Abstract
Campylobacteriosis is infection caused by the bacteria Campylobacter spp. and is considered a major public health concern. Campylobacter spp. have been identified as one of the most common causative agents of bacterial gastroenteritis. They are typically considered a foodborne pathogen and have been shown to colonise the intestinal mucosa of all food-producing animals. Much emphasis has been placed on controlling the foodborne pathway of exposure, particularly within the poultry industry, however, other environmental sources have been identified as important contributors to human infection. This paper aims to review the current literature on the sources of human exposure to Campylobacter spp. and will cover contaminated poultry, red meat, unpasteurised milk, unwashed fruit and vegetables, compost, wild bird faeces, sewage, surface water, ground water and drinking water. A comparison of current Campylobacter spp. identification methods from environmental samples is also presented. The review of literature suggests that there are multiple and diverse sources for Campylobacter infection. Many environmental sources result in direct human exposure but also in contamination of the food processing industry. This review provides useful information for risk assessment.
Keywords: campylobacteriosis, Campylobacter spp., C. jejuni, environmental reservoirs, risk assessment
1. Introduction
Campylobacter spp. are the most common cause of acute bacterial enteritis in humans [1,2,3]. They are typically considered a foodborne pathogen and have been identified as the leading cause of food poisoning in Europe [4], the United States [5], Canada [6] and Australia [7].
Campylobacteriosis refers to disease as a result of Campylobacter spp. infection [3]. The most common cause of human infection is Campylobacter jejuni, followed by Campylobacter coli, but Campylobacter lari, Campylobacter fetus and Campylobacter upsaliensis have also been reported to cause human infections [3,4,8]. Commonly reported symptoms of campylobacteriosis include diarrhoea, abdominal pain, fever, malaise and headaches [9]. Although it is generally self-limiting, approximately one tenth of laboratory confirmed cases require hospitalisation [10]. There are also some rare complications associated with Campylobacter infection. Guillain-Barré syndrome (GBS) is estimated to occur in one out of 1,000 cases of C. jejuni infection. It is a disease of the nervous system which can result in acute neuromuscular paralysis [11]. Reiter’s syndrome affects approximately 1% of campylobacteriosis causes. It is a reactive arthritis that can affect multiple joints causing pain and incapacitation [9]. Irritable bowel syndrome is anther sequel to campylobacteroisis that causes significant social and economic burden [11].
Campylobacter spp. are capable of zoonotic transfer through the faecal-oral route and have been reported to be unable to multiply outside warm-blooded host animals [9,12,13]. They colonise the intestinal mucosa of all food-producing animals and humans. However the favoured environmental niche is considered to be the intestinal tract of all avian species [14]. Thus, the primary risk factor for Campylobacter infection is considered to be exposure to be contaminated food of poultry origin [15,16,17].
Although Campylobacter spp. are unable to multiply outside a host, they can survive in different environmental sources [1]. The survival time is depended on the species and the environmental conditions including temperature, light, biotic interactions, oxygen and nutrient concentrations [18]. These environmental sources are also considered important contributors to human infection and include soil, manure, aquatic environments and water sources. The precise role that each environmental source plays in the complex epidemiology of Campylobacter infection is still unknown [19,20,21]. This review will therefore explore the current knowledge about environmental sources of Campylobacter spp.
2. Foodborne Pathogen
2.1. Poultry
Campylobacteriosis is largely considered as a foodborne disease with poultry considered the principle vehicle of transmission. Studies have identified eating or handling raw or undercooked chicken as a major risk factor campylobacteriosis in humans [11,13,16,17]. The percentage of human campylobacteriosis cases that are attributed to eating or handling raw poultry varies between countries and studies. Estimates of cases that have a foodborne origin range from 30% [22] to 58%–76% [17] and up to 80% may be attributed to the chicken reservoir as a whole [11]. In 2008, Stafford et al. estimated that 75% of campylobacteriosis was foodborne [15]. In 2009 Gillespie published a rebuttal of Stafford et al. stating they over-estimated the role of chicken consumption in cases of campylobacteriosis by a factor of 3.4 [23]. Often the source of notified cases is unable to be determined, which means that the actual number of foodborne cases is unknown [24].
2.2. Poultry Production
The percentage of chickens contaminated with Campylobacter spp. varies between countries. In the United States studies indicate that nearly 90% of flocks are colonised [25] which is supported by data from a European Union study that found and average 71.2% of broiler batches and 75.8% of broiler carcasses to be contaminated with Campylobacter spp. [26]. However, national surveys in Sweden indicate that less than 10% of broilers are contaminated with Campylobacter spp. It remains to be seen if there is similar variability throughout other regions of the World [14].
There are a range of environmental sources poultry are exposed to both on the farm and at the processing plant that can result in contamination with Campylobacter spp. The spread of Campylobacter spp. throughout flocks is extremely rapid, particularly amongst hatchlings. Studies have shown that three days contact with a single Campylobacter spp. infected bird is sufficient for the majority of the flock to be colonised [27]. Pearson et al. [28] also suggests that vertical transmission of Campylobacter spp. from breeder flocks to offspring is a source of contamination, but this is not widely accepted [5]. Horizontal transmission can occur via contaminated water, litter, faecal contact and other vectors such as insects [29,30], rodents and farm personnel [25]. Feed has not been implicated in the spread of Campylobacter spp. as it is too dry to facilitate its survival [31].
2.3. Poultry Processing
Within processing plants cross contamination is a significant problem particularly when Campylobacter spp. free flocks follow contaminated flocks [13]. Contaminated poultry that enters the processing plant can contain Campylobacter spp. populations ranging from 105 to 108 CFU/g of faeces. These high levels allow the bacteria to be easily spread throughout the plant [31]. The scalding and defeathering procedures have the potential for cross-contamination. Campylobacter spp. has periodically been removed from scald water and it has been postulated that the opening and closing of follicles may allow the retention of Campylobacter spp. within the carcass [25]. The use of recycled water throughout processing plant is another procedure that results in cross contamination. Campylobacter spp. may also be transported throughout a processing plant by personnel moving from one area of the plant to another [31].
Due to the numerous opportunities for cross contamination, processing plants employ a variety of physical, chemical and irradiation based treatments to reduce the microbial contamination of poultry carcasses [31,32]. These treatments significantly reduce the levels of Campylobacter spp. but they are unable to achieve complete removal and since dose levels as low as 500 organisms have been reported to cause illness, the contaminated carcasses will still pose a threat to public health [31]. Contaminated poultry can also result in cross contamination to other reservoirs, including food produce [33] and natural waters [34].
2.4. Other Foodborne Pathways
Consumption of unpasteurised milk and raw red meat, fruits and vegetables have also been identified as sources of foodborne campylobacteriosis in humans [35,36,37]. Campylobacter spp. may be present in milk from faecal contamination during the milking process or an udder infection [37]. Unpasteurised milk was first identified as a source of human campylobacteriosis in 1978 when four cases of C. fetus infection were identified within a three week period in a hospital in Los Angeles County. Three of the four patients had drunk large quantities of an identical brand of commercially available certified raw milk and had C. fetus subspecies jejuni isolated from their blood. A telephone survey, that was conducted to compare cases to controls, identified that the consumption of raw milk was a confirmed risk factor of C. fetus infection [38]. Information from two outbreaks of campylobacteriosis associated with drinking unpasteurised milk, one from the United Kingdom [39] and one from the Netherlands [40] have been used to derived a C. jejuni dose response model (Equation (1)) [41]:
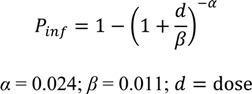 |
(1) |
A recent study from the United States found that 5% (12/262) of campylobacteriosis outbreaks from 1997–2008 were due to consumption of contaminated pork, beef or game [42]. In Europe, molecular typing studies of C. jejuni isolates from cattle have demonstrated a similarity with human strains [43,44]. Sporadic outbreaks of campylobacteriosis have been linked to contaminated red meat. In 1980 an outbreak in Dutch military barracks was associated with contaminated steak tartare and in 1979 an outbreak in a Japanese daycare centre was confirmed by culture to be caused by contaminated pork [45]. In 2004 a study in Canada investigated the prevalence of Campylobacter spp. throughout a cattle ranch. Sixty cows were tested over a 4-month period for the presence of Campylobacter spp. shed in faeces. The cows were restricted to individual pens to minimise transmission between animals. During the study, every cow tested positive for Campylobacter spp. at least once, although the survival time of Campylobacter spp. once excreted was not investigated [35].
Verhoeff-Bakkenes et al. identified consumption of raw fruits and vegetables contaminated with faecal matter as a possible source of campylobacteriosis [36]. In a study conducted in The Netherlands they found 13 out of 5,640 fruit and vegetables samples to be Campylobacter spp. positive, giving a prevalence of 0.23%. Packaged fruit and vegetables had a significantly higher prevalence of Campylobacter spp. (0.36%) when compared to unpackaged products (0.07%). However, recent case-control studies designed to identify risk factors for campylobacteriosis have found the consumption of fruit as a protective factor and not a risk [11,46].
2.5. Public Perception and Food Safety
Consumer knowledge on the potential risks associated with food preparation has been identified as a determining factor of foodborne infection [47]. In England and Wales it is estimated that at least 60 per cent of food poisoning is acquired in the home. Consumers are not fully aware of all the risks associated with food preparation and believe that the responsibility lies instead with food manufacturers [48]. Information provided to consumers on food safety including information on the dangers involved with raw meats, particularly the risk of cross-contamination and correct food preparation practices could help decrease the risk of foodborne campylobacteriosis [49].
3. Animal Vectors
3.1. Domestic Animals
A range of domestic animals have been identified as hosts for Campylobacter spp. [20,50,51,52]. In 1999 Baker et al. identified domesticated dogs and cats as a potential source of human campylobacteriosis with 55% (108/195) of cats and 49% (143/289) of dogs testing positive to Campylobacter spp. [53]. Further studies conducted on domestic dogs in Canada found that 58% (39/70) of healthy dogs’ faeces and 97% (63/65) of the diarrheic dogs’ faeces contained detectable levels of Campylobacter spp. Positive samples contained between 103 and 108 Campylobacter spp. per gram of faeces. In domestic dogs and cats, intensive housing and open drains have been shown to increase the risk of Campylobacter spp. carriage by 2 and 2.6 times, respectively. Feeding of raw meat to dogs has also been identified as a risk factor for dogs to become carriers of Campylobacter spp. [51]. Another study found C. jejuni in 5/70 (7%) healthy domestic dog faeces and 30/65 (46%) in diarrheic domestic dog faeces at concentrations up to 106 organisms/g [52]. As the infectious dose of C. jejuni is estimated to be 500 organisms [54], these high concentrations present in the faeces pose a risk for accidental exposure and possibly infection. A recent study by Gras et al. found 132/687 (19%) of domestic dogs and cat stools to be positive for Campylobacter spp. The detected C. jejuni and C. coli multilocus sequence type (STs) from pets and owners were compared. There were 2/68 (2.94%) cases where owner was infected with an identical ST to their pet (compared 0.134/68 (0.2%) expected to occur by chance). This study identified dog ownership, particularly puppy ownership, as a significant increase in risk for campylobacteriosis [55].
3.2. Wild Animal Faeces
The shedding of wild birds faeces into the environment has been identified as a significant reservoir of Campylobacter spp. [56]. Exposure to contaminated wild bird faeces in playgrounds has been recognised as an emerging environmental source of campylobacteriosis, particularly for children [19]. The frequent hand to mouth behaviour associated with children provides a mechanism for ingesting campylobacters [57]. Many playgrounds are natural habitats for a range of wild animals including birds, lizards, and stray cats and dogs. A New Zealand study tested avian faecal matter found in children’s playgrounds and discovered that a total of 12.5% (24/192) were positive for C. jejuni including 6.7% (4/60) dried samples and 15.2% (20/132) fresh samples. Three of these isolates also had indistinguishable genotypes to those isolates recovered from human clinical cases, which provides evidence to support the link between wild bird faeces in playgrounds and human campylobacteriosis [19].
The presence of contaminated reptile faeces in the environment is possibly another emerging source of campylobacteriosis. A recent study in the United States reported nine cases of infection with C. fetus subsp. testudinum subsp. nov. This is a recently discovered subspecies of Campylobacter that appears to have originated in reptiles [8]. This is supported by a study by Wang et al. that identified C. fetus in 6.7% (12/179) of reptile fecal samples collected from domestic and wild reptiles in Taiwan [58].
4. Density and Fate in Solids
Given the mesophilic and micro-aerophilic nature of many Campylobacter spp., this bacterium cannot survive outside the host for extended periods of time [9,21,59] and as such, densities found in human and animal (e.g., bovine) biosolids can be low [60,61] (see Table 1). The rapid inactivation of Campylobacter spp. was demonstrated by Sinton et al., who investigated the survival of laboratory-cultured C. jejuni in bovine faeces on pasture [62]. This work showed that in comparison to other indicator organisms (Escherichia coli, fecal streptococci, enterococci and Salmonella enterica), the inactivation of C. jejuni was rapid, even when the water content remained above 80%. T90 values were 16 days in winter, 2.7 days in spring, 1.2 days in summer and 4.7 days in autumn. It was concluded that temperature, rather than desiccation influenced survival and that post-excretion exposure to oxygen diffusing into the pat may have also increased the inactivation. Follow-up work by Gilipin et al. [23] also found that Campylobacter spp. naturally present in cow faeces exhibited a similar inactivation rate to that previously determined by Sinton et al. using laboratory-cultured strains [62]. Both studies are consistent with earlier work by Nicholson et al., who monitored the inactivation of Campylobacter spp. during the land application of farm yard manure to different soil types. At 15–20 °C, this work showed that Campylobacter spp. survived for up to 8 days after application to sandy arable soils and 8–32 days for clay loam grassland soils [63].
Table 1.
Density of C. jejuni in various sources of excrement.
More recently a study has demonstrated that Campylobacter spp. is able to persist and survive within compost, a relatively hostile environment, for up to 10 months. This study showed that the bacteria were able to survive within faeces from both untreated cattle and cattle treated with the antibiotics chlortetracycline and sulfamethazine. The antibiotics were added to cattle feed to improve weight gain, feed efficiency and to aid in the prevention of liver abscesses, bacterial diarrhoea, foot rot and bovine respiratory disease. Of the ingested chlortetracycline, 75% was excreted in cattle faeces; however, this had no effect on the presence and viability of Campylobacter spp. [35].
The discrepancy in survival noted between research by Sinton et al. [62] and Inglis et al. [35] could be attributed to the difference in culture and molecular identification techniques. Culture techniques often underestimate the number of bacteria in an environmental sample; the latter study used quantitative PRC in conjunction with ethidium monoazide treatment (EMA) which ensured only intact cells are amplified [67].
4.1. Storage of Biosolids and Manure
Ahmed and Sorensen [68] investigated the impact of temperature on inactivation rates of C. jejuni during the storage of dewatered biosolids. Compared to S. Typhimurium, C. jejuni was found to be more sensitive to heat. Results showed that 4.5 to >6 log10 reduction of C. jejuni occurred within one day at 49.5 °C. At cooler temperatures of 22 and 38 °C, 11 and six days were required to achieve a comparable log10 reduction. At 5 °C, C. jejuni were more persistent, whereby a 2 log10 reduction was observed within a 62 day period. Similar observations were noted during the storage of farmyard manure by Nicholson et al. This study reported a survival time of 2–5 days for both turned and unturned stockpiles of solid farmyard manure, when temperatures greater than 50 °C were obtained [63]. In contrast, Campylobacter spp. were shown to survive in stored slurries at temperatures of ca 15–20 °C for up to 32 days. In both instances, the inactivation of Campylobacter spp. was more rapid than Salmonella, Listeria and E. coli.
4.2. Anaerobic: Mesophilic Anaerobic Digestion
In contrast to aerobic environments, C. jejuni has been shown to be more resilient than indicator organisms during the anaerobic digestion of biosolids. During primary mesophilic anaerobic digestion of sewage sludge, Horan and his colleagues found no inactivation of C. jejuni within a operational period of 22 days and at a temperature of 35 °C [69]. Under the same conditions, a log removal of 1.66 was observed for E. coli, 2.23 for Listeria monocytogenes and 2.23 for Salmonella senftenberg respectively. The same study showed that a log10 C. jejuni removal of only 0.36 was achieved during a secondary sludge digestion stage, at cooler temperatures of 15 °C. This data is consistent with Kearney et al. who reported limited inactivation (1 log10 reduction in 793 days) of Campylobacter spp. during mesophilic anaerobic digestion [70].
5. Sewage
Studies have shown Campylobacter spp. to be ubiquitous in sewage [59,71,72,73,74]. Raw wastewater numbers of Campylobacter spp. can vary between 2–5 log10 per·L−1 [61,72].
The effectiveness of a sewage treatment plants in reducing Campylobacter spp. numbers during regular conditions depends on the complexity of the plant. Research by Arimi et al. found that primary sedimentation was able to reduce Campylobacter spp. numbers by 78%. Such a reduction is due to the fact that most micro-organisms are bound with solids [71]. In comparison to commonly used indicator organisms such as E. coli, Wéry et al. found C. jejuni were more resistant to biological treatment [61]. Nevertheless, biological treatment processes such as trickling filters, activated sludge plants and oxidation ponds are able to achieve a decimal reduction within the order of 0.6–2 log10 units [59,72,75].
Treated effluent is either pumped in to marine waters [21] or reused for irrigation [76,77] and the effective removal of Campylobacter spp. by tertiary treatments is crucial in preventing contamination of potential sources of human exposure [78].
6. Water Sources/Surface Waters
As previously mentioned, Campylobacter spp. are unable to grow outside warm blooded hosts [9] and can maintain long term contamination of environmental water sources [79]. There is great debate over the length of time that Campylobacter spp. can survive outside a host. Bushwell et al. demonstrated that Campylobacter spp. could survive only up to 29 days in water [80]; however, Rollins et al. demonstrated that they could survive for over 120 days in water [81]. The discrepancies between studies are thought to be caused by the bacteria entering a viable non-culturable state that prevents detection via traditional culture based techniques [81,82].
Environmental water sources have been associated with human campylobacteriosis. Campylobacter spp. have been isolated from a variety of environmental water sources including rivers [83], lakes [84], streams [85] and coastal waters [86]. Environmental waters can become contaminated through a variety of mechanisms including direct contamination with animal and avian faeces, agricultural run-off from farms, small holdings, slaughterhouses, slurry that is sprayed onto land and sewage effluent [21,83].
The incidence of campylobacteriosis follows a similar trend to most waterborne diseases, with a peak in incidence observed during late spring and early summer months [87]. This pattern, however, is not supported by quantitative studies of surface water which have shown there to be higher numbers of Campylobacter spp. present in surface waters during the winter months when compared to summer months. It has been postulated that the decrease in Campylobacter spp. numbers in the summer is due to elevated levels of UV light and higher temperatures [12].
In open waters, solar radiation is widely considered to be a dominant inactivation agent, which directly reduces the density of pathogens [88,89]. Sinton and his colleagues showed that sunlight insolation needed for 90% inactivation (S90 MJ·m−2) of Campylobacter spp. in river water and seawater augmented with STP effluent was 1.7 and 1.4 MJ m−2 respectively [62]. Under strong, optimal sunlight conditions in Spain and Bolivia (maximum global irradiance of ~1,050 W·m−2), Boyle et al. demonstrated that a 4 log10 unit reduction of C. jejuni was achieved in a short timeframe of 20 min, when using transparent water containers [90]. This resulted in a rapid solar inactivation rate (S90) of only 7 (±3) kJ/m−2. Under dark conditions, Sinton et al. found that T90 (time needed for 90% inactivation) values were a short 35 and 82.6 h for river water and seawater microcosms augmented with STP effluent. Inactivation of enteric microbes in dark, natural waters can largely attributed to the predatory, lytic, and grazing effects [91]. In the study by Sinton et al. the dark inactivation rates (T90) were thought to have been accelerated by the presence of residual predatory microbiota within the STP effluent. The significance of predatory biota on the survival of Campylobacter spp. in lake water was also noted in the work by Korhonen and Martikainen [92].
6.1. Groundwater
Groundwater is rarely considered as a reservoir for pathogenic microorganisms. The soils that bacteria must pass through to reach the surface generally function to attenuate microorganisms through simple filtration. Groundwater reaches the surface at boreholes, wells, springs and seeps. It is frequently used for irrigation and drinking water for livestock on farms [12,93]. However contaminated groundwater has been identified as a source of campylobacteriosis associated with outbreaks in poultry flocks [94] broiler chickens [95] and dairy farms [93]. Subsurface aquifers of groundwater provide favourable conditions for Campylobacter spp. survival, including constant temperature all year round, and protection from UV and desiccation. Similar conditions are found in larger groundwater aquifers that are used to deliver water to big cities and could be a potentially overlooked vehicle for transmission of campylobacteriosis in animals reared for food [12].
6.2. Drinking Water
Drinking water has been implicated in a number of sporadic outbreaks of campylobacteriosis [82,96,97,98,99]. Predominantly outbreaks are a result of consumption of untreated or contaminated water [12]. Private water supplies (PWS) are the number one source of contaminated drinking water. In England and Wales the majority of PWS are springs, boreholes and wells and a review conducted between 1992 and 2003 identified PWS as the source of 13 outbreaks of human campylobacteriosis [100]. Rainwater tanks specifically have been identified as a source of contamination in reported outbreaks of campylobacteriosis [101,102,103]. One study conducted in Australia found that 44% (12/27) rainwater tanks used for drinking water contained Campylobacter spp. with the major source of considered to be avian or possum faecal matter [99].
6.3. Campylobacter spp. and Water Disinfection
The contamination of water with Campylobacter spp. has been identified as both a direct source of contamination and an indirect source through the cross-contamination of drinking water and carcasses at processing plants. Studies have found that common disinfection processes designed to remove coliform bacteria from drinking water were sufficient to eliminate Campylobacter spp.. For example, Blaser et al. demonstrated that under a range of temperature and pH conditions, C. jejuni was more susceptible to chlorine and monochloramine disinfection than E. coli. This study showed that 2 log10 inactivation of C. jejuni was achieved after 15 min of contact with 1.0 mg L−1 of monochloramine or 5 min of contact with 0.1 mg L−1 of free chlorine [104]. Review data by Hijnen et al. also showed that C. jejuni was more susceptible to UV than E. coli. This review data showed that UV fluence of 3, 7 and 10 mJ/cm2 were required to permit a C. jejuni inactivation log10 credit of 1, 2 and 3 respectively, and that E. coli required an additional dose of ca. 2–4 mJ/cm2 to reach an equivalent inactivation credit. These results suggest that disinfection procedures which are commonly based on meeting E. coli targets are adequate to eliminate C. jejuni. This is supported by the absence of campylobacteriosis outbreaks associated with properly disinfected water [104].
A recent study has demonstrated a decrease in the susceptibility of Campylobacter spp. to disinfection. Snelling et al. demonstrated the ability of C. jejuni to become internalised by the waterborne protozoa Tetrahymena pyriformis and Acanthamoeba castellanii within broiler drinking water systems. This study also demonstrated that internalised Campylobacter spp. were significantly more resistant to disinfection than their planktonic counterparts [105]. The formation of protozoan cysts have been shown to provide internalised bacteria protection from a range of other unfavourable environmental conditions, such as low nutrients, heat, desiccation and osmotic stress. Internalised bacteria within the protozoa cysts are also provided with a method for further contamination as protozoan cyst can be dispersed through the air [106].
7. Detection Methods
The difficulties in identifying environmental reservoirs of Campylobacter spp. are amplified by inaccuracies surrounding detection methods. The three main methods of identification are: traditional culture method using selective agar, membrane filtration onto blood agar and real time PCR [107,108,109]. The most common selective agar used is charcoal cefaperazone desoxycholate agar containing 32 mg/L cefoperazone. Plates are incubated at 37 °C for two days in anaerobic campy jars. Selective culture is a quick, cheap and effective method for identifying C. jejuni and C. coli from faecal samples [108]. However plates are often overgrown by faster growing microorganisms and this method does not identify the less common species. Filtering samples through a cellulose triacetate membrane with 0.45 mm pores onto blood agar separates the Campylobacter spp. from other larger bacteria which could overgrow the agar. This method is able to detect all cultivable Campylobacter spp. as there is no antibiotic used in the medium. Plates are also able to be incubated for longer without being overgrown which allows for the isolation of slower growing species [108]. The sensitivity of the membrane filtration technique is less than the selective culture and both of these techniques are less effective when applied to isolating Campylobacter spp. from water samples [96]. The problems associated with isolating Campylobacter spp. from water samples it the bacteria’s tendency to enter a viable but non-culturable state in unfavourable environmental conditions. This includes starvation but also physical stress which can occur during the process of sampling and storing cells [110].
Real time PCR allows for the identification of Campylobacter spp. to the species level and results can be achieved in one day. However it does not provide an isolate for further research, it is expensive and also highly labour intensive [108]. Detection and enumeration of viable but non-culturable cells is achieved using real time PCR. The problem with this method is that total counts can be overestimated due to the amplification of non-viable or killed cells. DNA within environmental samples can be very stable and is able to persist for extended lengths of time [67]. Novitsky [111] demonstrated that in marine sediment and salt water only 60%–70% of the DNA from killed organisms was in 14 days. This is an important issue in detection due to the varying lifespan of Campylobacter spp. that is dependent on both the strain and a range of environmental conditions such as temperature and free oxygen [12]. Techniques such as the use of ethidium monoazide (EMA) can be used in tandem with real time PCR to ensure only whole intact cells are amplified. When exposed to light EMA binds to any DNA that is not protected by a cell membrane and hence prevents amplification and enumeration. EMA has been optimised for use in water but is yet to be optimised in other environmental samples and is still not widely applied [67,111,112,113].
rRNA specific fluorescently tagged probes have also been used to examine Campylobacter spp. within aquatic biofilms. This method is valuable in examining Campylobacter spp. in situ within a biofilm. They are not effective for detection as they does not allow for enumeration and specificity can be significantly reduced by background fluorescence [80].
7.1. Molecular Typing
Molecular typing is an emerging tool which has been used to enhance many epidemiological studies. It enables the source of a patient’s Campylobacter spp. isolate to be identified based on its genome [3]. Multi-locus sequence typing (MLST) of clinical isolates and food and environmental isolates has been used as an epidemiological tool to estimate the relative importance of each source of human campylobacteriosis in New Zealand [17], Finland [114], Canada [115] and Scotland [116]. Whole genome sequencing (WGS) is another nucleotide typing tool, the benefit of WGS is that it has the highest discriminatory power and can differentiate between a single base pair. The expensive nature and difficulties associated with managing large databases of information, means that it’s use is still not widespread. It remains to be seen if WGS can be used for large scale epidemiological surveillance or if there will continue to be a need for inexpensive front line sub-typing methods [117].
7.2. Comparison to Faecal Indicators
Indicators of faecal contamination such as faecal coliforms and streptococci are often used as indicator organisms for the presence of faecal pathogens [12]. However Carter et al. isolated Campylobacter spp. from a range of natural water sources in central Washington including ponds, lakes and mountain streams. Microbiological plate counts were conducted and the results established that there was a lack of significant correlation between the occurrence of Campylobacter spp. densities and faecal coliforms, total coliforms, faecal Streptococci and heterotrophic bacteria. This lack of correlation suggests that further studies are needed to identify possible indicator organisms which could predict the presence of Campylobacter spp. in water [85].
This review suggests that in comparison to indicators, Campylobacter spp. can potentially survive longer in environments that are low in oxygen (e.g., in stockpiled slurries and anaerobic digesters) and vice versa in aerobic environments (e.g., land application of solids, activated sludge) (see Table 2).
Table 2.
Campylobacter inactivation by environmental and treatment processes compared to faecal indicator organisms.
| Barrier | Source | Conditions | Inactivation | Units | Comparative inactivation | Reference |
|---|---|---|---|---|---|---|
| Solar inactivation | River water + STP effluent | Natural sunlight conditions | 1.65–1.68 | S90 (MJ·m−2) | Higher than E. coli and S. enterica | [62] |
| Sea water + STP effluent | Natural sunlight conditions | 1.28–1.38 | ||||
| Transparent water bottles | Optimal sunlight conditions | 7 (±3) | S90 (kJ·m−2) | Higher than S. epidermidis, E. coli and Y. enterocolitica and B. subtilis | [90] | |
| UV treatment | Potable water | UV fluence of 3 mJ/cm2 | 1 | Log10 | Higher than E. coli | [118] |
| UV fluence of 7 mJ/cm2 | 2 | |||||
| UV fluence of 10 mJ/cm2 | 3 | |||||
| Free chlorine | Potable water | 0.1 mg·L−1 after 5 min contact | 2 | Log10 | Higher than E. coli | [104] |
| Monochloramine | Potable water | 1.0 mg·L−1 after 15 min contact | 2 | |||
| Primary sedimentation | Sewage | 78 | % | [71] | ||
| Trickling filters | Sewage | 0.6 | [64] | |||
| Activated sludge + settling | Sewage | 1–2.5 | Log10 | Lower than E. coli | [61] | |
| 1 | [64] | |||||
| Dark inactivation | Unfiltered lake water | 14 days 4 °C | 100 | % | Higher than E. coli | [92] |
| 8 days at 25 °C | 100 | % | ||||
| 0.2 µm filter lake water | 27 days at 4 °C | 100 | % | |||
| 4 days at 25 °C | 100 | % | ||||
| River water + STP effluent | 120 L chambers | 82.6 | T90 (hours) | Higher than E. coli and S. enterica | [62] | |
| Seawater + STP effluent | 120 L chambers | 35 | T90 (hours) | |||
| Storage | Human biosolids | 49.5 °C for 1 day | 4.6–>6 | log10 | Higher than S. Typhimurium | [68] |
| 38 °C for 6 days | >6 | |||||
| 22 °C for 11 days | >6 | |||||
| 5 °C for 62 days | 2 | |||||
| Farmyard manure | 4 days at 50 °C | 3 | log10 | Higher than Salmonella, Listeria and E. coli | [63] | |
| 32 days 15–20 °C | 3 | |||||
| 4 and 17 °C for 112 days | 0 | log10 | Lower than E. coli, L. monocytogenes, Yersinia enterocolitica and S. typhimurium | [70] | ||
| Land application | Farmyard manure | Sandy arable soils. 4–8 days at 11–20 °C | >3 | log10 | Higher than Salmonella, Listeria and E. coli | [63] |
| Clay loam grassland soils. 8–32 days at 15–20 °C | 2 | |||||
| Bovine manure applied to pasture | Winter | 16 | T90 (days) | Higher than E. coli, fecal streptococci, enterococci and S. enterica | [62] | |
| Spring | 2.7 | |||||
| Summer | 1.2 | |||||
| Autumn | 4.7 | |||||
| Anaerobic digestion | Human biosolids | 22 days at 35 °C | 0 | log10 | Lower than E. coli, L. monocytogenes and S. senftenberg | [69] |
| 25 days at 15 °C | 0.36 | |||||
| Cattle slurry | 793 days at 35 °C | 1 | log10 | Lower than E. coli, L. monocytogenes, Yersinia enterocolitica and S. typhimurium | [70] |
8. Conclusions
This paper provides a review of environmental sources of human campylobacteriosis and a preliminary tool for risk assessment. Campylobacter spp. are present in a range of environmental reservoirs, which include faeces of wild and domestic animals and municipal sewage. Many of these are a direct cause of human infection but also cross-contaminate other environments such as recreational and source waters, which have also resulted in human infection. Our review shows that the foodborne pathway of human Campylobacter spp. exposure has previously been overestimated and can be reduced significantly by providing consumers with greater information on the risk of raw meat and correct handling procedures. Importantly, further research and greater consideration of non-foodborne environmental reservoirs as a source of campylobacteriosis is required. This requires a better understanding of the fate and transport of Campylobacter spp. in the environment to better develop exposure assessment. However, to facilitate this, standard detection methods of Campylobacter spp. need to be established and tailored to the environmental source sampled. This is especially important due to the lack of correlation between common faecal indicators organisms and Campylobacter spp., meaning that they cannot be used to predict the presence or fate of Campylobacter spp. in the environment. Recently, our understanding of campylobacteriosis epidemiology has increased due to the development of molecular typing methods to identify sources of infection. The future application of these molecular typing methods, particularly WGS, for epidemiological studies will continue to improve our understanding of transmission routes and improve management strategies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wilson D.J., Gabriel E., Leatherbarrow A.J.H., Cheesbrough J., Gee S., Bolton E., Fox A., Fearnhead P., Hart C.A., Diggle P.J. Tracing the source of campylobacteriosis. PLoS Genet. 2008;4:1–9. doi: 10.1371/journal.pgen.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strachan N.J., Gormley F.J., Rotariu O., Ogden I.D., Miller G., Dunn G.M., Sheppard S.K., Dallas J.F., Reid T.M., Howie H., et al. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J. Infect. Dis. 2009;199:1205–1208. doi: 10.1086/597417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheppard S.K., Dallas J.F., Strachan N.J.C., MacRae M., McCarthy N.D., Wilson D.J., Gormley F.J., Falush D., Ogden I.D., Maiden M.C.J., et al. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 2009;48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2005. EFSA J. 2006;94:4–288. [Google Scholar]
- 5.Altekruse S.F., Stern N.J., Fields P.I., Swerdlow D.L. Campylobacter jejuni—An emerging foodbourne pathogen. Emerg. Infect. Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen H. Acanthamoeba-Campylobacter Interactions. University of Ottawa; Ottowa, ON, Canada: 2001. [Google Scholar]
- 7.Australian National Notifiable Diseases Surveillance System . Number of Notifications for All Diseases by Year, Australia, 1991 to 2009 and Year-to-Date Notifications for 2010. Australian Department of Health and Aging; Adelaide, Australia: 2010. [Google Scholar]
- 8.Patrick M.E., Gilbert M.J., Blaser M.J., Tauxe R.V., Wagenaar J.A., Fitzgerald C. Human infections with new subspecies of Campylobacter fetus. Emerg. Infect. Dis. 2013;19:1679–1680. doi: 10.3201/eid1910.130883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ternhag A., Törner A., Svensson Å., Giesecke J., Ekdahl K. Mortality following Campylobacter infection: A registry-based linkage study. BMC Infect. Dis. 2005;5:1–5. doi: 10.1186/1471-2334-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie I.A., O’Brien S.J., Bolton F.J. Age patterns of persons with campylobacteriosis, England and Wales, 1990–2007. Emerg. Infect. Dis. 2009;15:2046–2048. doi: 10.3201/eid1512.090280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mughini Gras L., Smid J.H., Wagenaar J.A., de Boer A.G., Havelaar A.H., Friesema I.H., French N.P., Busani L., van Pelt W. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: A combined case-control and source attribution analysis. PLoS One . 2012;7:e42599. doi: 10.1371/journal.pone.0042599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones K. Campylobacters in water, sewage and the environment. Appl. Microbiol. 2001;90:68–79. doi: 10.1046/j.1365-2672.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- 13.Newell D.G., Shreeve J.E., Toszeghy M., Domingue G., Bill S., Humphrey T., Mead G. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 2001;67:2636–2640. doi: 10.1128/AEM.67.6.2636-2640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newell D.G., Fearnley C. Sources of Camplobacter colonization in broiler chickens. Appl. Environ. Microbiol. 2003;69:4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stafford R.J., Schluter P.J., Wilson A.J., Kirk M.D., Hall G., Unicomb L. Population-attributable risk estimates for risk factors associated with campylobacter infection, australia. Emerg. Infect. Dis. 2008;14:895–901. doi: 10.3201/eid1406.071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meldrum R.J., Griffiths J.K., Smith R.M.M., Evans M.R. The seasonality of human Campylobacter infection and Campylobacter isolates from fresh, retail chicken in Wales. Epidemiol. Infect. 2005;133:49–52. doi: 10.1017/S0950268804003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullner P., Spencer S.E.F., Wilson D.J., Jones G., Noble A.D., Midwinter A.C., Collins-Emerson J.M., Carter P., Hathaway S., French N.P. Assigning the source of human campylobacteriosis in New Zealand: A comparative genetic and epidemiological approach. Infect. Genet. Evol. 2009;9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Pitkänen T. Review of Campylobacter spp. in drinking and environmental waters. J. Microbiol. Methods. 2013;95:39–47. doi: 10.1016/j.mimet.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 19.French N.P., Midwinter A., Holland B., Collins-Emerson J., Pattison R., Colles F., Carter P. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl. Environ. Microbiol. 2009;75:779–783. doi: 10.1128/AEM.01979-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown P.E., Christensen O.F., Clough H.E., Diggle P.J., Hart C.A., Hazel S., Kemp R., Leatherbarrow A.J.H., Moore A., Sutherst J., et al. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 2004;70:6501–6511. doi: 10.1128/AEM.70.11.6501-6511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones A.B., O’Donohue M.J., Udy J., Dennison W.C. Assessing ecological impacts of shrimp and sewage effluent: Biological indicators with standard water quality analyses. Estuar. Coast. Shelf Sci. 2001;52:91–109. doi: 10.1006/ecss.2000.0729. [DOI] [Google Scholar]
- 22.Black A.P., Kirk M.D., Millard G. Campylobacter outbreak due to chicken consumption at an australian capital territory restaurant. CDI. 2006;30:373–377. [PubMed] [Google Scholar]
- 23.Gillespie I. Population-attributable risk estimates for Campylobacter infection, Australia. Emerg. Infect. Dis. 2009;15:850–851. doi: 10.3201/eid1505.081553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinersmann R.J., Phillips R.W., Hiett K.L., Fedorka-Cray P. Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 2005;71:6368–6374. doi: 10.1128/AEM.71.10.6368-6374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stern N.J., Fedorka-Cray P., Bailey J.S., Cox N.A., Craven S.E., Hiett K.L., Musgrove M.T., Ladley S., Cosby D., Mead G.C. Distribution of Campylobacter spp. In selected U.S. Poultry production and processing operations. J. Food Prot. 2001;64:1705–1710. doi: 10.4315/0362-028x-64.11.1705. [DOI] [PubMed] [Google Scholar]
- 26.European Food Safety Authority Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the eu, 2008—Part A: Campylobacter and Salmonella prevalence estimates. EFSA J. 2010;8:1503. doi: 10.2903/j.efsa.2010.1503. [DOI] [Google Scholar]
- 27.Shanker S., Lee A., Sorrell T.C. Horizontal transmission of Campylobacter jejuni amongst broiler chicks: Experimental studies. Epidemiol. Infect. 1990;104:101–110. doi: 10.1017/S0950268800054571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson A.D., Greenwood M.H., Feltham R.K.A., Healing T.D., Donaldson J., Jones D.M., Colwell R.R. Microbial ecology of Campylobacter jejuni in a united kingdom chicken supply chain: Intermittent common source, vertical transmission, and amplification by flock propagation. Appl. Environ. Microbiol. 1996;62:4614–4620. doi: 10.1128/aem.62.12.4614-4620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols G.L. Fly transmission of Campylobacter. Emerg. Infect. Dis. 2005;11:361–364. doi: 10.3201/eid1103.040460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Templeton J.M., Jong A.J.D., Blackall P.J., Miflin J.K. Survival of Campylobacter spp. in darkling beetles (Alphitobius diaperinus) and their larvae in australia. Appl. Environ. Microbiol. 2006;72:7909–7911. doi: 10.1128/AEM.01471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keener K.M., Bashor M.P., Curtis P.A., Sheldon B.W., Kathariou S. Comprehensive review of Campylobacter and poultry processing. Compr. Rev. Food Sci. Food Saf. 2004;4:105–116. doi: 10.1111/j.1541-4337.2004.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 32.Peyrat M.B., Soumet C., Maris P., Sanders P. Recovery of Campylobacter jejuni from surfaces of poultry slaughterhouses after cleaning and disinfection procedures: Analysis of a potential source of carcass contamination. Int. J. Food Microbiol. 2008;124:188–194. doi: 10.1016/j.ijfoodmicro.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 33.Luber P., Brynestad S., Topsch D., Scherer K., Bartelt E. Quantification of Campylobacter species cross-contamination during handling of contaminated fresh chicken parts in kitchens. Appl. Environ. Microbiol. 2006;72:66–70. doi: 10.1128/AEM.72.1.66-70.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vereen E., Jr., Lowrance R.R., Cole D.J., Lipp E.K. Distribution and ecology of Campylobacters in coastal plain streams (Georgia, United States of America) Appl. Environ. Microbiol. 2007;73:1395–1403. doi: 10.1128/AEM.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inglis G.D., Kalischuk L.D., Busz H.W. Chronic shedding of Campylobacter species in beef cattle. J. Appl. Microbiol. 2004;97:410–420. doi: 10.1111/j.1365-2672.2004.02313.x. [DOI] [PubMed] [Google Scholar]
- 36.Verhoeff-Bakkenes L., Jansen H.A.P.M., in ’t Veld P.H., Beumer R.R., Zwietering M.H., van Leusden F.M. Consumption of raw vegetables and fruits: A risk factor for Campylobacter infections. Int. J. Food Microbiol. 2011;144:406–412. doi: 10.1016/j.ijfoodmicro.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 37.Rapp D., Ross C.M., Pleydell E.J., Muirhead R.W. Differences in the fecal concentrations and genetic diversities of Campylobacter jejuni populations among individual cows in two dairy herds. Appl. Environ. Microbiol. 2012;78:7564–7571. doi: 10.1128/AEM.01783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor P.R., Weinstein W.M., Bryner J.H. Campylobacter fetus infection in human subjects: Association with raw milk. Am. J. Med. 1979;66:779–783. doi: 10.1016/0002-9343(79)91116-1. [DOI] [PubMed] [Google Scholar]
- 39.Evans M.R. A milk-borne Campylobacter outbreak following an educational farm visit. Epidemiol. Infect. 1996;117:457. doi: 10.1017/S0950268800059112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Brandhof W., Wagenaar J.A., van den Kerkhof H. An outbreak of campylobacteriosis after drinking unpasteurized milk, 2002, the Netherlands. Int. J. Med. Microbiol. 2003;293:548–549. [Google Scholar]
- 41.Teunis P., van den Brandhof W., Nauta M., Wagenaar J., van den Kerkhof H., van Pelt W. A reconsideration of the Campylobacter dose-response relation. Epidemiol. Infect. 2005;133:583. doi: 10.1017/S0950268805003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor E.V., Herman K.M., Ailes E.C., Fitzgerald C., Yoder J.S., Mahon B.E., Tauxe R.V. Common source outbreaks of Campylobacter infection in the USA, 1997–2008. Epidemiol. Infect. 2013;141:987–996. doi: 10.1017/S0950268812001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzgerald C., Stanley K., Andrew S., Jones K. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 2001;67:1429–1436. doi: 10.1128/AEM.67.4.1429-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouls L.M., Reulen S., Duim B., Wagenaar J.A., Willems R.J.L., Dingle K.E., Colles F.M., Embden J.D.A.V. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: Strain diversity, host range, and recombination. J. Clin. Microbiol. 2003;41:15–26. doi: 10.1128/JCM.41.1.15-26.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh T., Saito K., Maruyama T., Sakai S., Ohashi M., Oka A. An outbreak of acute enteritis due to Campylobacter fetus subspecies jejuni at a nursery school of tokyo. Microb. Immunol. 1980;24:371–379. doi: 10.1111/j.1348-0421.1980.tb02841.x. [DOI] [PubMed] [Google Scholar]
- 46.Doorduyn Y., van Den Brandhof W.E., van Duynhoven Y.T.H.P., Breukink B.J., Wagenaar J.A., van Pelt W. Risk factors for indigenous Campylobacter jejuni and Campylobacter coli infections in the Netherlands: A case-control study. Epidemiol. Infect. 2010;138:1391–1404. doi: 10.1017/S095026881000052X. [DOI] [PubMed] [Google Scholar]
- 47.Miles S., Braxton D.S., Frewer L.J. Public perceptions about microbiological hazards in food. Br. Food J. 1999;101:744–753. doi: 10.1108/00070709910293670. [DOI] [Google Scholar]
- 48.Worsfold D., Griffith C.J. Food safety behaviour in the home. Br. Food J. 1997;99:97–104. doi: 10.1108/00070709710168932. [DOI] [Google Scholar]
- 49.Todd E.C.D. Microbiological safety standards and public health goals to reduce foodborne disease. Meat Sci. 2004;66:33–43. doi: 10.1016/S0309-1740(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 50.Scott E. Food safety and foodborne disease in 21st century homes. Can. J. Infect. Dis. 2003;14:277–280. doi: 10.1155/2003/363984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenz J., Joffe D., Kauffman M., Zhang Y., LeJeune J. Perceptions, practices, and consequences associated with foodborne pathogens and the feeding of raw meat to dogs. CVJ. 2009;50:637–643. [PMC free article] [PubMed] [Google Scholar]
- 52.Chaban B., Ngeleka M., Hill J. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 2010;10:73. doi: 10.1186/1471-2180-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker J., Barton M., Lanser J. Campylobacter species in cats and dogs in south Australia. Aust. Vet. J. 1999;77:662–666. doi: 10.1111/j.1751-0813.1999.tb13159.x. [DOI] [PubMed] [Google Scholar]
- 54.Kothary M.H., Babu U.S. Infective dose of foodborne pathogens in volunteers: A review. J. Food Saf. 2001;21:49–68. doi: 10.1111/j.1745-4565.2001.tb00307.x. [DOI] [Google Scholar]
- 55.Gras L.M., Smid J.H., Wagenaar J.A., Koene M.G.J., Havelaar A.H., Friesema I.H.M., French N.P., Flemming C., Galson J.D., Graziani C., et al. Increased risk for Campylobacter jejuni and C. Coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol. Infect. 2013;141(12):2526–2535. doi: 10.1017/S0950268813000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waldenstrom J., Axelsson-Olsson D., Olsen B., Hasselquist D., Griekspoor P., Jansson L., Teneberg S., Svensson L., Ellstrom P. Campylobacter jejuni colonization in wild birds: Results from an infection experiment. PLoS One. 2010;5:1–8. doi: 10.1371/journal.pone.0009082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tulve N.S., SUGGS J.C., McCURDY T., HUBAL E.A.C., MOYA J. Frequency of mouthing behavior in young children. J. Expo. Anal. Environ. Epidemiol. 2002;12:259–264. doi: 10.1038/sj.jea.7500225. [DOI] [PubMed] [Google Scholar]
- 58.Wang C.-M., Shia W.-Y., Jhou Y.-J., Shyu C.-L. Occurrence and molecular characterization of reptilian Campylobacter fetus strains isolated in Taiwan. Vet. Microbiol. 2013;164:67–76. doi: 10.1016/j.vetmic.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Stampi S., de Luca G., Varoli O., Zanetti F. Occurrence, removal and seasonal variation of thermophilic Campylobacters and Arcobacter in sewage sludge. Zentralblatt für Hygiene und Umweltmedizin. 1999;202:19–27. [PubMed] [Google Scholar]
- 60.Stampi S., Varol O., Zanetti F., Luca G.D. Arcobacter cryaerophilus and thermophilic Campylobacters in a sewage treatment plant in Italy: Two secondary treatments compared. Epidemiol. Infect. 1993;110:633–639. doi: 10.1017/S0950268800051050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wéry N., Lhoutellier C., Ducray F., Delgenès J.-P., Godon J.-J. Behaviour of pathogenic and indicator bacteria during urban wastewater treatment and sludge composting, as revealed by quantitative PCR. Water Res. 2008;42:53–62. doi: 10.1016/j.watres.2007.06.048. [DOI] [PubMed] [Google Scholar]
- 62.Sinton L.W., Braithwaite R.R., Hall C.H., Mackenzie M.L. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl. Environ. Microbiol. 2007;73:7917–7925. doi: 10.1128/AEM.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicholson F.A., Groves S.J., Chambers B.J. Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 2005;96:135–143. doi: 10.1016/j.biortech.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 64.Koenraad P.M.F.J., Hazeleger W.C., van der Laan T., Beumer R.R., Rombouts F.M. Survey of Campylobacter spp. in sewage plants in the Netherlands. Food Microbiol. 1994;11:65–73. doi: 10.1006/fmic.1994.1009. [DOI] [Google Scholar]
- 65.Blaser M.J., Hardesty H.L., Powers B., Wang W.L. Survival of Campylobacter fetus subsp. jejuni in biological milieus. J. Clin. Microbiol. 1980;11:309–313. doi: 10.1128/jcm.11.4.309-313.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inglis G.D., Kalischuk L.D. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time quantitative PCR. Appl. Environ. Microbiol. 2004;70:2296–2306. doi: 10.1128/AEM.70.4.2296-2306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nocker A., Sossa P., Burr M., Camper A. Use of propidium monoazide for live-dead distinction in microbial ecology. Appl. Environ. Microbiol. 2007;73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmed A.U., Sorensen D.L. Kinetics of pathogen destruction during storage of dewatered biosolids. Water Environ. Res. 1995;67:143–150. doi: 10.2175/106143095X131286. [DOI] [Google Scholar]
- 69.Horan N.J., Fletcher L., Betmal S.M., Wilks S.A., Keevil C.W. Die-off of enteric bacterial pathogens during mesophilic anaerobic digestion. Water Res. 2004;38:1113–1120. doi: 10.1016/j.watres.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 70.Kearney T.E., Larkin M.J., Levett P.N. The effect of slurry storage and anaerobic digestion on survival of pathogenic bacteria. J. Appl. Microbiol. 1993;74:86–93. doi: 10.1111/j.1365-2672.1993.tb03000.x. [DOI] [PubMed] [Google Scholar]
- 71.Arimi S.M., Fricker C.R., Park R.W.A. Occurrence of “thermophilic” campylobacters in sewage and their removal by treatment processes. Epidemiol. Infect. 1988;101:279–286. doi: 10.1017/S0950268800054194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koenraad P.M.F.J., Ayling R., Hazeleger W.C., Romboutst F.M., Newell D.G. The speciation and subtyping of Campylobacter isolates from sewage plants and waste water from a connected poultry abattoir using molecular techniques. Epidemniol. Infect. 1995;115:485–494. doi: 10.1017/S0950268800058647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waage A.S., Vardund T., Lund V., Kapperud G. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested pcr assay. Appl. Environ. Microbiol. 1999;65:1636–1643. doi: 10.1128/aem.65.4.1636-1643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rechenburg A., Kistemann T. Sewage effluent as a source of Campylobacter sp. In a surface water catchment. Int. J. Environ. Health Res. 2009;19:239–249. doi: 10.1080/09603120802460376. [DOI] [PubMed] [Google Scholar]
- 75.Lauria-Filgueiras A., Hofer E. Diversity of Campylobacter isolates from three activated sludge systems. Mem Inst Oswaldo Cruz Rio de Janeiro. 1998;93:295–298. doi: 10.1590/S0074-02761998000300003. [DOI] [PubMed] [Google Scholar]
- 76.Smith C.J., Hopmans P., Cook F.J. Accumulation of Cr, Pb, Cu, Ni, Zn and Cd in soil following irrigation with treated urban effluent in Australia. Environ. Pollut. 1996;94:317–323. doi: 10.1016/S0269-7491(96)00089-9. [DOI] [PubMed] [Google Scholar]
- 77.Rattan R.K., Datta S.P., Chhonkar P.K., Suribabu K., Singh A.K. Long-term impact of irrigation with sewage effluents on heavy metal content in soils, crops and groundwater—A case study. Agric. Ecosyst. Environ. 2005;109:310–322. doi: 10.1016/j.agee.2005.02.025. [DOI] [Google Scholar]
- 78.Betaie M., Jones B.K. Thermophilic Campylobacters in two sewage treatment plants in Libya. Lett. Appl. Microbiol. 1990;11:93–95. doi: 10.1111/j.1472-765X.1990.tb01284.x. [DOI] [Google Scholar]
- 79.Murphy C., Carroll C., Jordan K.N. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 2006;100:623–632. doi: 10.1111/j.1365-2672.2006.02903.x. [DOI] [PubMed] [Google Scholar]
- 80.Buswell C.M. Extended survival and persistence of Campylobacter spp. In water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 1998;64:733–741. doi: 10.1128/aem.64.2.733-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rollins D.M., Colwell R.R. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cools I., Uyttendaele M., Caro C., D’Haese E., Nelis H.J., Debevere J. Survival of Campylobacter jejuni strains of different origin in drinking water. J. Appl. Microbiol. 2003;94:886–892. doi: 10.1046/j.1365-2672.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 83.Daczkowska-Kozon E., Brzostek-Nowakowska J. Campylobacter spp. In waters of three main western pomerania water bodies. Int. J. Hyg. Environ. Health. 2001;203:435–443. doi: 10.1078/1438-4639-00048. [DOI] [PubMed] [Google Scholar]
- 84.Horman A., Rimhanen-Finne R., Maunula L., Bonsdorff C.-H.V., Torvela N., Heikinheimo A., Hanninen M.-L. Campylobacter spp., Giardia spp., Cryptosporidium spp., Noroviruses, and indicator organisms in surface water in southwestern Finland, 2000–2001. Appl. Environ. Microbiol. 2004;70:87–95. doi: 10.1128/AEM.70.1.87-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carter A.M., Pacha R.E., Clark G.W., Williams E.A. Seasonal occurrence of Campylobacter spp. In surface waters and their correlation with standard indicator bacteria. Appl. Environ. Microbiol. 1987;53:523–526. doi: 10.1128/aem.53.3.523-526.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Obiri-Danso K., Jones K. The effect of a new sewage treatment plant on faecal indicator numbers, Campylobacters and bathing water compliance in morecambe bay. J. Appl. Microbiol. 1999;83:603–614. doi: 10.1046/j.1365-2672.1999.00703.x. [DOI] [PubMed] [Google Scholar]
- 87.Sari Kovats R. Climate variability and Campylobacter infection: An international study. Int. J. Biometeorol. 2005;49:207. doi: 10.1007/s00484-004-0241-3. [DOI] [PubMed] [Google Scholar]
- 88.Davies-Colley R.J., Bell R.G., Donnison A.M. Sunlight inactivation of Enterococci and fecal coliforms in sewage effluent diluted in seawater. Appl. Environ. Microbiol. 1994;60:2049–2058. doi: 10.1128/aem.60.6.2049-2058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sinton L.W., Davies-Colley R.J., Bell R.G. Inactivation of Enterococci and fecal coliforms from sewage and meatworks effluents in seawater chambers. Appl. Environ. Microbiol. 1994;60:2040–2048. doi: 10.1128/aem.60.6.2040-2048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boyle M., Sichel C., Fernández-Ibñez P., Arias-Quiroz G.B., Iriarte-Puña M., Mercado A., Ubomba-Jaswa E., McGuigan K.G. Bactericidal effect of solar water disinfection under real sunlight conditions. Appl. Environ. Microbiol. 2008;74:2997–3001. doi: 10.1128/AEM.02415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sinton L.W., Hall C.H., Lynch P.A., Davies-Colley R.J. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 2002;68:1122–1131. doi: 10.1128/AEM.68.3.1122-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Korhonen L.K., Martikalnon P.J. Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J. Appl. Microbiol. 1991;71:379–382. doi: 10.1111/j.1365-2672.1991.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 93.Stanley K., Cunningham R., Jones K. Isolation of Campylobacter jejuni from groundwater. J. Appl. Microbiol. 1998;85:187–191. doi: 10.1046/j.1365-2672.1998.00494.x. [DOI] [PubMed] [Google Scholar]
- 94.Pearson A.D., Greenwood M., Healing T.D., Rollins D., Shahamat M., Donaldson J., Colwell R.R. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl. Environ. Microbiol. 1993;59:987–996. doi: 10.1128/aem.59.4.987-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giessen A.W.V.D., Bloemberg B.P.M., Ritmeester W.S., Tilburg J.J.H.C. Epidemiological study on risk factors and risk reducing measures for Campylobacter infections in Dutch broiler flocks. Epidemiol. Infect. 1996;117:245–250. doi: 10.1017/S0950268800001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moore J., Caldwell P., Millar B. Molecular detection of Campylobacter spp. in drinking, recreational and environmental water supplies. Int. J. Hyg. Environ. Health. 2001;204:185–189. doi: 10.1078/1438-4639-00096. [DOI] [PubMed] [Google Scholar]
- 97.Kuusi M., Klemets P., Miettinen I., Laaksonen I., Sarkkinen H., Ha¨nninen M.L., Rautelin H., Kela E., Nuorti J.P. An outbreak of gastroenteritis from a non-chlorinated community water supply. J. Epidemiol. Community Health. 2004;58:273–277. doi: 10.1136/jech.2003.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith A., Reacher M., Smerdon W., Adak G.K., Nichols G., Chalmers R.M. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992–2003. Epidemiol. Infect. 2006;134:1141–1149. doi: 10.1017/S0950268806006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ahmed W., Huygens F., Goonetilleke A., Gardner T. Real-time PCR detection of pathogenic microorganisms in roof-harvested rainwater in southeast Queensland, Australia. J. Appl. Environ. Microbiol. 2008;74:5490–5496. doi: 10.1128/AEM.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Said B., Wright F., Nichols G.L., Reacher M., Rutter M. Outbreaks of infectious disease associated with private drinking water supplies in England and Wales 1970–2000. Epidemiol. Infect. 2003;130:469–479. [PMC free article] [PubMed] [Google Scholar]
- 101.Daoud A.K., Swaileh K.M., Hussein R.M., Matani M. Quality assessment of roof-harvested rainwater in the west bank, palestinian authority. J. Water Health. 2011;9:525–533. doi: 10.2166/wh.2011.148. [DOI] [PubMed] [Google Scholar]
- 102.Savill M.G., Hudson J.A., Ball A., Klena J.D., Scholes P., Whyte R.J., McCormick R.E., Jankovic D. Enumeration of Campylobacter in New Zealand recreational and drinking waters. Appl. Microbiol. 2001;91:38–46. doi: 10.1046/j.1365-2672.2001.01337.x. [DOI] [PubMed] [Google Scholar]
- 103.Merritt A., Miles R., Bates J. An outbreak of Campylobacter enteritis on an island resort, north Queensland. CDI. 1999;23:215–218. [PubMed] [Google Scholar]
- 104.Blaser M.J., Smith P.F., Wang W.L., Hoff J.C. Inactivation of Campylobacter jejuni by chlorine and monochloramine. Appl. Environ. Microbiol. 1986;51:307–311. doi: 10.1128/aem.51.2.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Snelling W.J., McKenna J.P., Lecky D.M., Dooley J.S.G. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 2005;71:5560–5571. doi: 10.1128/AEM.71.9.5560-5571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown M.R.W., Barker J. Unexplored reservoirs of pathogenic bacteria: Protozoa and biofilms. Trends Microbiol. 1999;7:46–50. doi: 10.1016/S0966-842X(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 107.Lawson A.J., Linton D., Stanley J., Owen R.J. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. Helveticus in human faeces and comparison with culture techniques. J. Appl. Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 108.Kulkarni S.P., Lever S., Logan J.M.J. Detection of Campylobacter species: A comparison of culture and polymerase chain reaction based methods. J. Clin. Pathol. 2002;55:749–753. doi: 10.1136/jcp.55.10.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maher M., Finnegan C., Collins E., Ward B., Carroll C., Cormican M. Evaluation of culture methods and a DNA probe-based pcr assay for detection of campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 2003;41:2980–2986. doi: 10.1128/JCM.41.7.2980-2986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baserisalehi M., Bahador N., Kapadnis B.P. A novel method for isolation of Campylobacter spp. From environmental samples, involving sample processing, and blood- and antibiotic-free medium. J. Appl. Microbiol. 2004;97:853–860. doi: 10.1111/j.1365-2672.2004.02375.x. [DOI] [PubMed] [Google Scholar]
- 111.Novitsky J.A. Degradation of dead microbial biomass in a marine sediment. Appl. Environ. Microbiol. 1986;52:504–509. doi: 10.1128/aem.52.3.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rudi K., Moen B., Dromtorp S.M., Holck A.L. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 2005;71:1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pan Y., Breidt F. Enumeration of viable Listeria monocytogenes cells by real-time PCR with Propidium monoazide and Ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 2007;73:8028–8031. doi: 10.1128/AEM.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kärenlampi R., Rautelin H., Schönberg-Norio D., Paulin L., Hänninen M.-L. Longitudinal study of finnish Campylobacter jejuni and C. Coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 2007;73:148–155. doi: 10.1128/AEM.01488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lévesque S., Frost E., Arbeit R.D., Michaud S. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 2008;46:3404–3411. doi: 10.1128/JCM.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sheppard S.K., Dallas J.F., MacRae M., McCarthy N.D., Sproston E.L., Gormley F.J., Strachan N.J.C., Ogden I.D., Maiden M.C.J., Forbes K.J. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 2009;134:96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taboada E.N., Clark C.G., Sproston E.L., Carrillo C.D. Current methods for molecular typing of Campylobacter species. J. Microbiol. Methods. 2013;95:24–31. doi: 10.1016/j.mimet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Hijnen W.A.M., Beerendonk E.F., Medema G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review. Water Res. 2006;40:3–22. doi: 10.1016/j.watres.2005.10.030. [DOI] [PubMed] [Google Scholar]


