Abstract
The development of improved methods for early detection and characterization of cancer presents a major clinical challenge. One approach that has shown excellent potential in preclinical and clinical evaluation is molecular imaging with small-scaffold, non-antibody based, engineered proteins. These novel diagnostic agents produce high contrast images due to their fast clearance from the bloodstream and healthy tissues, can be evolved to bind a multitude of cancer biomarkers, and are easily functionalized by site-specific bioconjugation methods. Several small protein scaffolds have been verified for in vivo molecular imaging including affibodies and their two-helix variants, knottins, fibronectins, DARPins, and several natural ligands. Further, the biodistribution of these engineered ligands can be optimized through rational mutation of the conserved regions, careful selection and placement of chelator, and modification of molecular size.
Introduction
Molecular imaging can provide critical clinical information regarding the presence, concentration, and localization of cancer biomarkers in vivo. The data, which can be obtained dynamically or longitudinally if desired, empowers early detection, patient stratification, and treatment monitoring. A major challenge in this field is the development of high affinity, specific ligands for the multitude of important biomarkers. Directed evolution and other strategies position engineered proteins to offer a robust, high throughput means for ligand generation (Figure 1). Many of these engineered proteins have shown promising results in preclinical evaluation as well as early clinical results. While antibodies and their fragments have been explored [1], their large size and slow clearance are not ideal due to the functional requirement of reduced background signal necessary for imaging contrast; moreover, for targeting of poorly vascularized tumors, such as those in early formation, large size slows extravasation and delivery [2]. For these reasons, as well as benefits in stability, production, and chemical conjugation, alternative protein topologies have been studied as scaffolds for molecular imaging. Short linear and cyclic peptides have exhibited significant success as molecular imaging agents [3], although their robustness in engineering high affinity toward novel targets is limited. Thus, this review will focus on advances in molecular imaging of cancer, particularly positron emission tomography (PET), single-photon emission computed tomography (SPECT), and gamma-camera imaging using engineered, folded, non-antibody proteins (Figure 2, Table 1). The majority of the research has been performed using subcutaneous xenografted tumors in mice. There are exceptions to these experimental systems, including clinical data, which will be noted.
Figure 1.
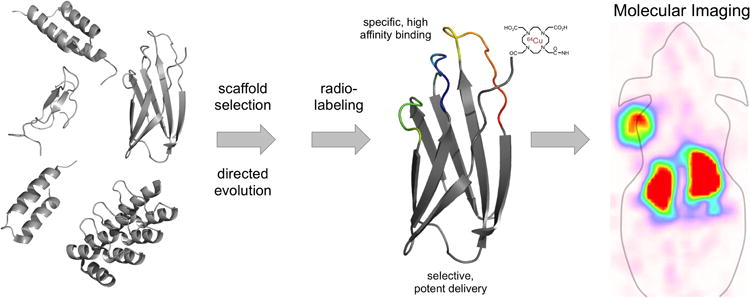
Schematic for molecular imaging agent development. Several scaffolds have been validated for in vivo molecular imaging. Selection of an appropriate scaffold, followed by directed evolution – for binding and delivery characteristics – and radiolabeling yield an imaging agent with potent delivery and retention properties capable of producing high contrast molecular images.
Figure 2.
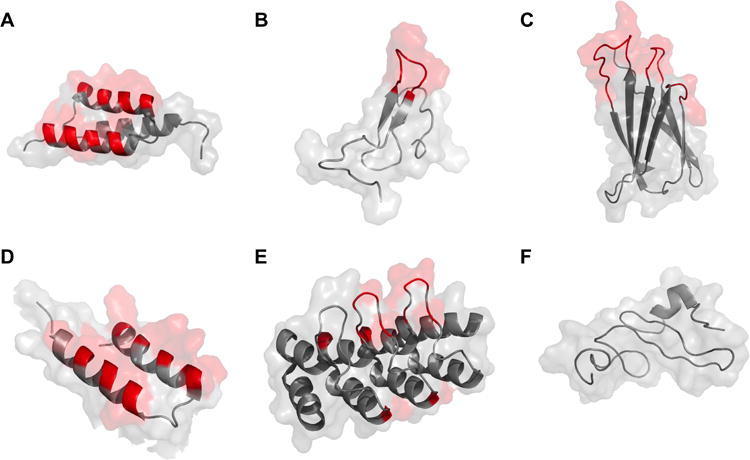
Cartoon structure with surface representation of protein scaffolds used in molecular imaging applications. Variable regions are highlighted in red. Conserved portions are shown in grey. A: affibody (PDB: 2B88); B: knottin (1HYK); C: fibronectin (1TTF); D: two-helix affibody (modified from three-helix affibody 2KZJ); E: DARPin (2JAB); F: natural ligand (EGF) (2KV4).
Table 1. Characteristics of non-antibody molecular imaging agents.
| Name | Topology | Amino Acids | Typical Paratope |
|---|---|---|---|
| Affibody | triple α-helical bundle | 58 | surface of helices 1 and 2 (13 amino acids) |
| Knottin | knotted 3-disulfide core | 30 - 50 | loop 1 |
| Fibronectin | β sandwich | 94 | 1-3 loops (10-24 amino acids) |
| DARPin | 4-6 repeats of: β turn + two α helices | 130 - 200 | β turn and first α helix (7 amino acids) |
| Two-Helix Affibody | 2 α-helices | 35 | surface of helices 1 and 2 (13 amino acids) |
| Natural Ligands | e.g. VEGF, EGF, annexin V | variable | natural interface |
EGF(R): epidermal growth factor (receptor)
EpCAM: epithelial cell adhesion molecule
HER2: human epidermal growth factor receptor type II
IGF1R: insulin-like growth factor type I receptor
VEGF(R): vascular endothelial growth factor (receptor)
Protein-Based Molecular Imaging Agents
Affibody
The affibody is a 58 amino acid, three helical bundle [4]. Typically, randomization of 13 amino acids on the surface of helices 1 and 2 is used to generate novel binding ligands. The most extensively studied class of affibodies is those targeting human epidermal growth factor receptor 2 (HER2). The second generation HER2-binding affibody, ZHER2:342, was engineered to bind with 22 pM affinity [5], and has successfully imaged HER2-expressing tumor xenografts in mice when labeled with 125I [5], 111In [6], 99mTc [7], 18F [8], 114mIn [9], 124I [10], 68Ga [11], and 11C [12]. Tumor targeting is effective across many mouse models with a median tumor uptake of 9 %ID/g at 4 h (range: 1 – 26 %ID/g), tumor-to-blood ratio median of 31 (range: 1.5 – 187), and tumor-to-muscle ratio median of 61 (range; 3 – 650). Kidney uptake is generally high with a median of 111 %ID/g (range 2 – 324 %ID/g). Hepatic retention is minimal with a median of 1.5 %ID/g (range 0.2 – 19 %ID/g). ZHER2:342 has been extensively modified to maximize imaging contrast and decrease signal in clearance organs. Clinical translation has been initiated as one derivative was labeled with 111In and 68Ga for SPECT and PET imaging of metastatic breast cancer patients [13]. The tracers effectively imaged metastases and were well tolerated.
Three different anti-epidermal growth factor receptor (EGFR) affibodies (0.9 – 50 nM affinity) labeled with 111In identified tumors using a gamma camera, exhibiting 2.4-3.4 %ID/g tumor with tumor-to-blood and tumor-to-muscle ratios of 7-15 and 18-27 [14-16]. One of these affibodies was labeled at a unique Cys residue with 64Cu via a 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) chelator. PET imaging in xenografted mice exhibited 12±2 %ID/g tumor but only 1.04±0.01 tumor-to-blood at 4 h. Pre-injection of 50 μg cold affibody elevated tumor uptake to 17±4 %ID/g with 10±3 tumor-to-blood and 49±11 tumor-to-muscle [17]. Separately, this affibody was labeled with an 18F precursor. PET imaging exhibited 8±1 %ID/g tumor with 2.6±0.5 tumor-to-blood and 37±13 tumor-to-muscle at 3 h [18].
An affibody engineered to 0.5 nM affinity for insulin-like growth factor type I receptor (IGF1R) labeled with 111In yielded 1.3±0.1 %ID/g tumor with 2.5±0.2 tumor-to-blood and 4.3±1.0 tumor-to-muscle at 4 h [19]. However, no significant contrast was observed for tumor relative to pancreas, spleen, stomach, and colon. Contrast was modestly improved using 99mTc labeling via an HEHEHE purification tag [20].
Two-helix affibody
A 36 amino acid two-helix derivative has been explored as a smaller alternative to the affibody via removal of the third helix. The resulting destabilization was partially compensated by stabilizing mutations and disulfide bonding [21,22]. The two-helix affibody has only been able to achieve low nanomolar affinity to date [21]. The two-helix affibody has been applied to molecular imaging in HER2-expressing tumor xenograft models successfully when labeled with 68Ga [23], 18F [24], and 111In [25]. In a comparative study, the two-helix affibody (2 nM affinity) showed higher tumor-to-blood ratio than the parental affibody (78 pM affinity) [25]. However, the tumor uptake of the two-helix derivative was 40% lower than the parental clone. Further stabilization and affinity maturation could make the two-helix affibody an advantageous option for high-contrast molecular imaging.
Knottin
Knottins are 30-50 amino acid polypeptides containing three disulfide bonds that form a knotted structure [26]. Binding motifs have been grafted into one to two loops to provide binding functionality. Randomizations of loop lengths, motif position, and surrounding amino acids have been used for affinity maturation. An integrin-binding motif was grafted into Ecballium elaterium trypsin inhibitor II and evolved for mid-nanomolar affinity to integrin αvβ3 with cross-reactivity for integrins α5β1 and αvβ5. 64Cu radiolabeling via DOTA enabled PET imaging with 4.5±1.2 %ID/g tumor, 6±1 tumor-to-blood, and 17±8 tumor-to-muscle at 1 h and 4.0±0.7 %ID/g tumor, 42±9 tumor-to-blood, and 21±13 tumor-to-muscle at 4 h [27]. Renal retention was 4±1 %ID/g at 1 h while hepatic signal was 2.2±0.2 %ID/g. Minimal degradation was observed in blood, minor breakdown in tumor, and >50% degradation in kidneys. A knottin variant with six loop mutations, 2.5D, was then labeled with 18F so that radioisotope kinetics would better match pharmacokinetics [28]. Effective tumor imaging was observed with 2.6±0.7 %ID/g tumor at 0.5 h. Kidney and liver uptake were similarly decreased relative to 64Cu-knottin values. In a comparative study in a transgenic mouse model with nascent lung tumors, the 64Cu-knottin tracer exhibited superior tumor-to-lung ratios (6.0±0.6 versus 4.4±0.7) relative to 18F-fluorodeoxyglucose [29]. The 2.5D knottin was also dually labeled with 64Cu and near infrared fluorophore as a multimodality probe, which improved tumor retention but elevated renal and hepatic signal [30].
Integrin-binding motifs were also grafted and evolved to mid-nanomolar affinity in the Agouti-related protein knottin to yield clone 7C. 64Cu labeling enabled effective PET imaging of xenografted tumors with rapid uptake including 2.7±0.9 %ID/g tumor at 1 h and 6.6±1.2 tumor-to-blood and 17±5 tumor-muscle by 2 h [31]. Kidney retention was higher (60±18 %ID/g at 2 h) than with the Ecballium elaterium trypsin inhibitor II scaffold. 111In labeling of this knottin yielded 5.7±1.6 %ID/g tumor at 0.5 h but rapid reduction in tumor signal (2.5±0.4 %ID/g tumor at 2 h) [32]. Renal retention was improved relative to the 64Cu probe (34±8 %ID/g). 18F radiolabeling via 18F-fluoropropionate yields effective PET imaging with a more translatable isotope: 2.5±0.2 %ID/g tumor, 20±2 %ID/g kidney, 7±1 tumor-to-muscle, and 4±1 tumor-to-blood at 1 h [33].
The breadth of knottin scaffolds and their targets was expanded as Momordica cochinchinensis trypsin inhibitor II was grafted with integrin αvβ6-binding motif and evolved to 3-6 nM affinity [34]. 64Cu labeling yielded 4.3±0.7 %ID/g tumor at 1 h with 9±2 tumor-to-muscle but 75±5 %ID/g kidney. Grafting this loop onto a serine-rich Lens culinaris trypsin inhibitor knottin reduced renal retention to 18±4 %ID/g, although tumor uptake was reduced two-fold. This tracer also effectively imaged orthotopic pancreatic tumors. Radiolabeling with 18F yielded effective PET imaging as early as 0.5 h with significant decline in renal retention [35]. In further evidence of the breadth of this topology, fluorophore-conjugated agatoxin was successfully able to image integrin αvβ3-expressing U87MG glioblastoma xenografts in mice [36].
Fibronectin Domain
The tenth type III domain of human fibronectin (also known as monobody or Adnectin) is a 94 amino acid β-sandwich protein. Diversification of 10-24 amino acids in one to three solvent-exposed loops imparts novel binding activity [37,38]. Recently, the fibronectin scaffold was validated for in vivo molecular PET imaging in mouse tumor xenograft models [39]. PET imaging with a 64Cu-DOTA conjugated EGFR-binding fibronectin showed good tumor uptake (3.4±1.0 %ID/g at 1 h), retention (2.7±0.6 %ID/g at 24 h), and contrast (8.6±3.0 tumor-to-muscle ratio, 8.9±4.7 tumor-to-blood ratio). Dynamic PET indicated effective contrast was present as early at 15 minutes post-injection. High renal retention was partially alleviated by 18F radiolabeling. Biodistribution was further modified by mutation of hydrophobic and charged residues [40]. A 64Cu-DOTA-fibronectin specific to CD20 has also been developed and tested in a humanized transgenic mouse model for B-cell imaging (Arut Natarajan, BJH, Sanjiv Gambhir, unpublished). Resulting PET images showed encouraging results for the application of this fibronectin in imaging non-Hodgkin's lymphoma. Both spleen uptake and spleen-to-blood ratio for the 64Cu-DOTA-fibronectin were significantly higher than 64Cu-DOTA-rituximab, showing that the biodistribution properties and imaging ability of the fibronectin domain are superior to monoclonal antibodies.
Designed Ankyrin Repeat Protein
The designed ankyrin repeat protein (DARPin) is composed of 4-6 repeat units of 33 amino acids making one β turn followed by 2 α-helices. Diversification of 7 amino acids per repeat unit yields novel binding activity [41]. 99mTc(CO)3-labeled DARPins with affinities between 90 pM and 270 nM have been applied to imaging HER2-expressing tumor xenografts in mice using SPECT/CT [42]. The tumor uptake ranged from 1.5±0.2 %ID/g for the weakest affinity to 9.1±1.8 %ID/g for the strongest affinity DARPin at 1 h. The tumor-to-blood ratio for the 90 pM agent is 27±15 at 4 h, however, kidney uptake is 239±33 %ID/g. Poly(ethylene glycol) (PEG) conjugation increased tumor uptake at late time points but reduced tumor-to-blood ratios; ratios above 1 were not observed until the 24 h post injection time point. Biodistribution for a fusion of Pseudomonas aeruginosa exotoxin A and an EpCAM-specific DARPin was determined in vivo with a Cy5.5-conjugated DARPin fusion construct [43]. Ex vivo analysis 48 h post injection shows the highest localization in the EpCAM-expressing tumor, providing greater signal than the clearance organs.
Natural Ligands
Targeting agents can also be derived from natural ligands as these molecules often have high specificity and affinity for their respective receptors. However, the major disadvantage of using these ligands for molecular imaging is their potential agonistic effects. Despite this, several natural ligands have been used. Vascular endothelial growth factor (VEGF) has been used to image angiogenesis in tumors using several different radioisotopes including 123I [44], 64Cu [45], and 68Ga [46]. A single chain analogue of VEGF has also been used to image VEGF receptor-expressing tumor xenografts [47,48]. Radiolabeled EGF has been applied for imaging of EGFR-expressing tumors in mouse models, although high liver signal is a noted problem in multiple studies [49,50]. Radiolabeled annexin V has been utilized for treatment monitoring applications. Annexin V, which binds the apoptosis marker phosphatidylserine, has been used to assess the effectiveness of radiation therapy [51] and doxorubicin-based therapy [52]. Annexin V effectively predicted response to chemotherapy with 94% accuracy in separating responders from non-responders [53].
Designing Delivery
It is evident that disparate protein topologies can be engineered for in vivo molecular recognition and delivery of radioisotopes to tumors. As the field continues to mature, we should strive to identify the ideal molecule for the application of interest, to maximize tumor uptake, minimize off-target retention, and tune kinetics for efficacy, logistics, and patient safety. Quantitative design rules would be tremendously useful to optimize performance. Select studies have been performed that initiate this effort.
Molecular Size
As previously stated, small size aids permeability across the vascular wall [2,54], which speeds tumor uptake. It also speeds clearance, which is beneficial for low background but reduces the input function available for tumor targeting. A mathematical compartmental model predicts uptake and specificity are maximized by small proteins [2], which is in agreement with experimental data. A HER2-targeted affibody exhibited ≥10-fold greater tumor-to-blood relative to the antibody trastuzumab at 6, 24, and 72 h [10] (Figure 3). 7 kDa affibody monomers yielded a median of 3.4-fold higher tumor uptake than 15 kDa affibody dimers despite blood clearance that was comparable to the monomer (median 1.0-fold) [5,14,55-57]. A two-helix affibody derivative yielded 2.7-fold higher tumor-to-blood and 1.8-fold higher tumor-to-muscle at 1 h relative to the larger three-helix affibody albeit with a 40% reduction in tumor uptake [25]. A 15 kDa DARPin also exhibited 2.3-fold greater tumor signal at 1 h and similar tumor signal at 4 h relative to a 35 kDa conjugate with PEG despite an increased blood concentration for the PEGylated domain [42].
Figure 3.
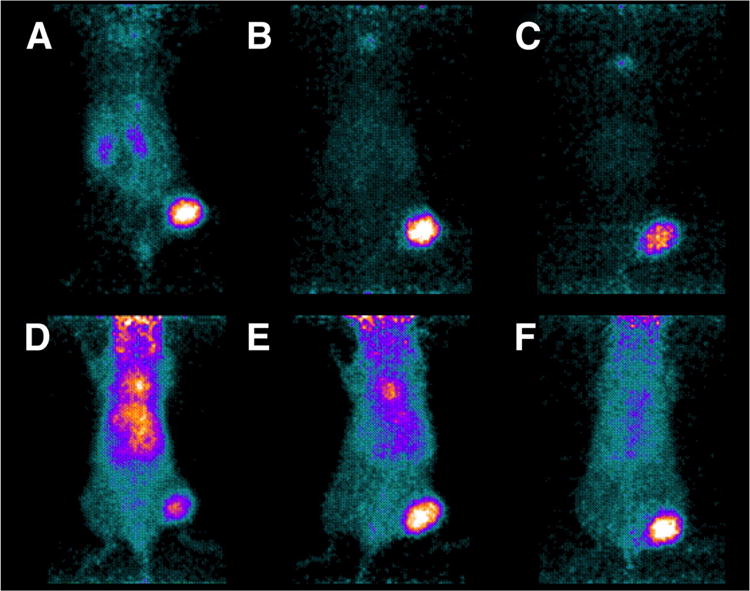
Female BALB/c nu/nu mice with HER2-expressing NCI-N87 xenografts were imaged via PET using 124I-PIB-ZHER2:342 affibody (A-C) and 124I-PIB-trastuzumab (D-F). Mice were sacrificed and imaged 6 (A and D), 24 (B and E), and 72 h (C and F) post injection. Urinary bladders were removed before scanning. Figure reproduced.[10]
Affinity
The affinity of the engineered probe for its target affects retention in the tumor site. 111In-DOTA-affibodies engineered to 117 pM, 157 pM, and 3.8 nM affinities for HER2 were compared in SKOV-3 (HER2high) and LS174T (HER2low) tumor xenograft models [58]. In the SKOV-3 model, tumor uptake was indistinguishable at 4 h (13±3, 16±7, and 14±1 %ID/g respectively), but at 24 h, the tumor signal was 2-fold higher for the picomolar affinity affibodies. In the LS174T model, the tumor uptake for the 3.8 nM affinity was only 1.7±0.2 %ID/g at 4 h compared to 7.4±1.6 and 10.8±1.3 %ID/g for the picomolar affibodies, respectively. 99mTc-DOTA-DARPins with 90 pM and 1.5 nM affinities for HER2 in SKOV-3 model showed similar results, with tumor uptakes being indistinguishable at 4 h, but the 90 pM affinity showing better retention at longer time points [42]. The tumor uptake of these DARPins became distinguishable when a 10 nM DARPin is compared to the 90 pM DARPin over all time points.
Hydrophilicity
Four fibronectin domains were engineered with a range of hydrophilicities to study the impact on biodistribution. Renal retention was observed to positively correlate with hydrophilicity whereas hepatic retention was inversely correlated [40]. Moreover, using phylogenetic sequence frequency, a metric was identified to select appropriate mutations to introduce hydrophilicity. Thus, protein hydrophilicity is an addressable modulator of physiological distribution. The decision between renal and hepatic retention can be made based on the application; else, hydrophilicity can be introduced to minimize hepatic retention and renal retention can be reduced by other means.
Molecular Charge
Modification of molecular charge has impacted renal retention in numerous studies, although the impacts of total and net charge are unclear. Charge removal from cystine knots reduced renal retention [34]. Removal of two or three lysines from an affibody chelator eliminated 74 and 86 % of renal retention [59]; removal of two or three basic residues eliminated 91 and 84 %ID/g tumor [7]. Removal of six acidic and five basic residues from fibronectin decreased renal retention by 34% [40]. In a separate fibronectin clone, removal of four acidic and two basic residues also reduced renal retention by 34%; however, removal of one or two more acidic residues re-elevated renal retention.
Chelator
Both chelator structure and location of ligand conjugation can be impactful on in vivo biodistribution. The comparison of three 111In-labeled macrocyclic chelators, DOTA, 1,4,7-triazacyclononane-N,N′,N″-triacetic acid (NOTA), and 1-(1,3-carboxypropyl)-4,7-carboxymethyl-1,4,7-triazacyclononane (NODAGA), using anti-HER2 affibodies showed NOTA to have a two-fold high liver signal than the others at 4 h [60]. NODAGA demonstrated the fastest clearance kinetics providing the highest tumor-to-blood imaging contrast whereas DOTA had a slightly higher tumor uptake due to longer plasma residence. In addition to ring structures, amino acid based chelators (3 residues in length) provide high labeling efficiency and may be chosen based on imaging background requirements. In particular, the kidneys and intestines are commonly sources for off-target uptake and a tradeoff may be made between the two with chelator choice. Positively or negatively charged chelators (Asp-Ser-Asp and Lys-Lys-Lys) showed high kidney signal (33 and 120 %ID/g) [59,61] whereas neutral chelators (Ser-Ser-Ser and Gly-Gly-Gly) reduced kidney uptake (19 and 6 %ID/g) [59,62]. The location of chelator conjugation is also impactful on biodistribution; C-terminal DOTA outperformed those placed N-terminal and mid-sequence with a two-fold improvement in tumor-to-blood signal [63].
In summary, several protein topologies provide intriguing preclinical results, and the initial clinical results with the affibody are encouraging. The field should aim to continue to broaden the target portfolio, elucidate generalizable means to optimize specific targeting – both in terms of tumor uptake and off-target retention – and translate agents into the clinic.
Highlights.
Molecular imaging is a valuable clinical tool for cancer detection and treatment.
Non-antibody protein scaffolds are an effective source of molecular imaging agents.
Affibodies, knottins, fibronectins, and DARPins have imaged many cancer biomarkers.
Ligand biophysical properties can be modulated to optimize in vivo performance.
Acknowledgments
This work was funded by the National Institutes of General Medical Science Biotechnology Training Grant (L.A.S.) and the University of Minnesota.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
- 1.Knowles SM, Wu AM. Advances in immuno-positron emission tomography: antibodies for molecular imaging in oncology. Journal of Clinical Oncology. 2012;30:3884–3892. doi: 10.1200/JCO.2012.42.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •2.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther. 2009;8:2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. A mechanistic mathematical model is developed and validated to predict the impact of molecular size and binding affinity on delivery to tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schottelius M, Wester HJ. Molecular imaging targeting peptide receptors. Methods. 2009;48:161–177. doi: 10.1016/j.ymeth.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Feldwisch J, Tolmachev V. Engineering of affibody molecules for therapy and diagnostics. Methods Mol Biol. 2012;899:103–126. doi: 10.1007/978-1-61779-921-1_7. [DOI] [PubMed] [Google Scholar]
- •5.Orlova A, Magnusson M, Eriksson TLJ, Nilsson M, Larsson B, Höidén-Guthenberg I, Widström C, Carlsson J, Tolmachev V, Ståhl S, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66:4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. An affibody, evolved to 22 pM affinity for HER2 and radiolabeled with 125I, exhibited excellent tumor targeting capabilities in a subcutaneously xenografted tumor model in mice as measured by excised tissue biodistribution and gamma scintigraphy. [DOI] [PubMed] [Google Scholar]
- 6.Orlova A, Tran T, Widström C, Engfeldt T, Eriksson Karlström A, Tolmachev V. Preclinical evaluation of [111In]-benzyl-DOTA-Z(HER2:342), a potential agent for imaging of HER2 expression in malignant tumors. Int J Mol Med. 2007;20:397–404. [PubMed] [Google Scholar]
- 7.Tran T, Engfeldt T, Orlova A, Sandstroem M, Feldwisch J, Abrahmsén L, Wennborg A, Tolmachev V, Karlstroem AE. Tc-99m-maEEE-Z(HER2: 342), an affibody molecule-based tracer for the detection of HER2 expression in malignant tumors. Bioconjug Chem. 2007;18:1956–1964. doi: 10.1021/bc7002617. [DOI] [PubMed] [Google Scholar]
- 8.Kramer-Marek G, Kiesewetter DO, Martiniova L, Jagoda E, Lee SB, Capala J. [18F]FBEM-ZHER2:342–Affibody molecule—a new molecular tracer for in vivo monitoring of HER2 expression by positron emission tomography. Eur J Nucl Med Mol Imaging. 2007;35:1008–1018. doi: 10.1007/s00259-007-0658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlova A, Rosik D, Sandstrom M, Lundqvist H, Einarsson L, Tolmachev V. Evaluation of [(111/114m)In]CHX-A“-”DTPA-ZHER2:342, an affibody ligand coniugate for targeting of HER2-expressing malignant tumors. Q J Nucl Med Mol Imaging. 2007;51:314–323. [PubMed] [Google Scholar]
- •10.Orlova A, Wallberg H, Stone-Elander S, Tolmachev V. On the Selection of a Tracer for PET Imaging of HER2-Expressing Tumors: Direct Comparison of a 124I-Labeled Affibody Molecule and Trastuzumab in a Murine Xenograft Model. J Nucl Med. 2009;50:417–425. doi: 10.2967/jnumed.108.057919. In a direct comparison, a HER2-targeted affibody provides far superior tumor selectivity relative to the antibody trastuzumab for PET imaging of subcutaneous tumor xenografts in mice. This highlights the rapid clearance benefits of small protein scaffolds. [DOI] [PubMed] [Google Scholar]
- 11.Kramer-Marek G, Shenoy N, Seidel J, Griffiths GL, Choyke P, Capala J. (68)Ga-DOTA-Affibody molecule for in vivo assessment of HER2/neu expression with PET. Eur J Nucl Med Mol Imaging. 2011;38:1967–1976. doi: 10.1007/s00259-011-1810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wållberg H, Grafström J, Cheng Q, Lu L, Martinsson Ahlzén HS, Samén E, Thorell JO, Johansson K, Dunås F, Olofsson MH, et al. HER2-positive tumors imaged within 1 hour using a site-specifically 11C-labeled Sel-tagged affibody molecule. J Nucl Med. 2012;53:1446–1453. doi: 10.2967/jnumed.111.102194. [DOI] [PubMed] [Google Scholar]
- •13.Baum RP, Prasad V, Müller D, Schuchardt C, Orlova A, Wennborg A, Tolmachev V, Feldwisch J. Molecular imaging of HER2-expressing malignant tumors in breast cancer patients using synthetic 111In- or 68Ga-labeled affibody molecules. J Nucl Med. 2010;51:892–897. doi: 10.2967/jnumed.109.073239. In the first published human use of an affibody, 111In and 68Ga labeling of a HER2-targeting ligand yield effective SPECT and PET imaging agents that are well tolerated and provide quality imaging. [DOI] [PubMed] [Google Scholar]
- 14.Tolmachev V, Friedman M, Sandström M, Eriksson TLJ, Rosik D, Hodik M, Ståhl S, Frejd FY, Orlova A. Affibody molecules for epidermal growth factor receptor targeting in vivo: aspects of dimerization and labeling chemistry. J Nucl Med. 2009;50:274–283. doi: 10.2967/jnumed.108.055525. [DOI] [PubMed] [Google Scholar]
- 15.Tolmachev V, Rosik D, Wållberg H, Sjöberg A, Sandström M, Hansson M, Wennborg A, Orlova A. Imaging of EGFR expression in murine xenografts using site-specifically labelled anti-EGFR 111In-DOTA-Z EGFR:2377 Affibody molecule: aspect of the injected tracer amount. Eur J Nucl Med Mol Imaging. 2010;37:613–622. doi: 10.1007/s00259-009-1283-x. [DOI] [PubMed] [Google Scholar]
- 16.Nordberg E, Orlova A, Friedman M, Tolmachev V, Ståhl S, Nilsson FY, Glimelius B, Carlsson J. In vivo and in vitro uptake of 111In, delivered with the affibody molecule (ZEGFR:955)2, in EGFR expressing tumour cells. Oncol Rep. 2008;19:853–857. doi: 10.3892/or.19.4.853. [DOI] [PubMed] [Google Scholar]
- 17.Miao Z, Ren G, HHongguang L, Jiang L, Cheng Z. Small-animal PET imaging of human epidermal growth factor receptor positive tumor with a 64Cu labeled affibody protein. Bioconjug Chem. 2010;21:947–954. doi: 10.1021/bc900515p. [DOI] [PubMed] [Google Scholar]
- 18.Miao Z, Ren G, Liu H, Qi S, Wu S, Cheng Z. PET of EGFR Expression with an 18F- Labeled Affibody Molecule. Journal of Nuclear Medicine. 2012;53:1110–1118. doi: 10.2967/jnumed.111.100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolmachev V, Malmberg J, Hofström C, Abrahmsén L, Bergman T, Sjöberg A, Sandström M, Gräslund T, Orlova A. Imaging of insulinlike growth factor type 1 receptor in prostate cancer xenografts using the affibody molecule 111In-DOTA-ZIGF1R:4551. J Nucl Med. 2012;53:90–97. doi: 10.2967/jnumed.111.090829. [DOI] [PubMed] [Google Scholar]
- 20.Orlova A, Hofström C, Strand J, Varasteh Z, Sandström M, Andersson K, Tolmachev V, Gräslund T. [99mTc(CO)3]+-(HE)3-ZIGF1R:4551, a new Affibody conjugate for visualization of insulin-like growth factor-1 receptor expression in malignant tumours. Eur J Nucl Med Mol Imaging. 2013;40:439–449. doi: 10.1007/s00259-012-2284-8. [DOI] [PubMed] [Google Scholar]
- 21.Honarvar H, Jokilaakso N, Andersson K, Malmberg J, Rosik D, Orlova A, Karlström AE, Tolmachev V, Järver P. Evaluation of backbone-cyclized HER2-binding 2-helix Affibody molecule for In Vivo molecular imaging. Nucl Med Biol. 2013;40:378–386. doi: 10.1016/j.nucmedbio.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Webster JM, Zhang R, Gambhir SS, Cheng Z, Syud FA. Engineered two-helix small proteins for molecular recognition. Chembiochem. 2009;10:1293–1296. doi: 10.1002/cbic.200900062. [DOI] [PubMed] [Google Scholar]
- 23.Ren G, Zhang R, Liu Z, Webster JM, Miao Z, Gambhir SS, Syud FA, Cheng Z. A 2-helix small protein labeled with 68Ga for PET imaging of HER2 expression. J Nucl Med. 2009;50:1492–1499. doi: 10.2967/jnumed.109.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Z, Ren G, Jiang L, HHongguang L, Webster JM, Zhang R, Namavari M, Gambhir SS, Syud F, Cheng Z. A novel 18F-labeled two-helix scaffold protein for PET imaging of HER2-positive tumor. Eur J Nucl Med Mol Imaging. 2011;38:1977–1984. doi: 10.1007/s00259-011-1879-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Rosik D, Orlova A, Malmberg J, Altai M, Varasteh Z, Sandström M, Karlström AE, Tolmachev V. Direct comparison of 111In-labelled two-helix and three-helix Affibody molecules for in vivo molecular imaging. Eur J Nucl Med Mol Imaging. 2011;39:693–702. doi: 10.1007/s00259-011-2016-5. A two-helix affibody derivative yields 2.7-fold higher tumor-to-blood and 1.8-fold higher tumor-to-muscle ratios relative to its three-helix parental molecule despite 27-fold lower affinity. [DOI] [PubMed] [Google Scholar]
- 26.Moore SJ, Cochran JR. Engineering knottins as novel binding agents. Meth Enzymol. 2012;503:223–251. doi: 10.1016/B978-0-12-396962-0.00009-4. [DOI] [PubMed] [Google Scholar]
- 27.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res. 2009;69:2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Z, Ren G, HHongguang L, Kimura RH, Jiang L, Cochran JR, Gambhir SS, Cheng Z. An engineered knottin peptide labeled with 18F for PET imaging of integrin expression. Bioconjug Chem. 2009;20:2342–2347. doi: 10.1021/bc900361g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Nielsen CH, Kimura RH, Withofs N, Tran PT, Miao Z, Cochran JR, Cheng Z, Felsher D, Kjær A, Willmann JK, et al. PET imaging of tumor neovascularization in a transgenic mouse model with a novel 64Cu-DOTA-knottin peptide. Cancer Res. 2010;70:9022–9030. doi: 10.1158/0008-5472.CAN-10-1338. An engineered knottin exhibits superior tumor selectivity relative to 18F-fluorodeoxyglucose for imaging nascent lung tumors in a transgenic mouse model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura RH, Miao Z, Cheng Z, Gambhir SS, Cochran JR. A Dual-Labeled Knottin Peptide for PET and Near-Infrared Fluorescence Imaging of Integrin Expression in Living Subjects. Bioconjug Chem. 2010;21:436–444. doi: 10.1021/bc9003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, Kimura RH, Miao Z, Silverman AP, Ren G, HHongguang L, Li P, Gambhir SS, Cochran JR, Cheng Z. Evaluation of a (64)Cu-labeled cystine-knot peptide based on agouti-related protein for PET of tumors expressing alphavbeta3 integrin. J Nucl Med. 2010;51:251–258. doi: 10.2967/jnumed.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang L, Miao Z, Kimura RH, Silverman AP, Ren G, HHongguang L, Lu H, Cochran JR, Cheng Z. 111In-labeled cystine-knot peptides based on the Agouti-related protein for targeting tumor angiogenesis. [Internet] Journal of Biomedicine and Biotechnology. 2012;2012:368075. doi: 10.1155/2012/368075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H, Moore SJ, Liu S, HHongguang L, Miao Z, Cochran FV, Liu Y, Tian M, Cochran JR, Zhang H, et al. A novel radiofluorinated agouti-related protein for tumor angiogenesis imaging. Amino Acids. 2013;44:673–681. doi: 10.1007/s00726-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 34.Kimura RH, Teed R, Hackel BJ, Pysz MA, Chuang CZ, Sathirachinda A, Willmann JK, Gambhir SS. Pharmacokinetically Stabilized Cystine Knot Peptides That Bind Alpha-v-Beta-6 Integrin with Single-Digit Nanomolar Affinities for Detection of Pancreatic Cancer. Clin Cancer Res. 2012;18:839–849. doi: 10.1158/1078-0432.CCR-11-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hackel BJ, Kimura RH, Miao Z, Liu H, Sathirachinda A, Cheng Z, Chin FT, Gambhir SS. 18F-Labeled Cystine Knot Peptides for PET Imaging of Integrin αvβ6. J Nucl Med. doi: 10.2967/jnumed.112.110759. [no date], Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore SJ, Leung CL, Norton HK, Cochran JR. Engineering agatoxin, a cystine-knot Peptide from spider venom, as a molecular probe for in vivo tumor imaging. PLoS ONE. 2013;8:e60498. doi: 10.1371/journal.pone.0060498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipovsek D. Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel. 2011;24:3–9. doi: 10.1093/protein/gzq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen TF, de Picciotto S, Hackel BJ, Wittrup KD. Engineering fibronectin-based binding proteins by yeast surface display. Meth Enzymol. 2013;523:303–326. doi: 10.1016/B978-0-12-394292-0.00014-X. [DOI] [PubMed] [Google Scholar]
- 39.Hackel BJ, Kimura RH, Gambhir SS. Use of 64Cu-labeled Fibronectin Domain with EGFR-Overexpressing Tumor Xenograft: Molecular Imaging. Radiology. 2012;263:179–188. doi: 10.1148/radiol.12111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •40.Hackel BJ, Sathirachinda A, Gambhir SS. Designed hydrophilic and charge mutations of the fibronectin domain: towards tailored protein biodistribution. Protein Eng Des Sel. 2012;25:639–647. doi: 10.1093/protein/gzs036. The fibronectin domain was further substantiated as an effective scaffold for molecular PET imaging. The ability to engineer hydrophilicity and charge to modulate off-target distribution was shown. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaskovic R, Simon M, Stefan N, Schwill M, Plückthun A. Designed ankyrin repeat proteins (DARPins) from research to therapy. Meth Enzymol. 2012;503:101–134. doi: 10.1016/B978-0-12-396962-0.00005-7. [DOI] [PubMed] [Google Scholar]
- •42.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, Dreier B, Schibli R, Binz HK, Waibel R, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer Res. 2010;70:1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. DARPins are validated for in vivo molecular imaging in a mouse tumor xenograft model. Benefits of small size and high affinity are demonstrated using DARPins with and without PEGylation and over an affinity range of 90 pM to 270 nM. [DOI] [PubMed] [Google Scholar]
- 43.Martin-Killias P, Patricia MK, Stefan N, Rothschild S, Plückthun A, Zangemeister-Wittke U. A novel fusion toxin derived from an EpCAM-specific designed ankyrin repeat protein has potent antitumor activity. Clin Cancer Res. 2011;17:100–110. doi: 10.1158/1078-0432.CCR-10-1303. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Peck-Radosavljevic M, Kienast O, Preitfellner J, Havlik E, Schima W, Traub-Weidinger T, Graf S, Beheshti M, Schmid M, et al. Iodine-123-vascular endothelial growth factor-165 (123I-VEGF165). Biodistribution, safety and radiation dosimetry in patients with pancreatic carcinoma. Q J Nucl Med Mol Imaging. 2004;48:198–206. [PubMed] [Google Scholar]
- 45.Chen K, Cai W, Li ZB, Wang H, Chen X. Quantitative PET Imaging of VEGF Receptor Expression. Mol Imaging Biol. 2008;11:15–22. doi: 10.1007/s11307-008-0172-1. [DOI] [PubMed] [Google Scholar]
- 46.Kang CM, Kim SM, Koo HJ, Yim MS, Lee KH, Ryu EK, Choe YS. In vivo characterization of 68Ga-NOTA-VEGF121 for the imaging of VEGF receptor expression in U87MG tumor xenograft models. Eur J Nucl Med Mol Imaging. 2012;40:198–206. doi: 10.1007/s00259-012-2266-x. [DOI] [PubMed] [Google Scholar]
- 47.Backer MV, Levashova Z, Patel V, Jehning BT, Claffey K, Blankenberg FG, Backer JM. Molecular imaging of VEGF receptors in angiogenic vasculature with single-chain VEGF-based probes. Nat Med. 2007;13:504–509. doi: 10.1038/nm1522. [DOI] [PubMed] [Google Scholar]
- 48.Eder M, Krivoshein AV, Backer M, Backer JM, Haberkorn U, Eisenhut M. ScVEGF-PEG-HBED-CC and scVEGF-PEG-NOTA conjugates: comparison of easy-to-label recombinant proteins for [68Ga]PET imaging of VEGF receptors in angiogenic vasculature. Nucl Med Biol. 2010;37:405–412. doi: 10.1016/j.nucmedbio.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Velikyan I, Sundberg AL, Lindhe O, Höglund AU, Eriksson O, Werner E, Carlsson J, Bergström M, Långström B, Tolmachev V. Preparation and evaluation of (68)Ga-DOTA-hEGF for visualization of EGFR expression in malignant tumors. Journal of Nuclear Medicine. 2005;46:1881–1888. [PubMed] [Google Scholar]
- 50.Li W, Niu G, Lang L, Guo N, Ma Y, Kiesewetter DO, Backer JM, Shen B, Chen X. PET imaging of EGF receptors using [18F]FBEM-EGF in a head and neck squamous cell carcinoma model. Eur J Nucl Med Mol Imaging. 2011;39:300–308. doi: 10.1007/s00259-011-1969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo MF, Zhao Y, Tian R, Li L, Guo L, Xu F, Liu YM, He YB, Bai S, Wang J. In vivo99mTc-HYNIC-annexin V imaging of early tumor apoptosis in mice after single dose irradiation. J Exp Clin Cancer Res. 2009;28:136. doi: 10.1186/1756-9966-28-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu S, Kiesewetter DO, Zhu L, Guo N, Gao H, Liu G, Hida N, Lang L, Niu G, Chen X. Longitudinal PET Imaging of Doxorubicin-Induced Cell Death with 18F-Annexin V. Mol Imaging Biol. 2012;14:762–770. doi: 10.1007/s11307-012-0551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rottey S, Slegers G, Van Belle S, Goethals I, Van de Wiele C. Sequential 99mTc-hydrazinonicotinamide-annexin V imaging for predicting response to chemotherapy. Journal of Nuclear Medicine. 2006;47:1813–1818. [PubMed] [Google Scholar]
- 54.Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP, Jain RK. Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size. Cancer Res. 1995;55:3752–3756. [PubMed] [Google Scholar]
- 55.Cheng Z, De Jesus OP, Namavari M, De A, Levi J, Webster JM, Zhang R, Lee B, Syud FA, Gambhir SS. Small-Animal PET Imaging of Human Epidermal Growth Factor Receptor Type 2 Expression with Site-Specific 18F-Labeled Protein Scaffold Molecules. J Nucl Med. 2008;49:804–813. doi: 10.2967/jnumed.107.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolmachev V, Mume E, Sjöberg S, Frejd FY, Orlova A. Influence of valency and labelling chemistry on in vivo targeting using radioiodinated HER2-binding Affibody molecules. Eur J Nucl Med Mol Imaging. 2009;36:692–701. doi: 10.1007/s00259-008-1003-y. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Z, Jesus OP, Kramer DJ, De A, Webster JM, Gheysens O, Levi J, Namavari M, Wang S, Park JM, et al. 64Cu-Labeled Affibody Molecules for Imaging of HER2 Expressing Tumors. Mol Imaging Biol. 2009;12:316–324. doi: 10.1007/s11307-009-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolmachev V, Tran TA, Rosik D, Sjöberg A, Abrahmsén L, Orlova A. Tumor targeting using affibody molecules: interplay of affinity, target expression level, and binding site composition. J Nucl Med. 2012;53:953–960. doi: 10.2967/jnumed.111.101527. [DOI] [PubMed] [Google Scholar]
- 59.Tran TA, Ekblad T, Orlova A, Sandström M, Feldwisch J, Wennborg A, Abrahmsén L, Tolmachev V, Eriksson Karlström A. Effects of lysine-containing mercaptoacetyl-based chelators on the biodistribution of 99mTc-labeled anti-HER2 Affibody molecules. Bioconjug Chem. 2008;19:2568–2576. doi: 10.1021/bc800244b. [DOI] [PubMed] [Google Scholar]
- 60.Malmberg J, Perols A, Varasteh Z, Altai M, Braun A, Sandström M, Garske U, Tolmachev V, Orlova A, Karlström AE. Comparative evaluation of synthetic anti-HER2 Affibody molecules site-specifically labelled with 111In using N-terminal DOTA, NOTA and NODAGA chelators in mice bearing prostate cancer xenografts. Eur J Nucl Med Mol Imaging. 2012;39:481–492. doi: 10.1007/s00259-011-1992-9. [DOI] [PubMed] [Google Scholar]
- 61.Ekblad T, Tran T, Orlova A, Widström C, Feldwisch J, Abrahmsén L, Wennborg A, Karlström AE, Tolmachev V. Development and preclinical characterisation of 99mTc-labelled Affibody molecules with reduced renal uptake. Eur J Nucl Med Mol Imaging. 2008;35:2245–2255. doi: 10.1007/s00259-008-0845-7. [DOI] [PubMed] [Google Scholar]
- 62.Engfeldt T, Tran T, Orlova A, Widström C, Feldwisch J, Abrahmsén L, Wennborg A, Karlström AE, Tolmachev V. 99mTc-chelator engineering to improve tumour targeting properties of a HER2-specific Affibody molecule. Eur J Nucl Med Mol Imaging. 2007;34:1843–1853. doi: 10.1007/s00259-007-0474-6. [DOI] [PubMed] [Google Scholar]
- 63.Perols A, Honarvar H, Strand J, Selvaraju R, Orlova A, Karlström AE, Tolmachev V. Influence of DOTA chelator position on biodistribution and targeting properties of (111)In-labeled synthetic anti-HER2 affibody molecules. Bioconjug Chem. 2012;23:1661–1670. doi: 10.1021/bc3002369. [DOI] [PubMed] [Google Scholar]


