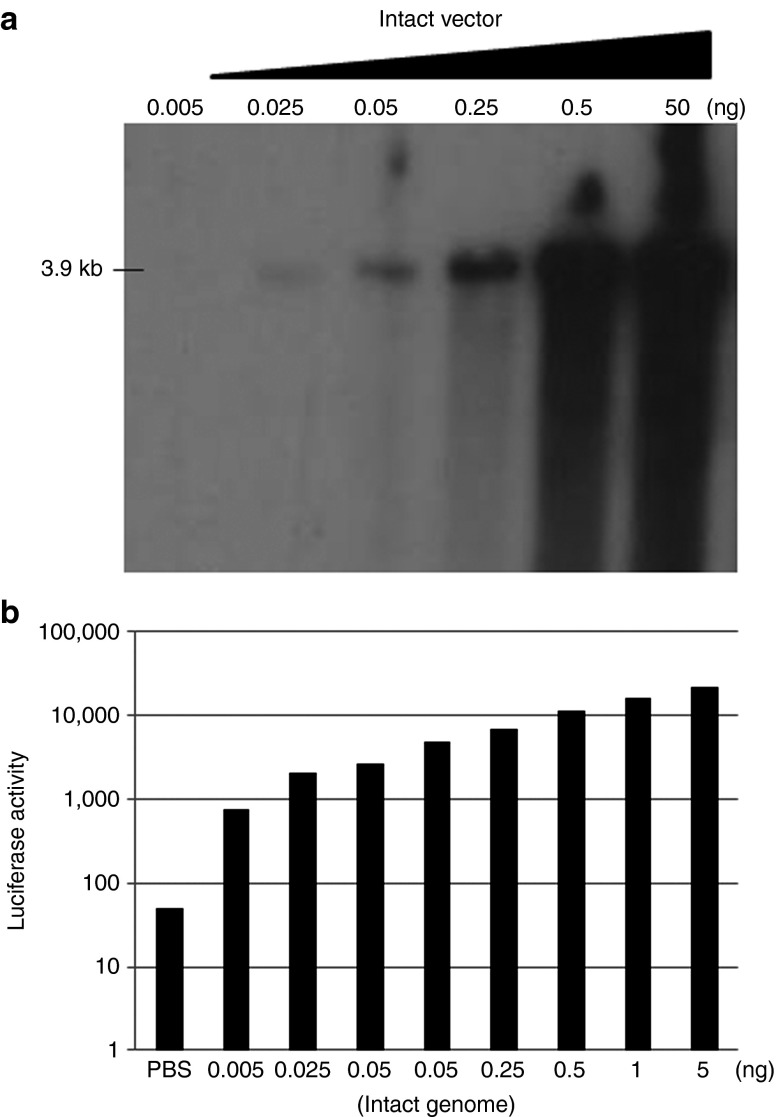
Thus far, AAV-mediated transduction of oversized cassettes has been primarily assumed to be mediated by AAV fragments based on the absence of large packaged genomes observed by Southern blotting (Figure 1b).18,19,20,23,24 However, it is possible that DNase-resistant genomes are packaged17 and mediate transduction yet are below the sensitivity of detection (Figure 1b). To define the sensitivity of detection of the Southern blotting protocol used herein, we diluted the intact AAV–luciferase (3.9 kb) vector beyond our detection and compared transduction efficiencies in 293 cells. The intact vector genome was detected on alkaline Southern gels in the range of 50 ng–50 pg using a 5-day exposure to film (Figure 5a). A viral genome amount of 25 pg defined the lower limit of our Southern detection; however, transduction by this amount of packaged AAV genomes resulted in luciferase activity 2-logs higher than that of the vehicle control (Figure 5b). An additional fivefold dilution to 5 pg of viral DNA still maintained transduction >tenfold higher than that of background (Figure 5). This result undermines the assumption that fAAV transduction is actually mediated by fragments because it is quite clear that viral DNA species not observed by alkaline Southern blots mediate efficient transduction.18,19,20,23,24
Figure 5.
Southern blot sensitivity and AAV transduction. (a) The indicated amount of packaged viral genomes (AAV–CBA–luciferase 3.9 kb “intact”) was analyzed by alkaline Southern blot using the luciferase coding sequence as a probe. (b) Intact vector transduction of 293 cells using intact particles harboring the indicated amount of viral genomes.
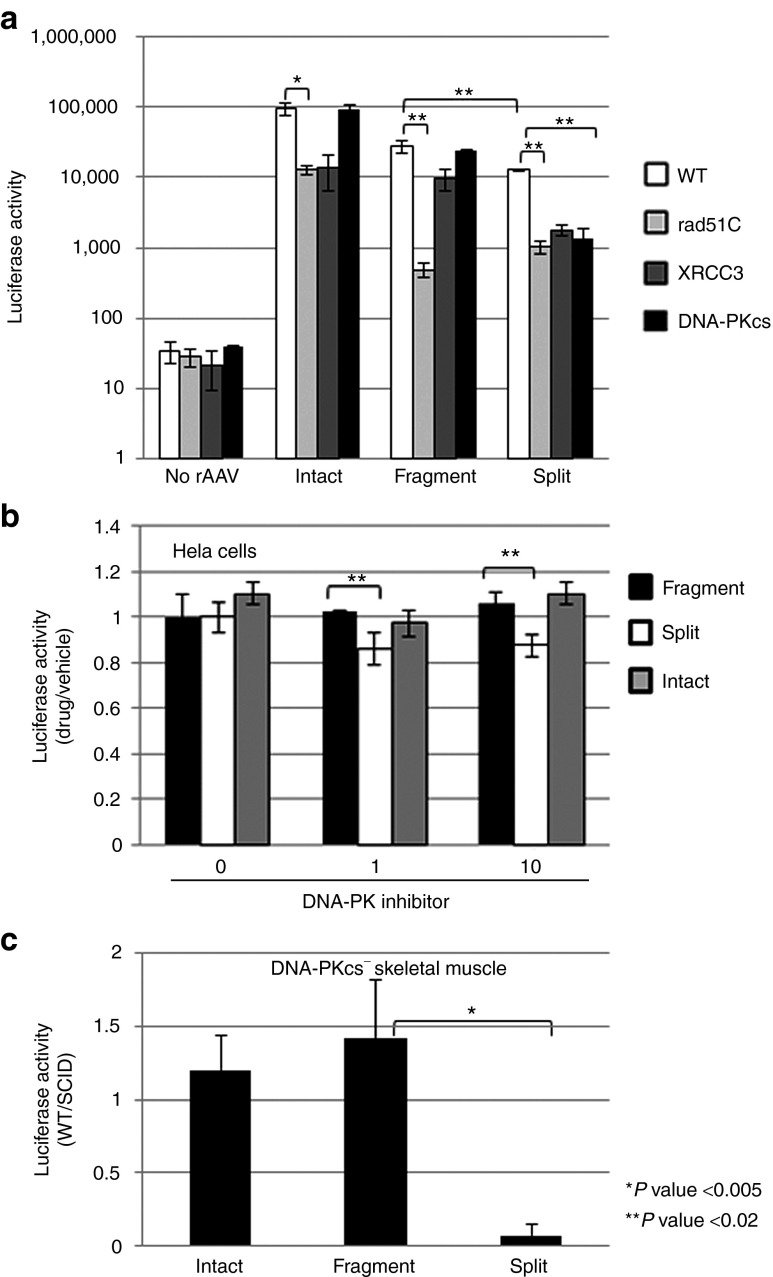
Considering the concerns of detection sensitivity using Southern analysis, a genetic approach was used to investigate the DNA repair mechanism (if one exists) responsible for the large-transgene reconstruction initially by using well-established Chinese hamster ovary (CHO) mutant cell lines.5 For these experiments, we used a V3 DNA-PKcs mutant that is defective for NHEJ.29,30 We also investigated rAAV transduction in irs3 and irs1SF cells, which are reported deficient in HR due to undetectable levels of Rad51C and XRCC3, respectively.31,32 For these experiments, AAV2 large-gene transduction was determined by luciferase activity estimation 2 days postadministration in the mutant strains and compared with the value determined for the WT parental lines.5 As the intact vector does not require DNA repair, other than second-strand synthesis, for luciferase expression, it serves as our reassembly-independent control vector. In the absence of DNA-PKcs, intact and fAAV transduction was not significantly altered (Figure 6a). In contrast, split AAV transduction decreased ninefold in these cells, indicating a role for DNA-PKcs in split-vector large-gene reconstruction (Figure 6a). The results obtained from intact vector transduction of the Rad51C-deficient cells demonstrate a nearly tenfold decrease in transduction, indicating a reassembly-independent role for Rad51C in AAV vector transduction (Figure 6a). A similar decrease in transduction in the Rad51C-mutant cells was noted for split-vector transduction. In contrast, fAAV transduction was disproportionately affected, demonstrating a 50-fold decrease in the absence of Rad51C (Figure 6a). Additionally, rAAV vector transduction was investigated in CHO cells containing the irs1SF mutation, which is complemented by the RecA/Rad51-related HR protein XRCC3. Similar to the results obtained in the Rad51c-deficient line, intact and split vector transduction decreased ~tenfold compared with the appropriate control strains.5 However, fAAV transduction was affected to a lesser degree, <threefold in the XRCC3 background (Figure 6a). These collective results provide genetic evidence indicating that the DNA repair factors that contribute to intact, split, and fAAV vector transduction are distinct: (i) split and intact AAV vector transduction relies to a similar extent on Rad51C and XRCC3; (ii) split transduction requires DNA-PKcs while intact and fAAV does not; and (iii) fAAV transduction disproportionately requires Rad51C, although the transduction deficit for intact and split vectors observed in the XRCC3 background is not observed (Figure 6a).
Figure 6.
rAAV vector transduction dependency on DNA repair proteins. (a) The indicated AAV–luciferase vector type was used for transduction in the appropriate WT, Rad51C-deficient (irs3), XRCC3-deficient (irs1SF), and DNA-PKcs-deficient (V3-3) CHO cells as described in text. Luciferase activity was determined 24 hours after virus administration, and statistical significance was determined a student t test followed by Bonferroni correction to account for the multiple comparisons. (b) The indicated rAAV reporter context was used for transduction in the increasing presence of a DNA-PK inhibitor II (µmol/l) as described in the text. Again, luciferase activity was assayed 24 hours after adding AAV, and the results are presented as the fold change in activity (drug/vehicle). (c) The indicated rAAV6 vector type was administered to WT or DNA-PKcs-deficient mice by muscle injection (N = 3) at a dose of 5e10 viral genomes. Luciferase activity was determined 20 days postinjection and is presented as the fold change by dividing the value determined for WT mice with that determined in the mutant mice. In (b,c) a student t test was used for statistical significance.
To further confirm the canonical NHEJ independence of fAAV transduction, the influence of DNA-PK inhibitor II was investigated in HeLa cells. Consistent with decreased split AAV transduction in the DNA-PKcs–deficient cells, both of the tested inhibitor concentrations (1 and 10 µmol/l) decreased luciferase activity following split AAV transduction (Figure 6b). In contrast, intact and fAAV transduction were not significantly altered at either concentration of the DNA–PK inhibitor (Figure 6b).
Mechanistic evaluation of AAV large-gene transduction strategies in vivo
Results from genetic and pharmacological analyses demonstrate a differing role for DNA-PKcs in AAV large-gene transduction; split AAV is DNA-PKcs dependent, whereas fAAV is Rad51C dependent and completely independent of DNA-PKcs. To observe whether this pattern holds true in vivo, AAV large-gene transduction was investigated in mice deficient for DNA-PKcs. For this experiment, 5e10 vg of either vector type packaged in AAV6 were administered directly to mouse skeletal muscle (N = 3). Twenty days postinjection, the transduction efficiency was determined by luciferase activity. Consistent with the results obtained in vitro, split AAV vectors showed tenfold decreased transduction in the DNA-PKcs background, whereas the transduction efficiency of fAAV and intact AAV was not significantly altered (Figure 6c).
Discussion
Thus far, therapeutic applications of rAAV-mediated gene therapy have met with encouraging success for genetic disorders, such as Leber congenital amaurosis and hemophilia B. These initial human results have increased the popularity of AAV for DNA delivery in the clinic and also present additional challenges, most of which can be addressed by vector enhancements that allow efficient transduction at lower doses. It was traditionally thought that rAAV was of little use for the treatment of genetic diseases requiring transgenic DNA >5 kb, a restriction imposed by its small viral capsid. However, recent work evaluating an earlier claim that AAV5 can package 8.9-kb genomes collectively concluded using Southern detection that no genomes, using vectors with different sequences, >5 kb were packaged.18,19,20,23,24 Instead, truncated large genome fragments were thought to be packaged, which mediated large-gene transduction at an unknown efficiency.22 Mechanistic evidence supporting this theory is yet to be shown, although, recent work based on Southern blotting concluded that AAV5-based oversized gene transduction in muscle fibers relies on HR, again suggesting that this process relies on fragmented AAV genomes.24 However, no direct evidence was provided demonstrating host-mediated HR of the presumed fragmented AAV genomes, and furthermore, this conclusion contradicts the long-held understanding that HR is restricted to dividing cells.24 The work herein provides genetic evidence demonstrating that the majority of oversized AAV vector transduction is mediated by fAAV genomes that show a disproportionate dependence on a unique DNA repair pathway (Figures 1,6).
Our analysis of the 6.2-kb AAV–luciferase vector preparations demonstrated that DNA genome species of ≈5 kb were packaged, as determined by Southern analysis (Figure 1b). Despite this result, the detection sensitivity of Southern blotting is a concern (Figure 5), and hence it remains a formal possibility that larger genomes could be packaged and perhaps mediate transduction as previously reported.17,33 Subsequent data rebuking the initial claim of large-gene packaging did not conclusively rule out this possibility, and neither does the work herein; however, our genetic analyses suggest that the majority of oversized transduction is mediated by AAV genome fragments (Figure 6).18,19,20,23,24 Specifically, the majority of the observed fAAV reporter activity requires Rad51C, a DNA strand–transfer protein involved in annealing during HR, at a level more than tenfold that observed for intact AAV (Figure 6a). Conceptually, as both small and large intact AAV genomes require second-strand synthesis and transcriptional activation (intact and fAAV both have an identical promoter), it is not immediately apparent how proteins involved in HR would differentially act on these templates that differ only by 2.5 kb (supporting the notion that fAAV transduction is mediated by DNA fragments). In addition, we have performed plasmid transfections in CHO lines using the constructs used to generate the intact and fAAV vectors. Importantly, no fAAV construct–specific effect was noted in the Rad51C or XRCC3 backgrounds, demonstrating that there is no transcription-specific effect of these repair proteins on the 3.9- or 6.2-kb cassette expression (DNS). Thus, the genetic data argue that Rad51C influences AAV transduction of intact, fAAV, and split vectors while an additional statistically significant effect (more than tenfold) is exerted on fAAV at a level other than transcription, presumably genome reconstruction. Therefore, our data imply that at least the majority of fAAV transduction herein is not mediated by large intact undetected genomes, but rather by genomes requiring DNA repair mechanisms other than those required by intact genomes (Figures 1b,5,6). However, fAAV transduction in the Rad51C-deficient cell line was not completely eliminated (Figure 6a). This result is not immediately understood but could be explained by an unknown Rad51C redundant function in this cell line, a Rad51C-independent pathway for fAAV transduction, or by the existence of intact AAV genomes that are not visible by Southern blotting yet mediate transduction (Figure 1d).
The preference to package discrete ssDNA species in the capsid is not understood, and we observed approximate sizes of 1.6, 2.4, and 5 kb while attempting to package a 6.2-kb genome (Figure 1). Interestingly, only gradient fraction containing the ≈5-kb DNA species demonstrated significant transduction. Current genome-sequencing efforts are yet to yield results. As the truncation mechanism is unknown, it remains possible that it occurs by either intracellular or extracellular pathways (during purification). Experiments testing these hypotheses are also underway. Currently, analyses of the replicative forms of our 6.2-kb luciferase reporter and the 7.5-kb dysf cassette suggest that genome fragmentation does not occur during replication, which is consistent with an earlier report (DNS).33 With reference to particle integrity, chloroform tolerance suggests that fAAV capsids are intact, unlike misformed AAV capsids that were capable of large-DNA transfer in cell culture (Figure 2b).26 The results of the ExoVII ssDNA digestion before fAAV transduction also support normal capsid integrity (Figure 2a).
In terms of transduction efficiency, fAAV showed decreased efficiency compared with intact AAV, anywhere from threefold to 50-fold depending on the transduced cell type/tissue and method of administration (Figures 2,3, Supplementary Figure S2b,c, online). An indirect correlation was noted between the size of the transduced compartment and the fold difference observed between fAAV and intact vectors (i.e., less than fivefold in isolated muscle following injections and >25-fold in the liver following intravenous injection). This observation is perhaps rationalized by requirement of multiple genome fragments for fAAV transduction although a single intact AAV genome can theoretically result in luciferase activity.
To determine the most efficient AAV large-gene delivery approach, a quantitative comparison was performed between fAAV and split AAV vectors. Our analyses in skeletal muscle are consistent with the results obtained in a variety of cell lines that demonstrate that fAAV is more efficient than the traditional split vector approach (Figure 4, Supplementary Figure S5, online). In addition, a comparison of large-gene transduction by these vectors in the mouse retina demonstrated a sevenfold increase in transduction efficiency for fAAV compared with split AAV (Figure 4d). However, it should be understood that these vectors, by nature, are not genetically identical. Specifically, split AAV vectors require a particular orientation of ITR-mediated concatemerization and transcription through a processed DNA hairpin, both of which inhibit the efficiency of this approach.10,34 The obtained results demonstrating that fAAV transduction is more efficient than split vectors makes sense theoretically as opposite strand polarities would be “directed” to the correct orientation by the overlapping and complementary truncated 5′ genome ends.18 In the case of split vectors, reconstruction of the desired transgene product only occurs with a likelihood of one in six, as concatemerization events are directed by homologous ITRs that flank all intact transduced genomes.10 Furthermore, AAV genomes prefer self-circularization.5 This speculation probably rationalizes the results in which the enhanced transduction by fAAV compared with split AAV was markedly increased in cell culture, skeletal muscle, and retina (Figure 4, Supplementary Figure S3, online). However, a recent report using “dual hybrid” trans-splicing AAV vectors, which contain additional “recombinogenic” sequences within the introns of split vectors A and B, demonstrated enhanced transduction, yet they were not tested herein.16
Our analysis of the genetic reconstruction of large genes mediated by AAV delivery suggested that the repair pathway dependence of intact, fragment, and split vectors is overlapping, yet distinct, in dividing cells (Figure 6): (i) intact vector transduction relies on Rad51C and XRCC3 while being independent of DNA-PKcs; (ii) fAAV vector transduction disproportionately requires Rad51C, and XRCC3 to a relatively lesser extent, while being DNA-PKcs independent; and (iii) split AAV requires Rad51C, XRCC3, and DNA-PKcs. This DNA repair requirement for split AAV genomes is consistent with previous reports that demonstrate split AAV genomes can be assembled by both HR and NHEJ DNA repair pathways.6,10,11 The results also demonstrate that although both intact and split AAV vectors are decreased in the XRCC3 background to a similar degree, there is significantly less of an effect on fAAV (Figure 6a). Thus, fAAV is highly dependent on Rad51C, and transduction is slightly enhanced in the absence of XRCC3 (compared with intact or split vector transduction) (Figure 6a). As Rad51C is a ssDNA-binding protein that promotes strand annealing during repair of double strand breaks, it seems logical that it may function on the overlapping and complementary free ssDNA ends of fAAV genomes (Figure 6a). Rad51C has been shown to be unique among the Rad51 paralog proteins in that it relocalizes to the nucleus after DNA damage, and it has been reported to transport Rad51 into the nucleus.35 In addition, Rad51C interacts with three paralog protein complexes involved in HR: (i) Rad51C/XRCC3, (ii) Rad51B/C, and (iii) Rad51-B-C-D/XRCC2.36,37,38 Given the diverse roles of Rad51C in HR, it perhaps is not surprising that fAAV transduction is disproportionately reduced, compared with intact and split vectors, in the deficient background. Furthermore, the modestly elevated levels of transduction in the XRCC3-deficient cells could suggest XRCC3-mediated inhibition of Rad51C-mediated fAAV genome reconstruction and, to a lesser extent, AAV transduction in general (Figure 6). Studies elucidating the precise mechanism of fAAV transduction are under investigation. Interestingly, fAAV transduction showed dependency on Rad51C in dividing cell culture; however, transduction was noted in contexts that are deficient for HR, including arrested cells and differentiated tissue (Figures 4,6). In these contexts, it is possible that noncanonical end-joining pathways reconstruct fAAV, and experiments testing this hypothesis are in progress. Canonical NHEJ, which relies on DNA-PKcs and largely mediates split AAV transduction, does not appear to influence fAAV, or intact AAV, transduction in vitro and in vivo under the tested conditions (Figure 6). In contrast, AAV vector concatemerization, split AAV, relies on the DNA-PKcs pathway, which has been previously reported.11 These experiments demonstrate the viral versatility of AAV vectors to use different host DNA repair pathways for natural persistence, which can then be exploited for diverse mechanistic and therapeutic applications (Figures 2c–d,6).17,23,24
In addition to elucidation of fragment vectors at the levels of transgenic DNA reconstruction and relative transduction efficiency, we extended our fAAV analysis to a CMV–Opt–dysf cassette in the skeletal muscle of the BlaJ Dysferlin-deficient mouse.28 Importantly, this extends our fAAV investigations to an additional promoter and transgene of a larger size (total cassette is of 7.5 kb; Figure 2c–e). Given previous immunological concerns with alternate open reading frames, dysferlin cDNA optimization, which changes the nucleotide codon sequence while preserving the native amino acid usage, was performed by GenScript.27 Despite several reports in which codon optimization resulted in increased protein abundance, we did not observe this scenario for Opt–dysf cDNA in vitro (Supplementary Figure S3, online).39,40 However, fAAV–Opt–dysf restored Dysferlin protein in deficient BlaJ muscle at a viral dose of 5e10 vg via intramuscular injection (Figure 2e). Previous experiments have demonstrated that restoration of dysferlin in a dysferlin-deficient mouse can partially relieve the dystrophic phenotypes.24,28,41 However, using a transgenic mouse approach, it was also observed that overexpression of this protein in muscle results in a progressive myopathy.42 Investigations are underway to define the efficacy levels of dysferlin and to regulate its cellular abundance following gene transfer experiments.
Of particular importance for therapeutic applications is the observation that the overall titer of fAAV is consistently fivefold to tenfold lower than intact AAV and, perhaps more troublesome, that it is contaminated by DNA species that do not mediate transduction (Figure 1, Supplementary Figure S1, online). In the past, we have noticed that different purification methods often favor certain vector genome species, and different column chromatographies are under investigation for fAAV particle isolation. Specifically, it is of interest to exclude the smaller fAAV genome fragments that were not transduction competent but contributed to our determination of overall vector titer (Figure 1b–d; Supplementary Figure S2, online). Thus, the effective titer of our fAAV preparations used herein are likely to be decreased more than tenfold compared with intact AAV and the efficiencies of transduction, if anything, are underestimates.
Collectively, the work herein is the first report providing mechanistic evidence that the majority of fAAV large-gene transduction is, indeed, mediated by genomes requiring a unique host DNA repair process. This is an important finding considering the assumptions based on the poor sensitivity of Southern blot detection (Figure 5) in light of a report that states that the AAV5 capsid, specifically, can package up to 8.9 kb.17,18,19,20,23,24 Given the cell/tissue targets used herein, we did not investigate oversized genome packaging in AAV5; however, AAV2, 6, and 8 capsids packaged DNA fragments of similar size. From a therapeutic standpoint, fAAV appears to be an efficient method for large-gene transduction given our head-to-head comparison with split AAV in vitro, in skeletal muscle, and in the retina. However, clinical translation concerns exist for fAAV applications, including the following: (i) the low production titer (Supplementary Figure S2a, online), (ii) the contamination of preparations by nontransducing DNA species (Figure 1b–d), and (iii) the potential production of aberrant genomes/proteins from nonconservative AAV genome repair. An additional concern following fAAV transduction is that by nature (as it is reported to have no ITR sequence at the 5′ end), it may be enhanced for host chromosome integration.18 However, preliminary studies in dividing cell culture suggest that the integration frequency is not enhanced relative to that of intact AAV vectors (DNS). Despite these concerns, fAAV transduction has already demonstrated preclinical success by alleviating symptoms of Stargardt syndrome and dysferlinopathy in mouse models.16,24 In addition, our group has demonstrated the ability to overcome the scalable production barriers for fAAV, by curing dogs with hemophilia A using a 5.8-kb therapeutic cassette.23 Therefore, from both mechanistic and therapeutic standpoints, fAAV represents a novel reagent to study the in vivo DNA damage response and exploit it for the generation of enhanced diseased therapies.
Materials and Methods
Maintenance of cells. Most cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and 1× Penn/Strep (DMEM+, Sigma). These cells include the lines CHO-K1, CHO AA8, CHO V79-4, CHO irs3, CHO irsf1, CHO V3-3, C2C12, HeLa, and HEK 293s.5,43,44 These cells were maintained at 37 °C in a 5% CO2 humidified incubator. C2C12 cells were differentiated into nondividing myotubules by growth in DMEM containing 2% fetal bovine serum for 2–3 weeks and myotubules were verified using microscopy (DNS). DNA-PK inhibitor II (SantaCruz, SantaCruz, CA) was used to treat cells at 1 and 10 µmol/l concentrations (in DMSO). For the comparison of protein production by weight or codon-optimized dysferlin cDNA, HEK293T cells were cultured in DMEM GlutaMax-I medium (Invitrogen) with 10% fetal bovine serum (Gibco, Invitrogen, Carlsbad, CA) and 10 μg/ml gentamicin (Gibco, Invitrogen) at 37 °C with 10% CO2. For transfection, 1.0E+07 cells were plated on a 10-cm dish (40–50% confluence), cultured for 24 hour, and transfected with 6 µg of plasmid DNA using Fugene (Roche, Indianapolis, IN) according to the recommended instruction. Three days after transfection, cells were harvested after trypsinization and collected into two tubes, for the western blot and the qPCR analysis.
Plasmid constructs. To generate the oversized AAV ITR plasmid (Figure 1a), the AvrII/SpeI fragment of pCINEO (Promega, Madison, WI) was cloned into the XbaI site of pTR2-CBA-luc by blunt ligation. The AAV genome size is 6.209 kb. For split luciferase vector construction, a PCR product was generated using a plasmid containing the β-globin intron II inserted into luciferase cDNA construct (RJS) as template. Terminal SphI and ClaI sites were introduced and used for cloning into the “split A” backbone pTR2–CBA–luc (“intact” 3.9-kb plasmid vector). The split B vector was generated using PCR cloning (of the same template) and inserted into the KpnI/NotI site of pTR2–egfp (RJS). Primer sequences are available on request. For the ITR plasmid that contains the fragment, the optimized dysferlin cDNA was cloned out of MH197 by XbaI and SphI digestions and inserted into the corresponding sites on pTR2–CMV–egfp. The comparison of expression of optimized dysferlin and wild form was performed using the following constructions: plasmid pCMV–eGFP_DysferlinFL_Isoforme8 (GENETHON) and plasmid CMV–eGFP_Opt–Dysferlin. The plasmid pCMV–eGFP_pOptiDysferlin was constructed by inserting a fragment containing the cytomegalovirus (CMV) promoter and the eGFP coding sequence into the plasmid pOpt–Dysferlin, upstream of the optimized dysferlin cDNA. Constructs were confirmed by DNA sequencing.
AAV production and characterization. All vectors used in this study were produced using a triple transfection protocol, followed by CsC1 gradient centrifugation and dialysis in 1× phosphate-buffered saline (PBS) solution as previously described.25 The pXR series plasmids 2, 6, and 8 were used to package genomes in capsids 2, 6, and 8, respectively. Final virus titer was determined after dialysis in 1× PBS by qPCR (described under qPCR analysis) and Southern dot blot, and the packaged species were analyzed by alkaline gel electrophoresis followed by Southern blotting.25 A plasmid cut fragment corresponding to the entire 3.9-kb intact luciferase genome was used as a template for P32-labeled probe synthesis by random primed labeling (Roche).
rAAV transduction. For experiments in vitro, 50,000 viable cells were seeded in a 24-well plate 24 hour before transduction. The next day, virus particles were added directly to the well at the dose described in the “Results.” At the indicated time points, cells were lysed directly in the well using a passive lysis buffer according to manufacturer's recommendation. Luciferase activity was then determined using a Victor 2 luminometer (Perkin Elmer, Waltham, MA) and normalized to total protein in the lysate determined by Pierce 660 nm protein assay reagent. All experiments were performed in triplicate on at least three different days, and the results are presented as averages of the experimental data. The ssDNA nuclease ExoVII (Epicentre) was used using the manufacturer's conditions for the vector pretreatment experiment. The negative control in these experiments was treated identically, however, without the addition of ExoVII. For rAAV transduction in vivo, experiments were carried out with 6- to 8-week-old female BALB/c, BALB/c DNA-PKcs, or the B6 BlaJ (dysferlin-deficient) mice (Jackson Labs, Bar Harbor, ME), unless otherwise indicated. All mice were maintained and treated in accordance with National Institutes of Health guidelines and as approved by IACUC at UNC–Chapel Hill. All intramuscular injections were performed following intraperitoneal injection of 2.5% avertin (250–500 mg/kg) or transient anesthetization using 2.5% isoflurane. Then the vector dose indicated in the text was injected directly into the muscle using a 30-gauge syringe in an equal volume of 80 µl for the gastrocnemius. For intravenous injections, the tail vein was used for virus or vehicle (in all cases, PBS) administration at the dose indicated in the results. For subretinal injection, a glass capillary pipette was used to pierce the sclera at the level of the limbus after the eye was dilated with tropicamide 1% (Falcon Pharmaceuticals, Abingdon, VA) and topical proparacaine (0.5%, Falcon Pharmaceuticals) was given for local anesthesia. A glass cover slip was utilized to visualize the needle in the vitreous cavity, and the contralateral retina was pierced for entry into the subretinal space. Approximately 1 µl of virus or PBS was injected into the eye using a microscope at the dose described in the text. A total of three eyes, in which injections were visible, considered equivalent, and without bleeding, were used for analysis. These injections were self-sealing and did not require sutures.
Animal imaging studies. Luciferase transgene expression in live animals was obtained using a Xenogen IVIS Lumina imaging system (Caliper Lifesciences, Hopkinton, MA). Mice were first anesthetized using 2.5% isoflurane in a gas chamber and then injected IP with excess d-luciferin (5 mg in PBS, Caliper). Five minutes after the injection, image analysis was carried out using the Living Image software (Caliper Lifesciences), and luciferase expression is reported in relative light units as described by manufacturer. As shown in Figure 3, the light intensity scale can be adjusted for image presentation purposes, which is independent of the data analysis by the Living Image software.
In vitro luciferase assay for eye and liver transduction. WT mice (N = 3) were injected with 1 × 1011 vg of intact or fragment AAV8 luciferase reporter vectors. At 15 days postinjection, an equivalent portion of the liver was harvested and analyzed for luciferase activity using standard conditions. Luciferase activity was normalized to the total recovered protein as determined by a Bradford assay. Noninjected mice (N = 3) were used as negative controls (no virus). For the eye injections described above, luciferase activity was determined (N = 3) 1 month postinjection in the total eye lysate. Luciferase activities were normalized to total recovered protein as described above.
Western blot analysis and immunoflourescence. Protein was extracted from transfected cells using cell lysis buffer (20 mmol/l Tris–HCl pH: 7.5, 150 mmol/l NaCl, 2 mmol/l EGTA, 0.1% Triton, and Proteases Inhibitors (E-64, Merck and Complete, Mini, Roche))[Manufacturer query should be added here], and protein concentrations were determined by the BCA Protein Assay kit (Pierce). The lysates were loaded at 215 μg total protein per lane. Detection of Dysferlin was performed using standard Odyssey protocol, with primary specific monoclonal anti-Dysferlin antibody (NCL-Hamlet-2, Novocastra) diluted 1:2,000. Blot was probed using the Odyssey secondary antibody DAM 800 diluted 1:5,000, scanned, and quantified using the Odyssey Application Software 2.1 (LI-COR). For in vivo western blot analysis (N = 3), total protein was recovered from the gastrocnemius muscle using the lysis solution M-PER (Pierce) and a TissueLyser LT (Qiagen, Redwood City, CA) as recommended by the manufacturers. Approximately 30 µg of total protein was separated on a NuPage 4–12% BisTris gel (Invitrogen) and transferred to a nitrocellulose membrane via i-Blot (Invitrogen). The NCL-Hamlet-2 antibody was used at 1:1,000 dilution overnight, and an anti-mouse IgG-horseradish peroxidase secondary antibody was used in conjunction with the Pierce West Pico chemiluminescent detection kit.
qPCR analysis. DNA was extracted from transfected cells by DNeasy Blood and Tissues Kit (Qiagen), according to the manufacturer's instructions, after homogenization with a Microtube Pellet Pestle Motor (Fisher Scientific, Waltham, MA). The quantity and quality of the isolated DNA was determined with NanoDrop ND 1000 (NanoDrop Technologies, Wilmington, DE). The primer pairs and Taqman probe used for enhanced green fluorescent protein (eGFP)-specific detection were as follows: 614GFP. Forward: 5′-AGTCCGCCCTGAGCAAAGA-3′ 680GFP. Reverse: 5′-GCGGTCACGAACTCCAGC-3′ and 636GFP_Probe: 5′-CAACGAGAAGCGCGATCACATGGTC-3′. The ubiquitous albumin protein was used to normalize the data across samples. Primers and probe used for albumin-specific detection were as follows: Alb. Forward: 5′-GCTGTCATCTCTTGTGGGCTGT-3′ Alb. Reverse: 5′-ACTCATGGGAGCTGCTGGTTC-3′ and Alb.Probe: 5′-CCTGTCATGCCCACACAAATCTCTCC-3′. The number of copies of eGFP and albumin in each sample was determined by a standard curve made by serial dilutions of a plasmid carrying the albumin–eGFP sequence.
For quantitation of the luciferase AAV preparations by qPCR using SYBR green staining, the following primers were used: Luciferase R: CCTTCGCTTCAAAAAATGGAAC, luciferase F: AAAAGCACTCTGATTGACAAATAC (5,105 and 1,093 bp from either ITR); SV40 polyA R: CCAGACATGATAAGATACATTGATGAGTT, SV40 polyA F: AGCAATAGCATCACAAATTTCACAA (233 and 5,954 bp from either ITR).
Statistical analysis. Student's t test was used to determine significant differences. A value of P < 0.05 was considered significant and, in most cases, the P value is indicated in each figure.
SUPPLEMENTARY MATERIAL Figure S1. Southern blot analysis of intact rAAV2 cesium chloride gradient fractions. Figure S2. Fragment AAV production and transduction evaluation. Figure S3. Comparison of wild type and optimized Dysferlin abundance following transfection. Figure S4. Transduction comparison of intact and fragment AAV vectors in the liver. Figure S5. Transduction comparison of the indicated AAV–CBA–luciferase vectors in: A) human 293 cells and B) Chinese hamster ovary cells (CHO).
Acknowledgments
We thank Camille Samulski for reviewing the text and Eric Horowitz for qPCR assistance. This work was supported by the Jain Foundation, the Northwest Genome Engineering Consortium and by the RO1 AI072176 awarded to MLH. NIH grants R01 AI080726, DK084033, AI072176, and U54 AR056953 awarded to RJS also supported this work.
Supplementary Material
Southern blot analysis of intact rAAV2 cesium chloride gradient fractions.
Fragment AAV production and transduction evaluation.
Comparison of wild type and optimized Dysferlin abundance following transfection.
Transduction comparison of intact and fragment AAV vectors in the liver.
Transduction comparison of the indicated AAV–CBA–luciferase vectors in: A) human 293 cells and B) Chinese hamster ovary cells (CHO).
References
- Samulski RJ, Berns KI, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AM, Nicolson SC, Warischalk JK, Samulski RJ. AAV's anatomy: roadmap for optimizing vectors for translational success. Curr Gene Ther. 2010;10:319–340. doi: 10.2174/156652310793180706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi VW, McCarty DM, Samulski RJ. Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol. 2006;80:10346–10356. doi: 10.1128/JVI.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentilin L, Marcello A, Giacca M. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J Virol. 2001;75:12279–12287. doi: 10.1128/JVI.75.24.12279-12287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Yue Y, Engelhardt JF. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J Virol. 2003;77:4751–4759. doi: 10.1128/JVI.77.8.4751-4759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi MP, McCarty DM. Differential effects of DNA double-strand break repair pathways on single-strand and self-complementary adeno-associated virus vector genomes. J Virol. 2010;84:8673–8682. doi: 10.1128/JVI.00641-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric J, Mano M, Zentilin L, Eulalio A, Zacchigna S, Giacca M. Terminal differentiation of cardiac and skeletal myocytes induces permissivity to AAV transduction by relieving inhibition imposed by DNA damage response proteins. Mol Ther. 2012;20:2087–2097. doi: 10.1038/mt.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch ML, Storici F, Li C, Choi VW, Samulski RJ. AAV recombineering with single strand oligonucleotides. PLoS ONE. 2009;4:e7705. doi: 10.1371/journal.pone.0007705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki K, Ma C, Storm TA, Kay MA, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81:11304–11321. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch ML, Fagan BM, Dumitru R, Bower JJ, Yadav S, Porteus MH, et al. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS ONE. 2011;6:e27520. doi: 10.1371/journal.pone.0027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj K, Ogston P, Beard P. Virus-mediated killing of cells that lack p53 activity. Nature. 2001;412:914–917. doi: 10.1038/35091082. [DOI] [PubMed] [Google Scholar]
- Duan D, Yue Y, Engelhardt JF. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Allen JM, Miller AD. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat Biotechnol. 2002;20:697–701. doi: 10.1038/nbt0702-697. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Yue Y, Duan D. Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum Gene Ther. 2011;22 1:77–83. doi: 10.1089/hum.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocca M, Doria M, Petrillo M, Colella P, Garcia-Hoyos M, Gibbs D, et al. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J Clin Invest. 2008;118:1955–1964. doi: 10.1172/JCI34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Yue Y, Duan D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome > or = 8.2 kb. Mol Ther. 2010;18:75–79. doi: 10.1038/mt.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Chen L, Dederich D, Dong B, He J, Steinmann KE, et al. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum Gene Ther. 2012;23:46–55. doi: 10.1089/hum.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch ML, Agbandje-McKenna M, Samulski RJ. Little vector, big gene transduction: fragmented genome reassembly of adeno-associated virus. Mol Ther. 2010;18 1:6–8. doi: 10.1038/mt.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan PE, Lothrop CD, Sun J, Hirsch ML, Kafri T, Kantor B, et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol Ther. 2010;18:1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose WE, Clark KR, Griffin D, Malik V, Shontz KM, Montgomery CL, et al. Homologous recombination mediates functional recovery of dysferlin deficiency following AAV5 gene transfer. PLoS ONE. 2012;7:e39233. doi: 10.1371/journal.pone.0039233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- Zhou X, Muzyczka N. In vitro packaging of adeno-associated virus DNA. J Virol. 1998;72:3241–3247. doi: 10.1128/jvi.72.4.3241-3247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Goudy K, Hirsch M, Asokan A, Fan Y, Alexander J, et al. Cellular immune response to cryptic epitopes during therapeutic gene transfer. Proc Natl Acad Sci USA. 2009;106:10770–10774. doi: 10.1073/pnas.0902269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostal W, Bartoli M, Bourg N, Roudaut C, Bentaïb A, Miyake K, et al. Efficient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer. Hum Mol Genet. 2010;19:1897–1907. doi: 10.1093/hmg/ddq065. [DOI] [PubMed] [Google Scholar]
- Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Samulski RJ. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Yue Y, Lai Y, Ye C, Qiu J, Pintel DJ, et al. Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum Gene Ther. 2004;15:896–905. doi: 10.1089/hum.2004.15.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem. 2009;284:31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J Biol Chem. 2002;277:8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci USA. 2001;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Sun J, Zhang T, Yin C, Yin F, Van Dyke T, et al. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol Ther. 2008;16:280–289. doi: 10.1038/sj.mt.6300355. [DOI] [PubMed] [Google Scholar]
- Foster H, Sharp PS, Athanasopoulos T, Trollet C, Graham IR, Foster K, et al. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol Ther. 2008;16:1825–1832. doi: 10.1038/mt.2008.186. [DOI] [PubMed] [Google Scholar]
- Millay DP, Maillet M, Roche JA, Sargent MA, McNally EM, Bloch RJ, et al. Genetic manipulation of dysferlin expression in skeletal muscle: novel insights into muscular dystrophy. Am J Pathol. 2009;175:1817–1823. doi: 10.2353/ajpath.2009.090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover LE, Newton K, Krishnan G, Bronson R, Boyle A, Krivickas LS, et al. Dysferlin overexpression in skeletal muscle produces a progressive myopathy. Ann Neurol. 2010;67:384–393. doi: 10.1002/ana.21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NJ, Cox R, Thacker J. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Caldecott K, Jeggo P. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern blot analysis of intact rAAV2 cesium chloride gradient fractions.
Fragment AAV production and transduction evaluation.
Comparison of wild type and optimized Dysferlin abundance following transfection.
Transduction comparison of intact and fragment AAV vectors in the liver.
Transduction comparison of the indicated AAV–CBA–luciferase vectors in: A) human 293 cells and B) Chinese hamster ovary cells (CHO).