Abstract
Peptide hormones and growth factors bind to membrane receptors and regulate a myriad of processes in insects and other metazoans. The evolutionary relationships among characterized and uncharacterized (“orphan”) receptors can provide insights into receptor-ligand biology and narrow target choices in deorphanization studies. However, the large number and low sequence conservation of these receptors make evolutionary analysis difficult. Here, we characterized the G-protein-coupled receptors (GPCRs), receptor guanylyl cyclases (RGCs), and protein kinase receptors (PKRs) of mosquitoes and select other flies by interrogating the genomes of Aedes aegypti, Anopheles gambiae, Culex quinquefasciatus, Drosophila melanogaster, and D. mojavensis. Sequences were grouped by receptor type, clustered using the program CLANS, aligned using HMMR, and phylogenetic trees built using PhyML. Our results indicated that PKRs had relatively few orphan clades whereas GPCRs and RGCs had several. In addition, more than half of the Class B secretin-like GPCRs and RGCs remained uncharacterized. Additional studies revealed that Class B GPCRs exhibited more gain and loss events than other receptor types. Finally, using the neuropeptide F family of insect receptors and the neuropeptide Y family of vertebrate receptors, we also show that functional sites considered critical for ligand binding are conserved among distinct family members and between distantly related taxa. Overall, our results provide the first comprehensive analysis of peptide hormone and growth factor receptors for a major insect group.
Keywords: GPCR, receptor tyrosine kinase, guanylyl cyclase, serine/threonine kinase, neuropeptide, protein hormone
Introduction
Neuropeptides, protein hormones, and growth factors regulate many processes by binding to three types of membrane receptors: G-protein-coupled receptors (GPCRs), receptor guanylyl cyclases (RGCs), and protein kinase receptors (PKRs). We use the term peptide hormones to encompass both larger protein hormones released into the hemolymph by glands, neuroendocrine cells, and midgut endocrine cells, and neuropeptides that function as neurotransmitters and modulators within the nervous system. The greatest number and diversity of peptide hormones are bound by GPCRs, whereas known growth factors interact almost exclusively with PKRs. All of these receptor proteins are post-translationally modified and transported to the cell membrane. They encode an extracellular amino (N)-terminal region, one or more transmembrane spans, and an intracellular carboxyl (C)-terminal region that transduces ligand binding through specific signaling and amplification pathways. However, most GPCRs that bind peptide hormones reside as monomers in the cell membrane, whereas most RGCs and PKRs form dimers prior to or during ligand binding (1–3).
The genomes of several insects and related arthropods (Aedes aegypti, Anopheles gambiae, Apis mellifera, Bombyx mori, Drosophila melanogaster, Nasonia vitripennis, Tribolium castaneum, Tetranychus urticae, and Daphnia pulex) have been sequenced and their peptide hormone genes analyzed (4–15). GPCRs known or predicted to bind specific peptide hormones have also been annotated for D. melanogaster (7, 16), Ap. mellifera (17), T. castaneum (18), B. mori (19, 20), and T. urticae (13). These studies provide important insights about peptide hormone and GPCR diversity among arthropods while also shedding light on the evolutionary history and function of certain genes. These analyses also indicate that several peptide hormones and GPCRs remain “orphans” (6, 9, 16, 18, 21, 22). In some cases, one-to-one orthologs of known ligand-receptor pairs in the genomes of related species have led to predicted pairing to an uncharacterized receptor (23). For others though, ligand-receptor relationships remain unclear because gene duplication or loss events either create uncertainties about the functional homology of in- and out-paralogs, or have resulted in clades that contain no characterized orthologs (7, 21, 24, 25). We also note that no lists have been published for RGCs or PKRs and their associated ligands in any insect.
Our interests primarily focus on mosquitoes (Diptera: Culicidae), which are critically important insects because of their ability to vector several disease-causing pathogens to humans and other mammals. Peptide hormones and growth factors are key regulators of many physiological processes in mosquitoes that affect disease transmission. Three vector species of mosquitoes have been sequenced: An. gambiae (26), Ae. aegypti (27), and Culex quinquefasciatus (28). Annotation of the An. gambiae and Ae. aegypti genomes identify 35 and 43 peptide hormone genes respectively that are expressed in the nervous system, isolated glands, or midgut endocrine cells (9, 10). The function of several of these hormones is known (Table S1 in Supplementary Material). Homology-based searches were previously used to characterize the GPCR superfamily in An. gambiae (24) and analysis of the Ae. aegypti genome also identified some predicted peptide hormone GPCRs (27). No recent collation of GPCRs or other receptor types, however, is available.
Here, we analyzed the GPCRs, PKRs, and RGCs that bind peptide hormones and growth factors in mosquitoes and select other Diptera to discern phylogenetic patterns of receptor evolution. Our results provide a number of new insights including the identification of several orphan receptors. Ligand binding studies are the only means to deorphanize a given receptor definitively. However, our results could greatly assist deorphanization studies because they identify evolutionary relationships between receptors and thus narrow the spectrum of candidate ligands that a given orphan most likely binds (29, 30).
Materials and Methods
Identification of peptide hormone receptors
Peptide hormone receptors were identified from previously published surveys of D. melanogaster (7), An. gambiae (24), and Ae. aegypti (27). These sequences were downloaded from NCBI and used to plumb the genomes of D. mojavensis and C. quinquefasciatus using BLASTp. Each set of peptide hormone receptors from an organism was searched against the other genomes. The program HMMscan (31) was used to independently verify the completeness of our gene sets. Each genome was scanned using the Pfam models for rhodopsin-like GPCRs (“7tm_1,” PF00001), secretin-like GPCRs (“7tm_2,” PF00002), and protein kinases (“Pkinase,” PF00069) for RGCs and PKRs. These protein sequences were also used to search OrthoDB (32) for orthologs that may have been missed by our homology-based searches. The lists of receptors identified by Pfam searches were compared to the lists generated by BLAST searches. Genes were retained for further analysis if they were identified in Pfam searches or had a BLAST hit that had >50% amino acid identity with a known GPCR, RGC, or PKR in one of the examined genomes. In some instances, multiple annotated genes were found to encode parts of a single receptor. We used publicly available RNAseq data where available to join separate genes. In other cases, multiple predicted genes aligned to successive regions of orthologs in genomes with superior annotations (D. melanogaster, An. gambiae), and this was taken as evidence that the genes had been split improperly during gene prediction. The split genes were concatenated and used for downstream analysis. The complete list of receptors and accessions for each species used in our study is presented in Table S1 in Supplementary Material.
Clustering of GPCR sequences
Attwood and Findlay (33) previously categorized GPCRs into six Classes (A–F) on the basis of shared sequence motifs and ligand binding affinities. Arthropod GPCRs reside in four of these classes: rhodopsin-like (Class A), secretin-like (Class B), metabotropic glutamate receptors (Class C), and frizzled/smoothened (Class F) (7, 13, 21, 34). Only Classes A and B contain receptors whose known or predicted ligands are peptide hormones. Our analysis likewise divided peptide hormone binding GPCRs into Class A rhodopsin-like GPCRs and Class B secretin-like GPCRs. We further analyzed the rhodopsin-like group using the CLANs program (35), which uses BLAST-search based similarity to create self-organizing maps (SOM). CLANs was run for 2000 rounds with an e-value cutoff of 10−5. Clusters were determined by bootstrap-based clustering with a minimum of 4 sequences per cluster and 500 rounds.
Alignment and phylogenetic tree construction
For GPCRs, protein sequence alignments were initially attempted using MAFFT (36) and MUSCLE (37). Multiple parameters were tested but failed to produce acceptable alignments. Due to an improved ability to align divergent sequences with conserved domains, the program HMMalign (31) was used to align sequences using the Pfams mentioned previously. The highly variable N- and C-terminal regions of the proteins were removed by implementing the – “trim” option and poorly aligned regions were manually removed using Jalview (38). Alignments are included in the Files S1–S5 in Supplementary Material.
Phylogenetic trees were constructed from alignments using PhyML (39) on the LIRMM server. Parameters were as follows: substitution model was LG, proportion of invariable sites was 0.0, four substitution categories were used, initial trees were constructed using BIONJ, tree improvement was done through NNI, and trees were optimized for topology and branch length. Likelihood scores were computed using aBayes. Trees were visualized in FigTree v1.3.1. For the GPCR orphans, we arbitrarily numbered each clade of orphans and included the letter, A or B, to indicate which GPCR class they belonged to. RGC and PKR orphan clades are indicated as OR# and OGC#, respectively.
Conservation of ligand binding domains
Deduction of receptor function from phylogenetic analysis is contingent upon conservation of the ligand binding region of the receptor and essential residues of the hormone. To examine whether functionally important regions of related receptors and ligands were conserved in divergent organisms, we examined neuropeptide F (NPF) in insects and its homologs in vertebrates, neuropeptide Y (NPY), pancreatic polypeptide (PP), and peptide YY (PYY). We aligned the NPF GPCRs from the five dipteran genomes with the NPY, PP, and PYY GPCRs from Homo sapiens, Mus musculus, and Danio rerio as described for the other GPCRs and visualized in Jalview (38). Previous targeted mutagenesis studies distinguished residues important for ligand binding (40–43) that were then identified and highlighted in the GPCR sequences.
Results
Database mining and phylogenetic analysis of dipteran receptors
Prior studies provide strong support for the monophyly of the Diptera while also showing that mosquitoes (Culicomorpha: Culicidae) are an early lineage that evolved ca. 225 Mya and drosophilids like D. melanogaster are a derived lineage (Ephydroidea: Drosophilidae) that emerged concurrently with other cyclorrhaphan flies 40–65 Mya (44). Prior studies also support the monophyly of the Culicidae, which consists of two subfamilies, the Anophelinae (as represented by An. gambiae) and the Culicinae (as represented by Ae. aegypti and C. quinquefasciatus) that diverged ca. 145–200 Mya (45, 46). We thus organized our study of peptide hormone and growth factor receptors to span both subfamilies of the Culicidae plus the phylogenetic breadth of the Diptera by including data from D. melanogaster and one other drosophilid (D. mojavensis). For each receptor group, we first summarize key features of our analysis.
We then address the receptors in specific clades and orphans of interest and the gene identification for the receptors in the five species. The known functions for peptide hormones in mosquitoes are summarized in Table S1 in Supplementary Material, along with the relevant references whereas the review by Nässel and Winther (58) provides an in depth summary of the function and signaling of peptide hormones in D. melanogaster. In addition, selected information and references for growth factors and their receptors and the RGCs characterized in D. melanogaster are provided in Table S1 in Supplementary Material. VectorBase or FlyBase accessions are also provided for all genes used in the study in Table S1 in Supplementary Material.
Peptide hormone GPCRs
We focused on the GPCRs first because their interactions with peptide hormones have been more extensively examined in insects than those of RGCs and PKRs. GPCRs are 40–60 kDa proteins, which contain an extracellular N-terminal region, seven transmembrane α-helices that form a ligand binding pocket, and an intracellular C-terminal region that mediates signaling through interactions with G-proteins. G-proteins consist of different subunits that interact with other proteins to produce a variety of intracellular signaling molecules including cAMP, cGMP, and calcium.
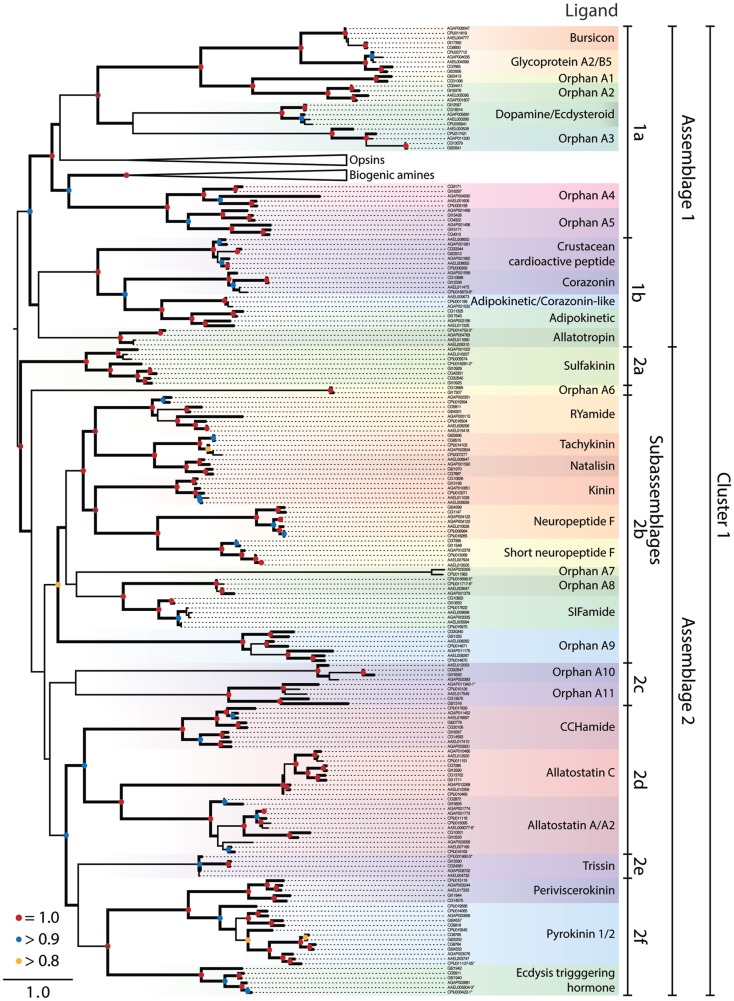
Most phylogenies generated previously for insect GPCRs utilized standard alignment algorithms (ClustalW or MegAlign) followed by neighbor-joining methods. Only rarely though have branch support values or other measures of robustness been reported (16). We initially used the full-length predicted amino acid sequence for each GPCR in our data set for tree building using the advanced alignment algorithm MAFFT. However, as found for vertebrates and other arthropods (17, 21, 30, 47), these approaches failed to produce suitable alignments due to extensive divergence in the N- and C-termini flanking the transmembrane domains of the predicted proteins (data not shown). We therefore used only the seven transmembrane α-helices for each GPCR followed by alignment using the domain-based algorithm HMMalign (31). This approach yielded suitable alignments for tree building. We sought to combine the previously divided GPCR subgroups as much as possible while retaining reasonable support at deep nodes within the trees. To achieve this, we used CLANS (35), which produced two distinct clusters (1 and 2) of Class A rhodopsin-like GPCRs with 352 and 41 proteins respectively (Table 1; Figures 1 and 2). Our analysis further indicated the Class B secretin-like GPCRs contained 82 proteins (Table 1; Figure 3).
Table 1.
Distribution of peptide hormone and growth factor receptors by type, species and characterization.
| Class A GPCR Cluster 1 | Class A GPCR Cluster 2 | All Class A GPCR | Class B GPCR | PKR | RGC | |
|---|---|---|---|---|---|---|
| Total genes | 204 | 41 | 245 | 82 | 93 | 31 |
| Clades | 34 | 7 | 41 | 11 | 17 | 6 |
| Characterized clades | 23 (68%) | 4 (57%) | 27 (66%) | 3 (27%) | 15 (88%) | 3 (50%) |
| Orphan clades | 11 (32%) | 3 (43%) | 14 (34%) | 8 (72%) | 2 (12%) | 3 (50%) |
| “In group” orphans | 33 (13%) | 1 (2.4%) | 27 (11%) | 0 (0%) | 9 (10%) | 0 (0%) |
| Ae. aegypti genes | 39 | 9 | 48 | 13 | 22 | 7 |
| An. gambiae genes | 41 | 6 | 47 | 12 | 16 | 6 |
| C. quinquefasciatus genes | 40 | 9 | 49 | 16 | 18 | 6 |
| D. melanogaster genes | 37 | 8 | 45 | 24 | 16 | 7 |
| D. mojavensis genes | 38 | 8 | 46 | 17 | 17 | 5 |
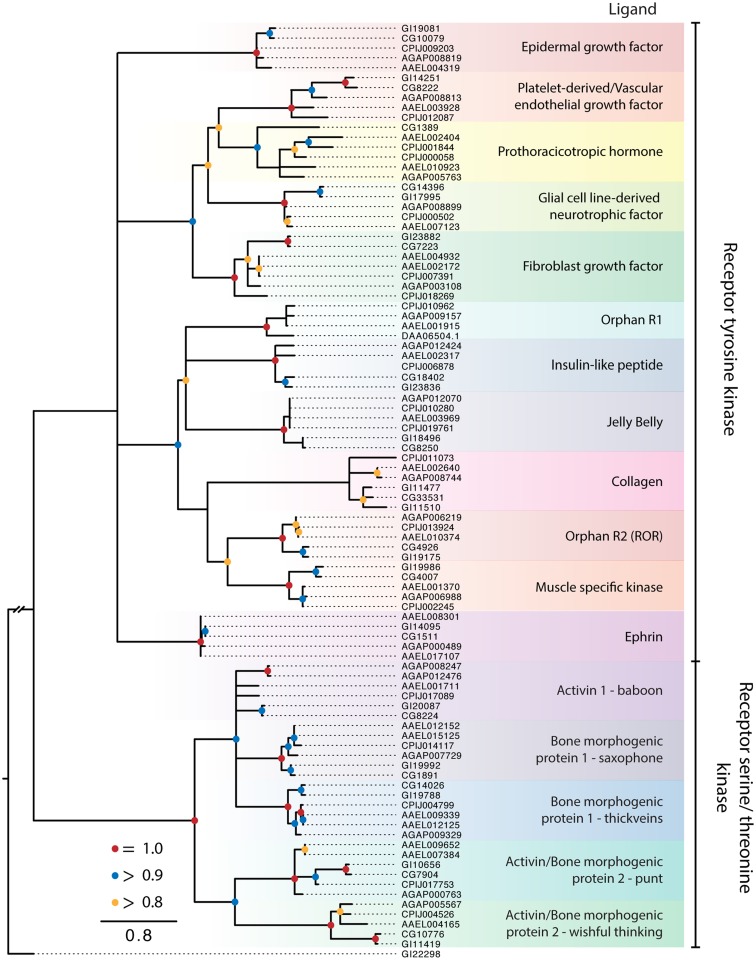
Figure 1.
Maximum-likelihood phylogeny of peptide hormone, Class A rhodopsin-like GPCRs in Cluster 1. The phylogeny presented is based on the seven transmembrane repeat region of proteins from Ae. aegypti, An. gambiae, C. quinquefasciatus, D. melanogaster, and D. mojavensis. Entries are identified by their VectorBase or FlyBase ID and accession number prefixes that indicate species: CG#, D. melanogaster; GI#, D. mojavensis; AAEL#, Ae. aegypti; AGAP#, An. gambiae; CPIJ#, C. quinquefasciatus. Assemblages and sub-assemblages are indicated to the right of the phylogeny. Each monophyletic clade of characterized or orphan GPCRs is highlighted with a different color and named after characterized ligands, where known. Clades containing the opsin and biogenic amine GPCRs have been collapsed. Nodes with likelihood scores >0.8 are denoted by thickened lines. Dots at nodes indicate level of support: 0.8–0.9 (yellow), 0.9–0.99 (blue), 1 (red); instances of split gene annotations are denoted by the inclusive range of accession numbers and marked with an asterisk. Isoforms of proteins were initially included in the tree but were removed if the two forms had a branch length of 0, and the tree was midpoint rooted.
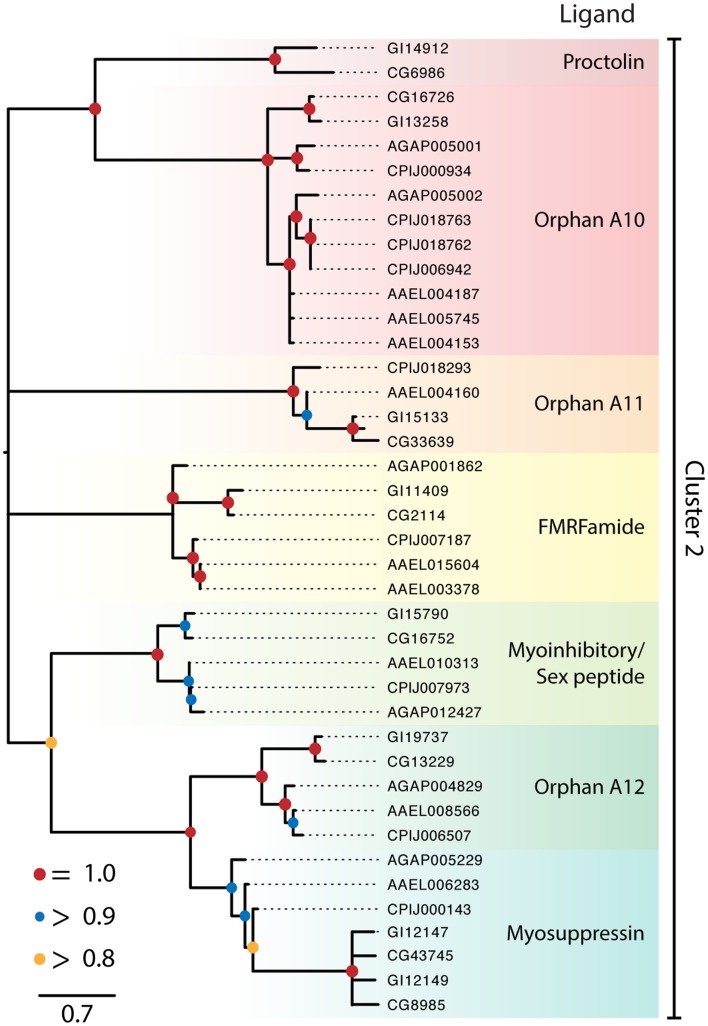
Figure 2.
Maximum-likelihood phylogeny of peptide hormone, Class A rhodopsin-like GPCRs in Cluster 2. Phylogenies were generated and named as described in Figure 1, except that nodes with likelihood scores <0.8 have been collapsed to polytomies. Isoforms of proteins were initially included in the tree but were removed if the two forms had a branch length of 0 and the tree was midpoint rooted.
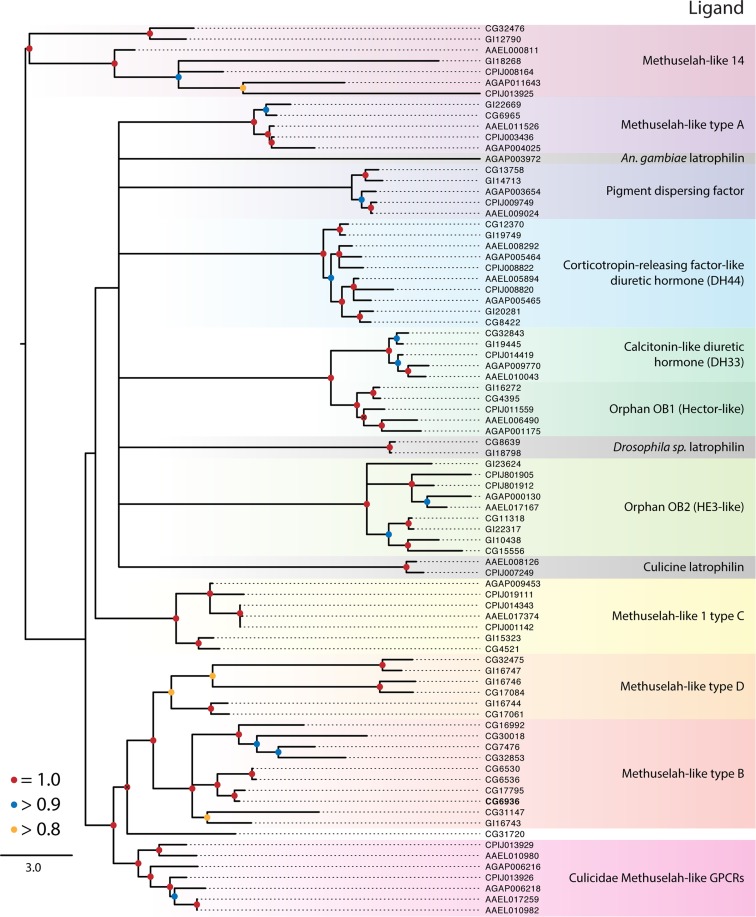
Figure 3.
Maximum-likelihood phylogeny of the Class B secretin GPCRs. Phylogenies were generated and named as described in Figure 2. The latrophilin receptors (gray shading) contain only a single transmembrane repeat, resulting in a poor alignment and phylogenetic placement (see text). Using full-length latrophilin sequences resurrects the monophyly of this group of unusual GPCRs (data not shown). Methuselah clades were named according to Li et al. (126). The D. melanogaster Methuselah receptor that binds Stunted is highlighted in bold. The tree was rooted at the midpoint.
Class a rhodopsin-like GPCRs
Cluster 1 for the Class A rhodopsin-like GPCRs contained peptide hormone receptors, opsins, and biogenic amine GPCRs that our analysis divided into two monophyletic clades we term assemblages 1 and 2 (Figure 1). We further divided the GPCR assemblages into sub-assemblages of nested, well-supported monophyletic clades, denoted by a letter after the assemblage they belong to. Assemblage 1 consisted of two deeply divergent sub-assemblages, 1a and b (Figure 1). Sub-assemblage 1a contained the bursicon, glycoprotein A2/B5, and dopamine/ecdysteroid receptors, three interspersed orphan clades, OA13, as well as two deeply diverging orphan clades named OA4–5 (Figure 1). Sub-assemblage 1a also contained the opsins and biogenic amine GPCRs. Sub-assemblage 1b contained the receptors for crustacean cardioactive peptide (CCAP), corazonin, adipokinetic/corazonin-like peptide (ACP), adipokinetic hormone (AKH), and allatotropin. Assemblage 2 consisted of 25 peptide hormone receptors and 5 orphan clades, which formed the following monophyletic sub-assemblages: (2a) sulfakinin; (2b) RYamide, tachykinin, natalisin, kinin, NPF, and short NPF (sNPF), and SIFamide; (2c) orphan clades A10–11; (2d) CCHamide, allatostatin A and C; (2e) trissin; (2f) periviscerokinin (PVK); pyrokinin (PK); and ecdysis-triggering hormone (ETH) (Figure 1).
Cluster 2 contained seven monophyletic clades of GPCRs that included receptors for proctolin, FMRFamide, myoinhibitory peptide (MIP)/sex peptide, and myosuppressin plus three orphan clades (Figure 2). Several of the Cluster 2 clades also contained orphan receptors. Some but not all of the clades generated by our analysis supported previously recognized lineages (16, 18). Key details for the two clusters and sub-assemblages are summarized below.
Cluster 1, assemblage 1
Sub-assemblage 1a: bursicon, GPA2/GPB5, and ecdysteroid receptors
Our analysis suggests the peptide hormone receptors of sub-assemblage 1b arose from a more recent ancestor of the biogenic amine/steroid hormone GPCRs than other peptide binding GPCRs. We thus refer to these as derived peptide GPCRs. Although we used only the transmembrane region in generating our GPCR phylogenies, all of the derived peptide GPCRs have an extended extracellular N-terminus in common with the leucine-rich repeat-containing G-protein-coupled receptors (LGRs) of which three types are conserved across vertebrates and invertebrates (48). With small exception, the vertebrate LGRs bind the cysteine-rich heterodimer glycoprotein hormones (i.e., GPA/GPB), which regulate gonadal and thyroid activity. This binding affinity is conserved in the dipteran receptors for GPA2/GPB5 (49). Another distinguishing feature of the derived peptide GPCRs is that their ligands are much larger than those of other GPCRs: the largest known ligand for other Class A GPCRs is only 36 amino acids (NPF), whereas the bursicon and GPA/GPB receptors bind ligands that exceed 100 amino acids and form 30 kDa dimers.
Bursicon activates cuticle tanning and hardening shortly after molting by insects (50, 51). Single orthologs of the bursicon receptor (LGR type B) are present in each mosquito and Drosophila species (Figure 1). Identification of a GPB ortholog in D. melanogaster led to its partner, GPA, and the deorphanization of the GPA2/GPB5 receptor [LGR type A, (49)]. Single orthologs of this receptor are also present in D. mojavensis and each mosquito species (Figure 1).
Sub-assemblage 1b also contains a GPCR that binds ecdysteroid hormones, which are major regulators of molting, egg development, and other processes. Most steroid hormones bind a nuclear receptor and potentiate changes in transcription, but some also bind to extracellular receptors including GPCRs (52). Srivastava and colleagues showed that CG18314 from D. melanogaster binds dopamine and ecdysteroids, including ecdysone and 20-hydroxyecdysone (53). Binding of dopamine, but not ecdysone, to the receptor leads to the phosphorylation of Akt, a kinase associated with insulin signaling. Ecdysteroid binding activates an extracellular signal-related kinase but no physiological function is currently known for this activity.
We identified five orphan clades (OA1–5) in sub-assemblage 1a (Figure 1). The OA1 and 2 receptors are sister to the bursicon and GPA2/GPB5 receptors and are possible orthologs of the vertebrate relaxin/insulin-like peptide LGRs (OA2). The OA2 orthologs are present in both Drosophila species, Ae. aegypti and An. gambiae, but the OA1 clade appears to have been lost in the Culicidae based on its presence in other insects (data not shown). OA3 is present in all five dipterans, as is OA4. OA5 encodes the D. melanogaster gene moody, which is implicated in the integrity of the blood-brain barrier (54), although its native ligand is unknown. Our results provide no insights regarding what kind of ligand(s) the OA4–5 orphans bind. However, a previous study that examined small clusters of GPCRs between humans and D. melanogaster suggested these orphan clades grouped most closely with peptide receptors (25).
Sub-assemblage 1b: CCAP, AKH, corazonin, ACP, and allatotropin receptors
Adipokinetic hormones, corazonin, and CCAP are structurally related but functionally distinct peptides (18). Corazonin and CCAP are cardioactive in An. gambiae (55, 56). In D. melanogaster, corazonin affects stress resistance and metabolism (57) and is involved in ecdysis, as is CCAP, which is also cardioactive (58). ACP and its receptor were recently identified in insects, including mosquitoes but not in D. melanogaster, and its function is unknown (59). Prior studies have also recognized a common evolutionary origin for this receptor group and coevolution of their associated ligands in mosquitoes (18, 59, 60).
Mosquitoes encode two different AKH genes (61) with AKH 1 mobilizing stored carbohydrate but not lipids, and AKH 2 having no effect on these processes or an identified receptor (62). Drosophila spp. in contrast encode only AKH 1, which plays a role in starvation-mediated behaviors and mobilization of lipid and carbohydrate stores (58). Recent studies confirm binding of AKH 1 to its predicted receptor in D. melanogaster and An. gambiae, while also identifying a splice variant receptor in Ae. aegypti (61, 63, 64). No AKH receptor was found in C. quinquefasciatus.
Allatotropin is not found in Drosophila, but in Ae. aegypti and other insects it stimulates juvenile hormone (JH) biosynthesis (65). Binding studies identify a single allatotropin receptor in Ae. aegypti (AAEL011680) for which a closely related paralog exists (AAEL005310) (65). We also identified one predicted allatotropin receptor in C. quinquefasciatus and An. gambiae.
Cluster 1, assemblage 2
Sub-assemblage 2a: sulfakinin related receptors
This sub-assemblage represents the earliest diverging GPCRs in assemblage 2. Sulfakinin regulates feeding behavior in D. melanogaster (66) and binds to two GPCR paralogs, CG32540 and CG42301 (67). Both Ae. aegypti and An. gambiae encode a single sulfakinin ortholog, while C. quinquefasciatus encodes two predicted sulfakinin receptor homologs: one that groups with the Drosophila sulfakinin receptors and a second that is more closely related to the mosquito sulfakinin orthologs.
Sub-assemblage 2b: RYamide, tachykinin, natalisin, NPF, short NPF, SIFamide, and related receptors
The RYamide gene was recently recognized in Diptera and other insects. It encodes two paracopies with no known function that were used to deorphanize a GPCR related to the tachykinin receptor (68, 69). Two copies of this gene are present in mosquitoes. A single gene encodes up to six tachykinin paracopies across the Diptera (13, 18, 70), which are multifunctional (71). D. melanogaster was initially thought to encode two tachykinin receptors (CG7887, CG6515) which exhibited preferential binding to different ligand paracopies (72–76). More recent studies show that CG6515 from D. melanogaster is not a tachykinin receptor but rather binds natalisin, a newly identified peptide hormone involved in mating (77). D. mojavensis, An. gambiae, and Ae. aegypti encode a single closely related gene to CG7887, whereas no ortholog is found in C. quinquefasciatus. We also detect orthologs of CG6515 in the genome of each mosquito, but the two partial sequences encoded by Ae. aegypti (AAEL017414 and AAEL017341) and one paralog in C. quinquefasciatus (CPIJ014103) were not long enough to be included in our phylogeny.
Kinins, which were originally identified as myotropic neuropeptides, are now considered key regulators of diuresis in mosquitoes and Drosophila (78–81). D. melanogaster encodes a single kinin receptor that has been functionally characterized (82). D. mojavensis, An. gambiae, and C. quinquefasciatus also encode a single predicted kinin receptor, whereas two copies are detected in Ae. aegypti. Notably, another type of diuretic peptide, inotocin, the homolog of vertebrate oxytocin/vasopressin, and its receptor are present in locusts and red flour beetles (T. castaneum), but both are absent from Drosophila spp. and mosquitoes (83).
Our phylogeny indicates the RYamide and tachykinin receptors are related to GPCRs that bind NPF, sNPF, and kinins. The relatedness of the NPF and sNPF receptors is surprising, given differences in ligand structure (36 vs. <11 AA respectively). A prior study also noted the shared homology between insect kinin and vertebrate tachykinin receptors (82). Taken together, we conclude this GPCR receptor cluster is an example of peptide ligand/receptor coevolution in insects.
Neuropeptide F is a member of the NPY/PP/PYY family conserved across vertebrates and higher invertebrates. It was first identified along with its receptor in D. melanogaster, where it affects behaviors related to feeding, learning, stress, and alcohol sensitivity (84). The unrelated sNPFs found only in arthropods are encoded by a different gene and processed into four peptides, which affect feeding, metabolism, and growth in D. melanogaster (57). The expression of NPF and sNPF is characterized for Ae. aegypti and An. gambiae, but little is known about their function (85–87). In Ae. aegypti but not in other mosquitoes or Drosophila spp. there is an apparent duplication of the sNPF gene, which encodes three identical “Aedes head peptides” (AHPs) isolated as non- and hydroxylated-proline forms (88, 89). Only the latter inhibits host seeking by females (90) and is also found in the accessory gland of males and transferred to females (91).
The single copy NPF receptor from D. melanogaster and one of two NPF copies in An. gambiae (AGAP004123) were identified and expressed to demonstrate NPF binding (85, 92). As in D. melanogaster, D. mojavensis and Ae. aegypti encode a single NPF receptor, whereas An. gambiae and C. quinquefasciatus encode two. The duplication of the NPF receptors in An. gambiae and C. quinquefasciatus appears to have occurred independently in each lineage. The two An. gambiae NPF receptors, AGAP004123 and AGAP004122, are highly similar at the nucleotide level (99% identical), yet AGAP004122 is truncated relative to AGAP004123 at its 3′ end. The two C. quinquefasciatus NPF receptors also exhibit a pattern of high similarity (99%) but truncation of one copy (CPIJ018265) is at the 5′ end. Further study will be needed to determine if these truncated genes are shorter forms of the NPF receptor are incompletely annotated or are pseudogenes.
The first sNPF receptor was identified as a single copy gene in D. melanogaster and expressed to confirm binding (93). An. gambiae also encodes a single sNPF receptor for which binding was confirmed (85). The structural relatedness of the sNPF and NPF receptors is born out by their shared inhibition of intracellular cAMP signaling (85, 92). Recently, the Ae. aegypti sNPF receptor was also demonstrated to bind AHPs, though silencing of the receptor did not impact host-seeking behavior (94).
SIFamide regulates adult courtship behavior in D. melanogaster (58), and a SIFamide homolog was previously identified in Ae. aegypti (9). A single SIFamide receptor (CG10823) was identified in D. melanogaster (95). An. gambiae and D. mojavensis encode one predicted SIFamide receptor, whereas Ae. aegypti and C. quinquefasciatus encode two. The SIFamide receptors are most closely related to the four receptors of clade OA8, present only in mosquitoes. Two mosquito receptors from An. gambiae and C. quinquefasciatus form the orphan clade OA7, which is an exceptionally long branch. Annotation suggests that these may be odorant-binding receptors, although no experimental evidence supports this. OA9 is the outgroup to this large clade of receptors, and is found in all five genomes including duplicate copies in the culicine mosquitoes. Two orphan receptors are found in clade OA6, which forms an outgroup to sub-assemblages 2b–f. These genes reside on a long branch with nothing known about their function.
Sub-assemblage 2c: orphan receptors A10 and 11
Two orphan clades, A10 and 11, comprise sub-assemblage 2c. While monophyletic in our analysis, support values for this sub-assemblage are weak. Homologs of A11 are found in all five dipterans, but there is no ortholog of A10 in C. quinquefasciatus.
Sub-assemblage 2d: CCHamide, allatostatin C and A receptors
All insects appear to encode two CCHamide genes (1 and 2) that arose by duplication after divergence of the hexapods from other arthropods (11, 96). The function of CCHamide 1 and 2 is unknown but binding studies identify two GPCRs from D. melanogaster (CG14593 and CG30106) as CCHamide receptors (96). Single copy orthologs of both D. melanogaster receptors are found in all species except C. quinquefasciatus, which lacks an ortholog of CG14593.
Three structurally distinct types of neuropeptides are called allatostatins in the literature because they inhibit JH synthesis by the corpora allata (CA) in different insects (58, 97). Mosquitoes and Drosophila spp. encode: (1) allatostatin A paracopies (FGLamides), (2) allatostatin B paracopies [9–13 AA; herein classified as MIP or Wx(6)Wamides], and (3) a single allatostatin C, also known as PISCF (98–101).
Two allatostatin C receptor paralogs were identified in mosquitoes and Drosophila spp. (102, 103) with some studies also suggesting these receptors are related to tachykinin receptors (16, 18) (see below). Our analysis, however, identifies the allatostatin C receptors as a separate clade from the tachykinin receptors, which is consistent with differences in ligand motifs.
Although allatostatin A paracopies inhibit JH biosynthesis in some insects, they have no effect on CA from D. melanogaster or female Ae. aegypti (104). Recent work suggests a role in foraging behavior of D. melanogaster larvae (105). Ast A paracopies bind to two receptor paralogs, CG2872 and CG10001, in D. melanogaster (16). We note that D. mojavensis and Ae. aegypti each encode two receptors for allatostatin A, whereas An. gambiae and C. quinquefasciatus encode three. The putative allatostatin A receptors from mosquitoes have not been functionally characterized, but their number suggests either all bind one or more allatostatin As or some are orphans.
Sub-assemblage 2e: trissin receptor
Trissin is another peptide hormone with no known function that binds a GPCR in D. melanogaster (69). We detected single orthologs of the D. melanogaster trissin receptor in the other genomes.
Sub-assemblage 2f: periviscerokinin, pyrokinin, and ecdysis triggering hormone receptors
Peptide ligands for the receptors in this clade share a similar C-terminal motif (PRL/Vamide) and are processed from precursors encoded by three genes in all dipteran species examined to date. The capability gene yields two PVKs ending with PRVamide and one PK ending with WFGPRLamide. The hugin gene produces two to four PK paracopies (7, 106) whereas the ETH gene produces two paracopies (9, 18, 58, 60, 107, 108). The receptor for ecdysis-triggering hormone (ETHR) has duplicated in D. mojavensis, but exists as a single copy gene in the other genomes.
Drosophila melanogaster has three PK GPCRs – CG9918 as the PK1 receptor, and CG8784 and CG8795 as PK2 receptors (109, 110) – and a single PVK receptor (CG14575). Single copy orthologs of the PVK receptor are present in all the species, while variable numbers of PK receptors were found among species. Initially, two were identified in An. gambiae, and the expressed CG9918-like receptor (AGAP000658) preferentially binds the PK1 peptides and the other receptor (AGAP003076), the PK2 peptides (108). The same study identified the related receptor in An. gambiae as a homolog of the PVK receptor in D. melanogaster. Four PK receptors are found in C. quinquefasciatus, though only a single PK receptor is found in Ae. aegypti.
Both splice variants of ETHR in D. melanogaster bind both forms of ETH (18, 60, 100, 107, 108). Both ETH paracopies also regulate ecdysis in D. melanogaster and Ae. aegypti (60, 107). Two ETHR splice variants were identified in D. melanogaster and Ae. aegypti that are more selective for ETHs than other related peptides (107, 111–113).
Cluster 2
Proctolin receptor
Proctolin was the first myostimulatory peptide isolated from an insect, and its receptor was first identified in D. melanogaster (58) and an ortholog was identified in D. mojavensis (Figure 2). In contrast no gene encoding this peptide or a receptor ortholog is known in mosquitoes, which is consistent with proctolin having no effect on mosquito tissues (114).
FMRFamide receptors
The single FMRFamide gene in dipterans encodes multiple paracopies that bind to the expressed D. melanogaster GPCR (115, 116) and are myostimulatory in D. melanogaster and An. gambiae (117, 118). The FMRFamide receptor is represented by a single ortholog in each of the Drosophila and mosquito genomes examined (Figure 2).
Myoinhibitory/sex peptide receptor
Myoinhibitory peptides were isolated based on their myoinhibitory activity. They were later shown to inhibit JH biosynthesis (so named, allatostatin B) in different insects and to be structurally related to the sex peptides in Drosophila (119). Binding studies established that MIPs and sex peptides both bind the D. melanogaster MIP receptor (119, 120). A single ortholog of this receptor is also present in D. mojavensis and each of the mosquito species (Figure 2). No sex peptide gene is known for mosquitoes, but one study showed that sex peptide from D. melanogaster bound to the Ae. aegypti MIP receptor (91).
Myosuppressin receptors
Myosuppressin inhibits gut and heart contraction in D. melanogaster and binds to duplicate GPCRs deorphanized for this species (15, 121). This duplication appears to have occurred prior to the divergence of D. mojavensis and D. melanogaster. We identified a single ortholog in each mosquito species (Figure 2). Specific binding of myosuppressin was also previously shown for the expressed An. gambiae ortholog (23). A related orphan receptor, OA14, is represented as a single ortholog in the Drosophila, Anopheles, and Culex spp. while two orthologs were identified in Ae. aegypti (Figure 2).
Other orphan clades
In addition to OA14, Cluster 2 contains two other orphan clades designated as OA12 and 13. OA12 is most closely related to the proctolin receptor but has expanded in the culicine mosquitoes, with Ae. aegypti encoding three copies and C. quinquefasciatus encoding four. OA13 has an uncertain position in Cluster 2 but at least one copy of this gene is found in each of the genomes with the exception of An. gambiae.
Class B secretin GPCRs
The balance of dipteran GPCRs that bind peptide hormones belong to the Class B secretin family (7). Our analysis distinguished nine clades within this class (Table 1; Figure 3), of which three have identified ligands: (1) calcitonin-like diuretic hormone (CT-DH), (2) corticotropin-releasing factor-like diuretic hormone (CRF-DH), and (3) pigment dispersing factor (PDF) (Figure 3). We included the latrophilin GPCRs that are conserved across invertebrates and vertebrates (122, 123) and bind latrotoxins from spiders in the genus Latrodectus (124). The endogenous ligand for the vertebrate receptors is teneurin-2, a glycoprotein displayed on the surface of cells (125), however the endogenous ligand in insects in unknown. Class B secretin GPCRs also include the Methuselah and Methuselah-like genes, which are grouped into four classes, A–D, based on the disulfide bridges present in their extracellular domain (126). Other phylogenetic data for Class B GPCRs have been produced for vertebrates and invertebrates (127) and insects alone (126).
Pigment dispersing factor receptor
This peptide was first identified by its stimulation of pigment dispersal in some arthropods, but in insects PDF was the first neuropeptide shown to regulate circadian activity in D. melanogaster (58). Its receptor was subsequently identified, and we found single orthologs of this receptor in D. mojavensis and each of the mosquito species. It is not known whether PDF has a role in circadian activity in insects generally, but host-seeking behavior is one circadian behavior that may be regulated by PDF in mosquitoes (128).
Corticotropin-releasing factor-like diuretic hormone receptor
Our phylogeny suggests the ancestor of this receptor underwent a duplication event prior to the divergence of the Culicidae and Drosophilidae. Both orthologs from D. melanogaster bind CRF-DH (129). The diuretic activity of CRF-DH is also well characterized in mosquitoes and D. melanogaster (130, 131).
Calcitonin-like diuretic hormone receptor
These ligands and their associated receptors mediate diuresis by Malpighian tubules (132). The activity and expression of CT-DH and its receptor are well characterized in mosquitoes and D. melanogaster (132–134). The CT-DH receptors are single copy in each genome and are sister to the orphan Hector-like GPCRs (Figure 3).
Methuselah-like receptors
Studies in D. melanogaster described mutations in a GPCR named Methuselah (mth), which conferred increased longevity and stress resistance (135). Additional mth-like genes were thereafter identified (34), while genome studies indicate that mth genes are likely present in all insects. Four major groups (A–D) of Mth receptors have been proposed on the basis of the disulfide bridges present in their extracellular domains (126, 136). While present in all insects examined in this study, D. melanogaster encodes many more mth-like genes (16) than D. mojavensis (8), C. quinquefasciatus (8), Ae. aegypti (6), or An. gambiae (4). We further note the Drosophila Mth receptors are widely distributed across the tree and are not monophyletic. Only three clades of Mth receptors include members from the Culicidae: the C type Mth, Mth-like 14, and a clade that is sister to the B and D type Mth receptors (Figure 3). Cvejic et al. (137) reported that the endogenous ligand for the Mth receptor in D. melanogaster was a peptide encoded by the gene stunted, and that disruption of its expression has a similar phenotypic effect as reduced mth expression. Others, however, report that Mth binds Drosophila sex peptide and other synthetic peptides and suggest this receptor may be a promiscuous GPCR with diverse ligands (138).
Latrophilin receptor
Each genome contains a single copy of the latrophilin GPCR. Our analysis suggests these receptors do not form a monophyletic group in the Diptera, but this conclusion is based on poor alignment due to the loss of six of the seven transmembrane domains in the Culicidae. The mosquito receptors, however, retain N-terminal domains that are similar to those of Drosophila spp. (data not shown). Truncation of the C-terminal region of latrotoxin-binding GPCRs in mammals does not impede responses to latrotoxin in cell culture (139), which suggests the culicid orthologs may be functional.
Orphan Class B/secretin receptors
In addition to the Mth-like and latrophilin GPCRs, our results identify two clades of orphan Class B secretin GPCRs. The Hector group (OB1) is sister to the CT-DH receptor and appears to have arisen by a duplication event in Diptera that predates the divergence of the Drosophilidae from the Culicidae. Although orphans, mutations in the Hector GPCRs have been shown affect mating behavior in D. melanogaster (140). OB2 was identified as a distant ortholog of the human epididymis six GPCR, itself an orphan, shortly after the publication of the D. melanogaster genome (34).
Receptor guanylyl cyclases
Receptor guanylyl cyclases are conserved homodimeric membrane proteins (~200 kDa) with intracellular protein kinase and guanylyl cyclase domains that catalyze cGMP formation (2). We identified six RGC clades in Diptera (Figure 4), two of which have characterized ligands: eclosion hormone (EH) and neuropeptide-like peptide 1-VQQ (NPLP1). Eleven forms of NPLP1 are encoded by the genes identified for D. melanogaster and other insects (141). The NPLP1 receptor was also shown to be an RGC in D. melanogaster, while functional studies showed that NPLP1 stimulates fluid transport in the midgut and Malpighian tubules, modulates stress, and affects immune responses (142). EH is a co-activator with ETH of ecdysis behavior in insects. Expression of the Drosophila EH receptor in a mammalian cell line followed by incubation with EH from D. melanogaster or another EH paracopy from the oriental fruit fly (Bactrocera dorsalis) elicited a strong cGMP response, which provided evidence this RGC functions as an EH receptor (143).
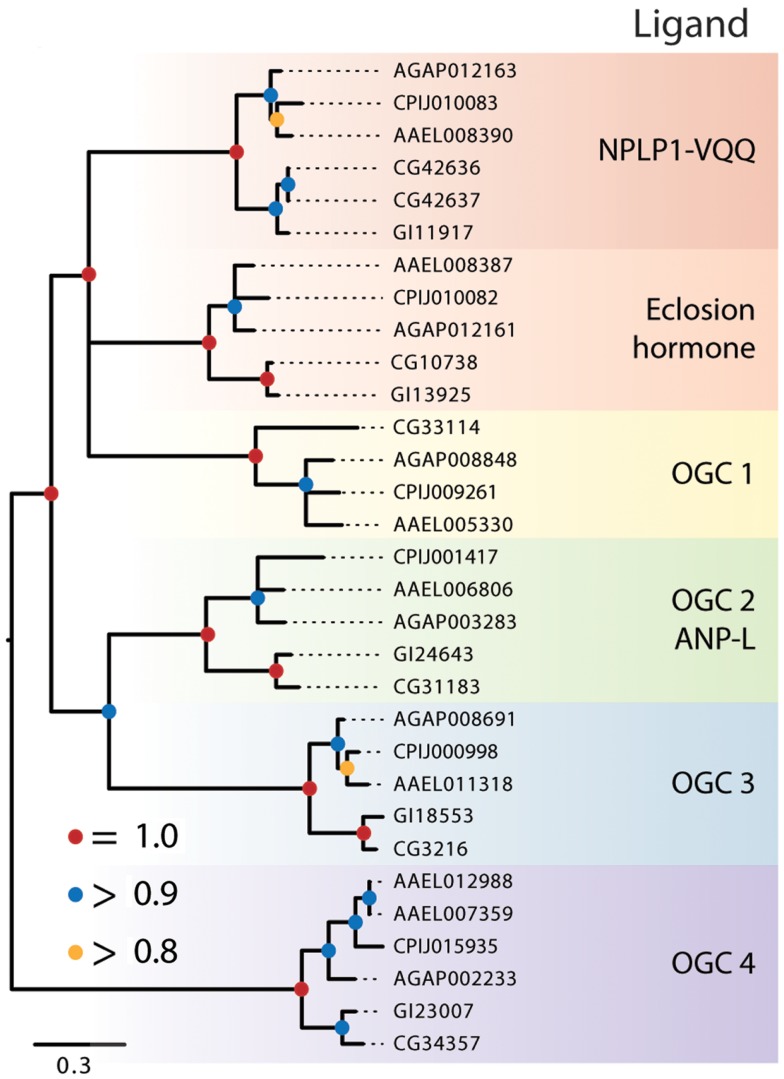
Figure 4.
Maximum-likelihood tree of membrane-bound RGCs using the five dipteran species described in Figure 2 with alignments made using the protein kinase domain of each receptor. Branches with maximum-likelihood support values <0.8 have been collapsed to polytomies. The tree was rooted at the midpoint.
The remaining dipteran RGCs resided in orphan clades, and to further characterize their relationships to other RGCs, we constructed a phylogenetic tree that included these RGCs and the five RGCs from H. sapiens, A–E (data not shown). This analysis indicated that two of the dipteran orphan clades (OGC1 and 3) have no vertebrate ortholog. In contrast, OGC2 appears orthologous to vertebrate RGC-A and -B that bind natriuretic peptides, while OGC3 is homologous to the vertebrate retinal RGC. Overall, our analysis indicates that dipteran RGCs have undergone few changes, with the possible exceptions of an apparent loss of OGC1 from D. mojavensis, the duplication of the NPLP receptor in D. melanogaster, and the duplication of the OGC4 homolog in Ae. aegypti.
Protein kinase receptors
Protein kinase receptors form non-covalently bound dimers upon reaching the cell surface or in response to ligand binding (2). The insulin receptor is an exception in that its monomers undergo proteolysis into α and β subunits that are linked by intra- and inter-subunit disulfide bonds to form a covalent stabilized heterotetramer (144). Peptide hormones, growth factors, and membrane proteins are ligands for these receptors, and their interactions, signaling, and function are well characterized for mammalian systems, intermediately characterized for D. melanogaster, and poorly characterized in other insects. Ligand binding to specific extracellular domains activates intracellular kinase domains that autophosphorylate either tyrosine or serine and threonine residues, presenting docking sites for cytoplasmic effectors, adaptors, and scaffold proteins that in turn activate one or more signal pathways (3). Activated PKRs are also targets of protein phosphatases that modulate and even block signaling in mammals. Our analysis of dipteran PKRs produced a phylogeny with two major branches: (1) an assemblage of 12 clades consisting of primarily characterized receptor tyrosine kinases (RTKs) and (2) five clades related to mammalian transforming growth factor beta (TGF-β) receptors, which contain a distinctive serine-threonine kinase domain (Figure 5). Similar to the RGCs but unlike the GPCRs, dipteran PKRs have undergone relatively few lineage-specific gains or losses, suggesting that most members were likely present in the most recent common ancestor of culicids and drosophilids.
Figure 5.
Maximum-likelihood tree of PKRs using the protein kinase domains of PKRs from the five dipteran species described in Figure 2. The tree was rooted using a guanylyl cyclase receptor GI22298 from D. mojavensis. Branches with maximum-likelihood support values <0.8 have been collapsed to polytomies. The C. quinquefasciatus gene CPIJ006878 has been placed within the clade it presumably belongs to, though it lacks the protein kinase region used for alignment. The D. mojavensis OR1 ortholog is not annotated in FlyBase, but EST evidence supports its presence (160). Clades are named after their characterized ligands, if known.
Receptor tyrosine kinases
The RTK clades we identified were conserved across the five dipteran species (Figure 5). Ten of these clades are also represented among the 20 recognized subfamilies of human RTKs, which have been classified on the basis of ligand interactions and other features (3). The remaining two RTK clades are known only from insects and other arthropods. One of these is an orphan receptor (OR1), which contains a venus flytrap domain, while the other is the prothoracicotropic hormone (PTTH) receptor. Most of the dipteran RTK clades are encoded by a single ortholog in each species. For the 10 clades with apparent human homologs, only OR2 (ROR) is an orphan. Five others interact with secreted ligands [epidermal growth factor (EGF), platelet-derived/vascular endothelial growth factor, fibroblast growth factor (FGF), insulin-like peptides (ILPs), and low-density lipoprotein repeat-containing factor], whereas the remaining four form activating complexes with unrelated membrane proteins (glial cell line-derived neurotrophic factors, collagen, muscle specific kinase, and ephrin).
ILP and related receptors
The ILP receptor (IR) was the first RTK identified in insects when cloned from D. melanogaster (145, 146). These studies showed this RTK bound mammalian insulin, and much later, the discovery of multiple ILP genes in D. melanogaster was taken as evidence that the encoded peptides were the endogenous ligands of the IR (147). Up to eight ILPs encoded by different genes are known for the dipteran species in our study (148), and all but the ILP6 subfamily are likely processed into disulfide-linked dimers (6–8 kDa). Only one study from Ae. aegypti confirms the high affinity binding of an endogenous insect ILP to its cognate IR (149), while another shows ILP displacement of human insulin bound to the D. melanogaster IR (150). Several studies also show the IR activates the canonical insulin signaling pathway in Ae. aegypti (151–153) and that ILPs have a diversity of functions in mosquitoes (148, 154).
The two RTKs that group with the IR include a homolog of human anaplastic lymphoma RTK (ALK) and leukocyte tyrosine kinase (LTK), and the orphan OR1. Both ALK and LTK play key roles in human cancers, whereas the ALK homolog in D. melanogaster directs gut and nervous system development and is associated with ethanol sensitivity and learning (155–158). No specific ligand has been identified for mammalian ALK or LTK, although a 61 kDa secreted protein ligand named Jelly Belly has been identified as an ALK ligand in D. melanogaster (159). However, Jelly Belly does not interact with mammalian ALK (159). Our analysis supports the presence of OR1 in mosquitoes, other insects, and other invertebrates as well as its loss from the D. melanogaster subgroup as first reported by Ahier et al. (160). Curated, partial mRNAs indicate this receptor is present in D. mojavensis, which was included in the phylogenetic tree, but the genome lacks an annotated receptor. The extracellular domain of OR1 contains a venus flytrap domain, which is also found in some GPCRs and RGCs, and has been implicated in binding amino acids and other small molecules (160, 161).
PTTH receptor
Prothoracicotropic hormone regulates molting and development in insects by stimulating the prothoracic glands (PGs) to produce ecdysteroid hormones (162–164). Expression of the RTK Torso is required for PTTH action in D. melanogaster. Torso was also shown to interact with Trunk, which is a protein growth factor that regulates early embryonic development. PTTH and Trunk are structurally related to other protein and growth factors that contain a cysteine knot motif and bind RTKs and TGF-β receptors. Although Trunk and PTTH differ in tissue and temporal expression, their interaction with Torso activates the Ras/mitogen-activated protein kinase pathway and calcium signaling. These shared features and actions support the designation of Torso as the PTTH receptor. However, direct binding of PTTH to Torso has not been demonstrated.
We identified duplicate PTTH RTK genes in C. quinquefasciatus and Ae. aegypti, but not in An. gambiae or D. melanogaster. The absence of a PTTH RTK ortholog in D. mojavensis is likely due to our inability to identify it rather than its loss given that orthologs of the trunk and PTTH genes are present (163). PTTH expression has been profiled in mosquito larvae (165–167), but it is not known to regulate ecdysteroid production, which occurs in unidentified cells in the abdominal or thoracic wall, not the PGs (168).
Other RTKs
The functional significance and signaling of the other RTK clades has been examined in D. melanogaster. The best characterized of them is the EGF receptor, which interacts with one secreted ligand (Vein) plus three other ligands (Spitz, Keren, and Gurken) derived from enzymatic cleavage of inactive membrane-bound precursors. Typically, these ligands regulate the trajectory of specific embryonic and tissue stem cell types (169, 170). An EGF receptor exists in each mosquito species, and its expression has been characterized in An. gambiae (171). However, the putative ortholog in C. quinquefasciatus was a partial annotation that lacked the protein kinase domain and was therefore not included in the phylogeny. The platelet-derived/vascular endothelial growth factor receptor and its three secreted ligands are involved in the regulation of hemocyte and midgut stem cell fate in D. melanogaster (172, 173). The FGF receptor is duplicated in each dipteran species. Functional studies in D. melanogaster show the two receptors differ in their structure and ligand interactions (174). Homolog 1 (Heartless, CG7223) interacts only with two of the three related secreted FGFs (Pyramus and Thisbe) to direct mesoderm migration and heart muscle differentiation of the dorsal vessel, whereas homolog 2 (Breathless, CG32134) is activated by its ligand (Branchless) to define branching of the tracheal system. Breathless appears to be incompletely annotated, and was removed from our alignments. Less is known about the RET RTK (175, 176), ROR RTK (177), and ephrin RTK (178) in the development of D. melanogaster, while the collagen and muscle specific kinase RTKs remain unstudied. We note that D. mojavensis encodes duplicate collagen receptors, while the D. melanogaster paralog CG34380 lacks the protein kinase domain used for alignment.
TGF-β-like receptors
The different elements of the TGF-β signaling pathway are well characterized in mammals and D. melanogaster, because they regulate many of the same developmental and cellular processes as RTKs (179–181). In common with secreted RTK ligands, TGF-β ligands are disulfide-linked homodimers (~30 kDa) of N-terminal regions cleaved from precursors. They are subdivided into the TGF-β/activin orthologs, Activin, Dawdle, Myoglianin, and Maverick, and bone morphogenic protein (BMP) orthologs, Decapentaplegic, Glass Bottom Boat/60A, and Screw, in Drosophila (182) and Anopheles spp. (183). In general, these ligands act as growth factors and cytokines in both mammals and insects (182).
Types I and II TGF-β receptors are single pass-transmembrane serine/threonine kinases, and ligand binding results in the formation of a heterotetramer of types I and II receptor dimers with the type II dimer phosphorylating the type I dimer, which in turn binds and phosphorylates SMAD or MAD proteins that activate other elements in a signaling pathway. In agreement with these studies, we identified three type I receptor clades (Baboon, Saxophone, Thickveins) and two type II receptor clades (Punt and Wishful Thinking). Each clade is represented as a single ortholog in the two Drosophila spp. and C. quinquefasciatus. Duplications have arisen in Ae. aegypti in the saxophone, thickveins, and punt genes. In contrast, An. gambiae has paralog copies of the baboon receptor.
The TGF-β ligands are not considered peptide hormones, per se, but activin may be an exception in that it is widely expressed in the nervous system and in neurosecretory cells associated with endocrine glands in D. melanogaster (182, 184). Mammalian activin homologs are also expressed in neuroendocrine cells in the pituitary gland, where they regulate the expression and release of gonadotropic hormones (185), and in endocrine cells in the pancreas islets, where they modulate insulin secretion (186). Other studies show that TGF-β signaling plays an important role in mosquito immunity and is responsive to mammalian TGF-β proteins ingested in blood meals (187, 188).
Conservation of active sites in receptors and hormones
An underlying assumption in the study of peptide hormone receptor evolution is that related receptors exhibit a degree of conservation in their active site, as do their peptide ligands. To test this, we compared the sequences of related NPFs and their GPCRs for the five dipterans to the NPY/PP/PYYs and their receptors in three vertebrates: zebrafish (D. rerio), mouse (M. musculus), and human (H. sapiens) (Figure 6). Insect NPF is a member of the NPY family, and evidence suggests that the neuropeptides regulate related behaviors across these groups (58).
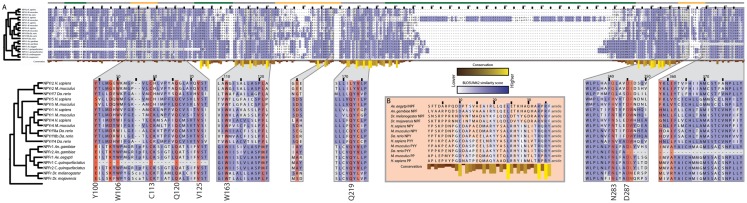
Figure 6.
Conservation of peptide active sites and receptor binding regions across the Bilateria. (A) Alignment of neuropeptide Y GPCRs subtypes (NPYr) that also bind pancreatic peptide (PP) and peptide YY (PYY) from M. musculus, Da. rerio, and H. sapiens with neuropeptide F GPCRs (NPFr) from the five dipteran genomes. Blue shading indicates amino acid similarity between residues according to BLOSUM62 scores; darker shades indicate increased similarity. Yellow bars below residues indicate conservation across the alignment. Position relative to the cell membrane is indicated by the colored line above the alignment: intracellular: green, extracellular: yellow, transmembrane: gray. Zoomed regions show domains that have been confirmed experimentally to be important for ligand binding by NPYrs. Residues highlighted in red indicate those determined to be involved in ligand binding in H. sapiens NPYr1 and 2. Binding residues that are highly conserved are listed next to the alignment with their amino acid in human NPYr1 or 2. (B) Alignment of vertebrate NPY, PP, and PYY with dipteran NPF sequences. Only the mature peptide is shown. Coloring is the same as in (A).
Mutagenesis studies have revealed a number of receptor residues that are involved in NPY binding (189, 190). We examined the corresponding locations in the dipteran receptors and found that many key binding residues are conserved, including several that are perfectly conserved, since the divergence of protostomes and deuterostomes 650 Mya. Loops 2–4 comprise a hydrophobic ligand binding pocket in NPY receptors (191), and within this pocket, multiple residues have been implicated in ligand-receptor interactions (41–43, 192, 193) (Figure 6). Six of the residues identified by mutagenesis of human NPY receptors are perfectly conserved in the full-length dipteran NPF receptors: W106, C113, Q120, V125, W163 and D287 (numbering refers to human NPYr1/2 positions). The first four residues are absent in the N-terminal truncated C. quinquefasciatus copy of the NPF receptor, while D287 is absent in the C-terminal truncated An. gambiae NPF receptor. Q219 is conserved in all receptors examined except H. sapiens and M. musculus NPYr1. Similarly, N283 is invariant in the dipteran NPF receptors, but differs between vertebrate forms of the receptor. The hydroxyl group of Y100 is implicated in NPY binding by the NPYr1 from H. sapiens (192). In the dipteran receptors, the homologous position is invariantly a glutamic acid with the exception of the truncated C. quinquefasciatus receptor, where it is absent. Although the amino acid at this position is different between the vertebrate and dipteran lineages, the glutamic acid residue still contains a free hydroxyl group, suggesting that the functionality of this position is conserved.
Among the NPF/NPY family, 3 of 36 residues in the mature peptide are perfectly conserved in the species examined: L24, R33, and R35 (Figure 6). The C-terminal A/TRXRY/Famide motif along with a D/E at position 10, and a leucine residue at position 24 of the mature peptide were also conserved. These data overall strongly suggested functional sites are conserved between related but distinct peptides and their associated receptors across hundreds of millions of years of evolution.
Conclusion
This study represents the first comprehensive analysis of the three receptor types that bind peptide hormones and growth factors in insects. Our primary motivation for undertaking this study was to develop a robust phylogenetic framework to study the function of particular orphan receptors in mosquitoes. However, by taking advantage of the comparative genomic data available for the Culicidae and Drosophilidae, our results also provide comprehensive phylogenetic information for the GPCRs, RGCs, and PKRs across the breadth of the Diptera (Nematocera and Brachycera).
The absence of any individual orphans within the clades of RGCs with characterized ligands suggests these receptors are stable within the Diptera and that diversification of RGCs occurred prior to the evolution of the order several hundred million years ago (44). Dipteran Class A and B GPCRs are more evolutionarily labile with each experiencing several instances of duplication and loss. For Class A GPCRs, most of the orphans in characterized clades are single duplication events that have occurred in a particular mosquito species. The functional significance of these duplications is in most cases unclear. Since many peptide hormones exist in multiple forms derived from a single propeptide, we speculate that different forms of a given receptor may preferentially interact with different forms of their cognate peptide hormone. In some cases, paralog receptors both bind a single hormone but vary in their binding affinity and phenotypic effect, as is seen in the two PK2 GPCRs of D. melanogaster (194). On the other hand, most of the GPCR orphan clades are Methuselah-like Class B secretin GPCRs, which are overrepresented in drosophilids generally and D. melanogaster in particular. Currently, the literature offers no insights into why this bias exists or what the functional significance of so many Methuselah-like GPCRs might be.
We tried to characterize the evolution of gains and losses of peptide hormone receptors in the Diptera but in many cases it was not possible to discern such events with certainty due to issues with annotation. Often what initially appeared as a gene duplication event was in fact two separate gene annotations for a single gene. This was verified in some cases by available RNAseq data covering the genes of interest, whereas in others, alignment to a single ortholog demonstrated that the actual gene had been divided during gene prediction. This was particularly true in the Class A rhodopsin-like GPCRs of Cluster 1. We had to omit some GPCRs from the analyses due to absence of the domain needed for identification and alignment. Several potential PKRs also lacked a complete kinase domain and were therefore removed from our analysis. Though care was taken to correct these annotation errors in our analysis, it is possible that some duplication events we report are artifacts of gene prediction algorithms.
Studies across a range of organisms have shown the utility of comparing orphan to characterized receptors. Hansen and colleagues (59) demonstrated that an orphan GPCR was sister to the AKH receptor, and these two receptors were sister to the corazonin receptor. Subsequent efforts to deorphanize the receptor identified a structurally intermediate peptide encoded in the genome of insects. Our results illuminate several orphan clades and individual receptors that are sister to characterized receptors, suggesting that these orphans may bind similar ligands. Additionally, the species distribution of orphan receptors and patterns of tissue and temporal expression may also reduce the target ligand pool, and hopefully aid in deorphanization. While GPCRs evolve at a fast rate (195), binding sites in both the receptor and peptide ligand can be conserved across hundreds of millions of years as demonstrated here for the vertebrate NPY/PP/PYY and dipteran NPFs and their GPCRs. Conservation of functional residues between receptors that bind related peptides suggest that phylogenetic position of related orphan receptors can aid the identification of ligands and provide insights into their function, because in many instances it is also conserved to a high degree, as demonstrated for the NPF/NPY superfamily (84). Thus, our results could assist in deorphanizing receptors in newly sequenced mosquito genomes by identifying their ligands. On the other hand, the literature is more equivocal in regard to sequence similarity between species also resulting in functional similarity. Thus, in addition to deorphanization, considerable work remains in understanding the physiological function of many peptide hormones and growth factors among different species of mosquitoes and other insects.
Author Contributions
Kevin J. Vogel, Mark R. Brown, and Michael R. Strand designed the study, Kevin J. Vogel performed the analyses, and Kevin J. Vogel, Mark R. Brown, and Michael R. Strand wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fendo.2013.00193/abstract
Acknowledgments
The authors thank Joe W. Crim for helpful comments on earlier versions of the manuscript. This work was funded by the Georgia Agricultural Experiment Station and a National Institutes of Health award R01 AI033108 to Mark R. Brown and Michael R. Strand.
References
- 1.Biarc J, Chalkley RJ, Burlingame AL, Bradshaw RA. Receptor tyrosine kinase signaling – a proteomic perspective. Adv Enzyme Regul (2011) 51:293–305 10.1016/j.advenzreg.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potter LR. Regulation and therapeutic targeting of peptide-activated receptor guanylyl cyclases. Pharmacol Ther (2011) 130:71–82 10.1016/j.pharmthera.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell (2010) 141:1117–34 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerjan B, Cardoen D, Bogaerts A, Landuyt B, Schoofs L, Verleyen P. Mass spectrometric profiling of (neuro)-peptides in the worker honeybee, Apis mellifera. Neuropharmacology (2010) 58:248–58 10.1016/j.neuropharm.2009.06.026 [DOI] [PubMed] [Google Scholar]
- 5.Coates D, Siviter R, Isaac RE. Exploring the Caenorhabditis elegans and Drosophila melanogaster genomes to understand neuropeptide and peptidase function. Biochem Soc Trans (2000) 28:464–9 10.1042/0300-5127:0280464 [DOI] [PubMed] [Google Scholar]
- 6.Hauser F, Neupert S, Williamson M, Predel R, Tanaka Y, Grimmelikhuijzen CJ. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J Proteome Res (2010) 9:5296–310 10.1021/pr100570j [DOI] [PubMed] [Google Scholar]
- 7.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res (2001) 11:1126–42 10.1101/gr.169901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science (2006) 314:647–9 10.1126/science.1124128 [DOI] [PubMed] [Google Scholar]
- 9.Predel R, Neupert S, Garczynski SF, Crim JW, Brown MR, Russell WK, et al. Neuropeptidomics of the mosquito Aedes aegypti. J Proteome Res (2010) 9:2006–15 10.1021/pr901187p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riehle MA, Garczynski SF, Crim JW, Hill CA, Brown MR. Neuropeptides and peptide hormones in Anopheles gambiae. Science (2002) 298:172–5 10.1126/science.1076827 [DOI] [PubMed] [Google Scholar]
- 11.Roller L, Yamanaka N, Watanabe K, Daubnerová I, Žitnan D, Kataoka H, et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol (2008) 38:1147–57 10.1016/j.ibmb.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 12.Southey BR, Sweedler JV, Rodriguez-Zas SL. Prediction of neuropeptide cleavage sites in insects. Bioinformatics (2008) 24:815–25 10.1093/bioinformatics/btn044 [DOI] [PubMed] [Google Scholar]
- 13.Veenstra JA, Rombauts S, Grbic M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem Mol Biol (2012) 42:277–95 10.1016/j.ibmb.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 14.Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res (2008) 18:113–22 10.1101/gr.6714008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickerson M, McCormick J, Mispelon M, Paisley K, Nichols R. Structure-activity and immunochemical data provide evidence of developmental- and tissue-specific myosuppressin signaling. Peptides (2012) 36:272–9 10.1016/j.peptides.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser F, Williamson M, Cazzamali G, Grimmelikhuijzen CJ. Identifying neuropeptide and protein hormone receptors in Drosophila melanogaster by exploiting genomic data. Brief Funct Genomic Proteomic (2006) 4:321–30 10.1093/bfgp/eli003 [DOI] [PubMed] [Google Scholar]
- 17.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJ. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol (2006) 80:1–19 10.1016/j.pneurobio.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 18.Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol (2008) 29:142–65 10.1016/j.yfrne.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Huang H, Deng X, He X, Yang J, Yang H, et al. Identification and functional characterization of two orphan G-protein-coupled receptors for adipokinetic hormones from silkworm Bombyx mori. J Biol Chem (2011) 286:42390–402 10.1074/jbc.M111.275602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka N, Yamamoto S, Žitnan D, Watanabe K, Kawada T, Satake H, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One (2008) 3:e3048. 10.1371/journal.pone.0003048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Y, Sun P, Wang Y, He X, Deng X, Chen X, et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem Mol Biol (2010) 40:581–91 10.1016/j.ibmb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 22.Taghert PH, Veenstra JA. Drosophila neuropeptide signaling. Adv Genet (2003) 49:1–65 10.1016/S0065-2660(03)01001-0 [DOI] [PubMed] [Google Scholar]
- 23.Schöller S, Belmont M, Cazzamali G, Hauser F, Williamson M, Grimmelikhuijzen CJ. Molecular identification of a myosuppressin receptor from the malaria mosquito Anopheles gambiae. Biochem Biophys Res Commun (2005) 327:29–34 10.1016/J.Bbrc.2004.11.139 [DOI] [PubMed] [Google Scholar]
- 24.Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, et al. G protein coupled receptors in Anopheles gambiae. Science (2002) 298:176–8 10.1126/Science.1076196 [DOI] [PubMed] [Google Scholar]
- 25.Metpally RP, Sowdhamini R. Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: application to functional annotation of orphan receptors. BMC Genomics (2005) 6:106. 10.1186/1471-2164-6-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science (2002) 298:129–49 10.1126/Science.1076181 [DOI] [PubMed] [Google Scholar]
- 27.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science (2007) 316:1718–23 10.1126/Science.1138878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science (2010) 330:86–8 10.1126/Science.1191864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardoso JC, Felix RC, Fonseca VG, Power DM. Feeding and the rhodopsin family g-protein coupled receptors in nematodes and arthropods. Front Endocrinol (Lausanne) (2012) 3:157. 10.3389/fendo.2012.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joost P, Methner A. Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol (2002) 3(11):RESEARCH006. 10.1186/gb-2002-3-11-research0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol (2011) 7:e1002195. 10.1371/Journal.Pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res (2011) 39:D283–8 10.1093/Nar/Gkq930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng (1994) 7:195–203 10.1093/protein/7.2.195 [DOI] [PubMed] [Google Scholar]
- 34.Brody T, Cravchik A. Drosophila melanogaster G protein-coupled receptors. J Cell Biol (2000) 150:F83–8 10.1083/Jcb.150.2.F83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics (2004) 20:3702–4 10.1093/Bioinformatics/Bth444 [DOI] [PubMed] [Google Scholar]
- 36.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res (2005) 33:511–8 10.1093/Nar/Gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res (2004) 32:1792–7 10.1093/Nar/Gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics (2009) 25:1189–91 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol (2010) 59:307–21 10.1093/Sysbio/Syq010 [DOI] [PubMed] [Google Scholar]
- 40.Merten N, Beck-Sickinger A. Molecular ligand-receptor interaction of the NPY/PP peptide family. In: Zukowska Z, Feuerstein G, editors. NPY Family of Peptides in Neurobiology, Cardiovascular and Metabolic Disorders: from Genes to Therapeutics (Vol. 95). Basel: Birkhäuser; (2006). p. 35–62 [Google Scholar]
- 41.Åkerberg H, Fällmar H, Sjödin P, Boukharta L, Gutiérrez-de-Terán H, Lundell I, et al. Mutagenesis of human neuropeptide Y/peptide YY receptor Y2 reveals additional differences to Y1 in interactions with highly conserved ligand positions. Regul Pept (2010) 163:120–9 10.1016/j.regpep.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 42.Berglund MM, Fredriksson R, Salaneck E, Larhammar D. Reciprocal mutations of neuropeptide Y receptor Y2 in human and chicken identify amino acids important for antagonist binding. FEBS Lett (2002) 518:5–9 10.1016/S0014-5793(02)02534-6 [DOI] [PubMed] [Google Scholar]
- 43.Fällmar H, Åkerberg H, Gutiérrez-de-Terán H, Lundell I, Mohell N, Larhammar D. Identification of positions in the human neuropeptide Y/peptide YY receptor Y2 that contribute to pharmacological differences between receptor subtypes. Neuropeptides (2011) 45:293–300 10.1016/j.npep.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 44.Wiegmann BM, Yeates DK, Thorne JL, Kishino H. Time flies, a new molecular time-scale for brachyceran fly evolution without a clock. Syst Biol (2003) 52:745–56 10.1080/10635150390250965 [DOI] [PubMed] [Google Scholar]
- 45.Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol (2006) 39:417–23 10.1016/J.Ympev.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 46.Reidenbach KR, Cook S, Bertone MA, Harbach RE, Wiegmann BM, Besansky NJ. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol (2009) 9:298. 10.1186/1471-2148-9-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassilatis DK, Hohmann JG, Zeng H, Li FS, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A (2003) 100:4903–8 10.1073/Pnas.0230374100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Hiel MB, Vandersmissen HP, Van Loy T, Broeck JV. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides (2012) 34:193–200 10.1016/J.Peptides.2011.11.004 [DOI] [PubMed] [Google Scholar]
- 49.Sudo S, Kuwabara Y, Park JI, Hsu SY, Hsueh AJ. Heterodimeric fly glycoprotein hormone-α2 (GPA2) and glycoprotein hormone-β5 (GPB5) activate fly leucine-rich repeat-containing G protein-coupled receptor-1 (DLGR1) and stimulation of human thyrotropin receptors by chimeric fly GPA2 and human GPB5. Endocrinology (2005) 146:3596–604 10.1210/En.2005-0317 [DOI] [PubMed] [Google Scholar]
- 50.Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, et al. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci U S A (2005) 102:2820–5 10.1073/Pnas.0409916102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, et al. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett (2005) 579:2171–6 10.1016/J.FEBSlet.2005.03.006 [DOI] [PubMed] [Google Scholar]
- 52.Zhu Y, Rice CD, Pang Y, Pace M, Thomas P. Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc Natl Acad Sci U S A (2003) 100:2231–6 10.1073/pnas.0336132100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava DP, Yu EJ, Kennedy K, Chatwin H, Reale V, Hamon M, et al. Rapid, nongenomic responses to ecdysteroids and catecholamines mediated by a novel Drosophila G-protein-coupled receptor. J Neurosci (2005) 25:6145–55 10.1523/JNEUROSCI.1005-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody Encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell (2005) 123:145–56 10.1016/J.Cell.2005.07.029 [DOI] [PubMed] [Google Scholar]
- 55.Hillyer JF, Estévez-Lao TY, Funkhouser LJ, Aluoch VA. Anopheles gambiae corazonin: gene structure, expression and effect on mosquito heart physiology. Insect Mol Biol (2012) 21:343–55 10.1111/J.1365-2583.2012.01140.X [DOI] [PubMed] [Google Scholar]
- 56.Estévez-Lao TY, Boyce DS, Honegger HW, Hillyer JF. Cardioacceleratory function of the neurohormone CCAP in the mosquito Anopheles gambiae. J Exp Biol (2013) 216:601–13 10.1242/Jeb.077164 [DOI] [PubMed] [Google Scholar]
- 57.Kapan N, Lushchak OV, Luo JN, Nässel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci (2012) 69:4051–66 10.1007/S00018-012-1097-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nässel DR, Winther AM. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol (2010) 92:42–104 10.1016/J.Pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 59.Hansen KK, Stafflinger E, Schneider M, Hauser F, Cazzamali G, Williamson M, et al. Discovery of a novel insect neuropeptide signaling system closely related to the insect adipokinetic hormone and corazonin hormonal systems. J Biol Chem (2010) 285:10736–47 10.1074/Jbc.M109.045369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park Y, Filippov V, Gill SS, Adams ME. Deletion of the ecdysis-triggering hormone gene leads to lethal ecdysis deficiency. Development (2002) 129:493–503 [DOI] [PubMed] [Google Scholar]
- 61.Kaufmann C, Merzendorfer H, Gäde G. The adipokinetic hormone system in Culicinae (Diptera: Culicidae): molecular identification and characterization of two adipokinetic hormone (AKH) precursors from Aedes aegypti and Culex pipiens and two putative AKH receptor variants from Ae. aegypti. Insect Biochem Mol Biol (2009) 39:770–81 10.1016/J.Ibmb.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 62.Kaufmann C, Brown MR. Regulation of carbohydrate metabolism and flight performance by a hypertrehalosaemic hormone in the mosquito Anopheles gambiae. J Insect Physiol (2008) 54:367–77 10.1016/J.Jinsphys.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caers J, Peeters L, Janssen T, De Haes W, Gäde G, Schoofs L. Structure-activity studies of Drosophila adipokinetic hormone (AKH) by a cellular expression system of dipteran AKH receptors. Gen Comp Endocrinol (2012) 177:332–7 10.1016/J.Ygcen.2012.04.025 [DOI] [PubMed] [Google Scholar]
- 64.Mugumbate G, Jackson GE, van der Spoel D. Open conformation of adipokinetic hormone receptor from the malaria mosquito facilitates hormone binding. Peptides (2011) 32:553–9 10.1016/J.Peptides.2010.08.017 [DOI] [PubMed] [Google Scholar]
- 65.Nouzova M, Brockhoff A, Mayoral JG, Goodwin M, Meyerhof W, Noriega FG. Functional characterization of an allatotropin receptor expressed in the corpora allata of mosquitoes. Peptides (2012) 34:201–8 10.1016/J.Peptides.2011.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Söderberg JA, Birse RT, Nassel DR. Insulin production and signaling in renal tubules of Drosophila is under control of tachykinin-related peptide and regulates stress resistance. PLoS One (2011) 6:e19866. 10.1371/journal.pone.0019866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kubiak TM, Larsen MJ, Burton KJ, Bannow CA, Martin RA, Zantello MR, et al. Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem Biophys Res Commun (2002) 291:313–20 10.1006/Bbrc.2002.6459 [DOI] [PubMed] [Google Scholar]
- 68.Collin C, Hauser F, Krogh-Meyer P, Hansen KK, de Valdivia EG, Williamson M, et al. Identification of the Drosophila and Tribolium receptors for the recently discovered insect RYamide neuropeptides. Biochem Biophys Res Commun (2011) 412:578–83 10.1016/J.Bbrc.2011.07.131 [DOI] [PubMed] [Google Scholar]
- 69.Ida T, Takahashi T, Tominaga H, Sato T, Kume K, Yoshizawa-Kumagaye K, et al. Identification of the endogenous cysteine-rich peptide trissin, a ligand for an orphan G protein-coupled receptor in Drosophila. Biochem Biophys Res Commun (2011) 414:44–8 10.1016/J.Bbrc.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 70.Siviter RJ, Coast GM, Winther AM, Nachman RJ, Taylor CA, Shirras AD, et al. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J Biol Chem (2000) 275:23273–80 10.1074/Jbc.M002875200 [DOI] [PubMed] [Google Scholar]
- 71.Birse RT, Söderberg JA, Luo JL, Winther AM, Nässel DR. Regulation of insulin-producing cells in the adult Drosophila brain via the tachykinin peptide receptor DTKR. J Exp Biol (2011) 214:4201–8 10.1242/Jeb.062091 [DOI] [PubMed] [Google Scholar]
- 72.Birse R, Winther AM, Nässel DR. Role of tachykinin-related peptide signaling in response to metabolic stress. J Neurogenet (2006) 20(3–4):89 [Google Scholar]
- 73.Li XJ, Wolfgang W, Wu YN, North RA, Forte M. Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J (1991) 10:3221–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poels J, Birse RT, Nachman RJ, Fichna J, Janecka A, Broeck JV, et al. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides (2009) 30:545–56 10.1016/J.Peptides.2008.10.012 [DOI] [PubMed] [Google Scholar]
- 75.Poels J, Verlinden H, Fichna J, Van Loy T, Franssens V, Studzian K, et al. Functional comparison of two evolutionary conserved insect neurokinin-like receptors. Peptides (2007) 28:103–8 10.1016/J.Peptides.2006.06.014 [DOI] [PubMed] [Google Scholar]
- 76.Van Loy T, Vandersmissen HP, Poels J, Van Hiel MB, Verlinden H, Vanden Broeck J. Tachykinin-related peptides and their receptors in invertebrates: a current view. Peptides (2010) 31:520–4 10.1016/J.Peptides.2009.09.023 [DOI] [PubMed] [Google Scholar]
- 77.Jiang H, Lkhagva A, Daubnerová I, Chae HS, Simo L, Jung SH, et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc Natl Acad Sci U S A (2013) 110:3526–34 10.1073/pnas.1310676110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beyenbach KW, Piermarini PM. Transcellular and paracellular pathways of transepithelial fluid secretion in Malpighian (renal) tubules of the yellow fever mosquito Aedes aegypti. Acta Physiol (2011) 202:387–407 10.1111/J.1748-1716.2010.02195.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cabrero P, Richmond L, Nitabach M, Davies SA, Dow JA. A biogenic amine and a neuropeptide act identically: tyramine signals through calcium in Drosophila tubule stellate cells. Proc Biol Sci (2013) 280:20122943. 10.1098/Rspb.2012.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kersch CN, Pietrantonio PV. Mosquito Aedes aegypti (L.) leucokinin receptor is critical for in vivo fluid excretion post blood feeding. FEBS Lett (2011) 585:3507–12 10.1016/J.FEBSlet.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 81.Lu HL, Kersch C, Pietrantonio PV. The kinin receptor is expressed in the Malpighian tubule stellate cells in the mosquito Aedes aegypti (L.): a new model needed to explain ion transport? Insect Biochem Mol Biol (2011) 41:135–40 10.1016/J.Ibmb.2010.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Radford JC, Davies SA, Dow JA. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J Biol Chem (2002) 277(41):38810–7 10.1074/Jbc.M203694200 [DOI] [PubMed] [Google Scholar]
- 83.Stafflinger E, Hansen KK, Hauser F, Schneider M, Cazzamali G, Williamson M, et al. Cloning and identification of the first oxytocin/vasopressin-like receptor and its ligand from insects. Proc Natl Acad Sci U S A (2008) 105:3262–7 10.1073/Pnas.0710897105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides (2011) 32:1335–55 10.1016/J.Peptides.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 85.Garczynski SF, Crim JW, Brown MR. Characterization of neuropeptide F and its receptor from the African malaria mosquito, Anopheles gambiae. Peptides (2005) 26:99–107 10.1016/J.Peptides.2004.07.014 [DOI] [PubMed] [Google Scholar]
- 86.Onken H, Moffett SB, Moffett DE. The anterior stomach of larval mosquitoes (Aedes aegypti): effects of neuropeptides on transepithelial ion transport and muscular motility. J Exp Biol (2004) 207:3731–9 10.1242/Jeb.01208 [DOI] [PubMed] [Google Scholar]
- 87.Stanek DM, Pohl J, Crim JW, Brown MR. Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides (2002) 23:1367–78 10.1016/S0196-9781(02)00074-8 [DOI] [PubMed] [Google Scholar]
- 88.Matsumoto S, Brown MR, Suzuki A, Lea AO. Isolation and characterization of ovarian ecdysteroidogenic hormones from the mosquito, Aedes aegypti. Insect Biochem (1989) 19:651–6 10.1016/0020-1790(89)90100-5 [DOI] [Google Scholar]
- 89.Stracker TH, Thompson S, Grossman GL, Riehle MA, Brown MR. Characterization of the AeaHP gene and its expression in the mosquito Aedes aegypti (Diptera: Culicidae). J Med Entomol (2002) 39:331–42 10.1603/0022-2585-39.2.331 [DOI] [PubMed] [Google Scholar]
- 90.Brown MR, Klowden MJ, Crim JW, Young L, Shrouder LA, Lea AO. Endogenous regulation of mosquito host-seeking behavior by a neuropeptide. J Insect Physiol (1994) 40:399–406 10.1016/0022-1910(94)90158-9 [DOI] [Google Scholar]
- 91.Naccarati C, Audsley N, Keen JN, Kim JH, Howell GJ, Kim YJ, et al. The host-seeking inhibitory peptide, Aea-HP-1, is made in the male accessory gland and transferred to the female during copulation. Peptides (2012) 34:150–7 10.1016/J.Peptides.2011.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides (2002) 23:773–80 10.1016/S0196-9781(01)00647-7 [DOI] [PubMed] [Google Scholar]
- 93.Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun (2002) 297:1140–8 10.1016/S0006-291X(02)02351-3 [DOI] [PubMed] [Google Scholar]
- 94.Liesch J, Bellani LL, Vosshall LB. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis (2013) 7:e2486. 10.1371/journal.pntd.0002486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jørgensen LM, Hauser F, Cazzamali G, Williamson M, Grimmelikhuijzen CJ. Molecular identification of the first SIFamide receptor. Biochem Biophys Res Commun (2006) 340:696–701 10.1016/J.Bbrc.2005.12.062 [DOI] [PubMed] [Google Scholar]
- 96.Hansen KK, Hauser F, Williamson M, Weber SB, Grimmelikhuijzen CJ. The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem Biophys Res Commun (2011) 404:184–9 10.1016/J.Bbrc.2010.11.089 [DOI] [PubMed] [Google Scholar]
- 97.Stay B, Tobe SS. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu Rev Entomol (2007) 52:277–99 10.1146/Annurev.Ento.51.110104.151050 [DOI] [PubMed] [Google Scholar]
- 98.Bendena WG, Donly BC, Tobe SS. Allatostatins: a growing family of neuropeptides with structural and functional diversity. Ann N Y Acad Sci (1999) 897:311–29 10.1111/J.1749-6632.1999.Tb07902.X [DOI] [PubMed] [Google Scholar]
- 99.Lenz C, Williamson M, Grimmelikhuijzen CJ. Molecular cloning and genomic organization of a second probable allatostatin receptor from Drosophila melanogaster. Biochem Biophys Res Commun (2000) 273:571–7 10.1006/Bbrc.2000.2964 [DOI] [PubMed] [Google Scholar]
- 100.Nässel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog Neurobiol (2002) 68:1–84 10.1016/S0301-0082(02)00057-6 [DOI] [PubMed] [Google Scholar]
- 101.Veenstra JA, Pattillo JM, Petzel DH. A single cDNA encodes all three Aedes leucokinins, which stimulate both fluid secretion by the Malpighian tubules and hindgut contractions. J Biol Chem (1997) 272:10402–7 10.1074/jbc.272.16.10402 [DOI] [PubMed] [Google Scholar]
- 102.Kreienkamp HJ, Larusson HJ, Witte I, Roeder T, Birgul N, Hönck HH, et al. Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, from Drosophila melanogaster and their ligands by reverse pharmacology. J Biol Chem (2002) 277:39937–43 10.1074/Jbc.M206931200 [DOI] [PubMed] [Google Scholar]
- 103.Mayoral JG, Nouzova M, Brockhoff A, Goodwin M, Hernandez-Martinez S, Richter D, et al. Allatostatin-C receptors in mosquitoes. Peptides (2010) 31:442–50 10.1016/J.Peptides.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li YP, Hernandez-Martinez S, Noriega FG. Inhibition of juvenile hormone biosynthesis in mosquitoes: effect of allatostatic head factors, PISCF- and YXFGL-amide-allatostatins. Regul Pept (2004) 118:175–82 10.1016/J.Regpep.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 105.Wang C, Chin-Sang I, Bendena WG. The FGLamide-allatostatins influence foraging behavior in Drosophila melanogaster. PLoS One (2012) 7:e36059. 10.1371/journal.pone.0036059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kean L, Cazenave W, Costes L, Broderick KE, Graham S, Pollock VP, et al. Two nitridergic peptides are encoded by the gene capability in Drosophila melanogaster. Am J Physiol (2002) 282:R1297–307 10.1152/ajpregu.00584.2001 [DOI] [PubMed] [Google Scholar]
- 107.Dai L, Adams ME. Ecdysis triggering hormone signaling in the yellow fever mosquito Aedes aegypti. Gen Comp Endocrinol (2009) 162:43–51 10.1016/J.Ygcen.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olsen SS, Cazzamali G, Williamson M, Grimmelikhuijzen CJ, Hauser F. Identification of one capa and two pyrokinin receptors from the malaria mosquito Anopheles gambiae. Biochem Biophys Res Commun (2007) 362:245–51 10.1016/J.Bbre.2007.06.190 [DOI] [PubMed] [Google Scholar]
- 109.Cazzamali G, Torp M, Hauser F, Williamson M, Grimmelikhuijzen CJ. The Drosophila gene CG9918 codes for a pyrokinin-1 receptor. Biochem Biophys Res Commun (2005) 335:14–9 10.1016/J.Bbrc.2005.07.038 [DOI] [PubMed] [Google Scholar]
- 110.Jurenka R, Nusawardani T. The pyrokinin/pheromone biosynthesis-activating neuropeptide (PBAN) family of peptides and their receptors in Insecta: evolutionary trace indicates potential receptor ligand-binding domains. Insect Mol Biol (2011) 20:323–34 10.1111/J.1365-2583.2010.01065.X [DOI] [PubMed] [Google Scholar]
- 111.Iversen A, Cazzamali G, Williamson M, Hauser F, Grimmelikhuijzen CJ. Molecular identification of the first insect ecdysis triggering hormone receptors. Biochem Biophys Res Commun (2002) 299:924–31 10.1016/S0006-291x(02)02798-5 [DOI] [PubMed] [Google Scholar]
- 112.Park Y, Kim YJ, Dupriez V, Adams ME. Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J Biol Chem (2003) 278:17710–5 10.1074/Jbc.M301119200 [DOI] [PubMed] [Google Scholar]
- 113.Roller L, Zitnanová I, Dai L, Šimo L, Park Y, Satake H, et al. Ecdysis triggering hormone signaling in arthropods. Peptides (2010) 31:429–41 10.1016/J.Peptides.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Messer AC, Brown MR. Nonlinear dynamics of neurochemical modulation of mosquito oviduct and hindgut contractions. J Exp Biol (1995) 198:2325–36 [DOI] [PubMed] [Google Scholar]
- 115.Johnson EC, Bohn LM, Barak LS, Birse RT, Nässel DR, Caron MG, et al. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J Biol Chem (2003) 278:52172–8 10.1074/Jbc.M306756200 [DOI] [PubMed] [Google Scholar]
- 116.Meeusen T, Mertens I, Clynen E, Baggerman G, Nichols R, Nachman RJ, et al. Identification in Drosophila melanogaster of the invertebrate G protein-coupled FMRFamide receptor. Proc Natl Acad Sci U S A (2002) 99:15363–8 10.1073/Pnas.252339599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duttlinger A, Mispelon M, Nichols R. The structure of the FMRFamide receptor and activity of the cardioexcitatory neuropeptide are conserved in mosquito. Neuropeptides (2003) 37:120–6 10.1016/S0143-4179(03)00023-4 [DOI] [PubMed] [Google Scholar]
- 118.Nichols R. Signaling pathways and physiological functions of Drosophila melanogaster FMRFamide-related peptides. Annu Rev Entomol (2003) 48:485–503 10.1146/Annurev.Ento.48.091801.112525 [DOI] [PubMed] [Google Scholar]
- 119.Poels J, Van Loy T, Vandersmissen HP, Van Hiel B, Van Soest S, Nachman RJ, et al. Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cell Mol Life Sci (2010) 67:3511–22 10.1007/S00018-010-0393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim YJ, Bartalska K, Audsley N, Yamanaka N, Yapici N, Lee JY, et al. MIPs are ancestral ligands for the sex peptide receptor. Proc Natl Acad Sci U S A (2010) 107:6520–5 10.1073/Pnas.0914764107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Egerod K, Reynisson E, Hauser F, Cazzamali G, Williamson M, Grimmelikhuijzen CJ. Molecular cloning and functional expression of the first two specific insect myosuppressin receptors. Proc Natl Acad Sci U S A (2003) 100:9808–13 10.1073/Pnas.1632197100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Knipper M, Madeddu L, Breer H, Meldolesi J. Black widow spider venom-induced release of neurotransmitters: mammalian synaptosomes are stimulated by a unique venom component (alpha-latrotoxin), insect synaptosomes by multiple components. Neuroscience (1986) 19:55–62 10.1016/0306-4522(86)90005-9 [DOI] [PubMed] [Google Scholar]
- 123.Silva JP, Ushkaryov YA. The latrophilins, “split-personality” receptors. Adv Exp Med Biol (2010) 706:59–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rohou A, Nield J, Ushkaryov YA. Insecticidal toxins from black widow spider venom. Toxicon (2007) 49:531–49 10.1016/j.toxicon.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, et al. Latrophilin 1 and its endogenous ligand lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A (2011) 108:12113–8 10.1073/pnas.1019434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li CJ, Chen M, Sang M, Liu X, Wu W, Li B. Comparative genomic analysis and evolution of family-B G protein-coupled receptors from six model insect species. Gene (2013) 519:1–12 10.1016/J.Gene.2013.01.061 [DOI] [PubMed] [Google Scholar]
- 127.Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM. Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol (2006) 6:108. 10.1186/1471-2148-6-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Das S, Dimopoulos G. Molecular analysis of light pulse stimulated blood feeding inhibition in Anopheles gambiae. Am J Trop Med Hyg (2008) 79:333. 10.1186/1472-6793-8-23 [DOI] [Google Scholar]
- 129.Hector CE, Bretz CA, Zhao Y, Johnson EC. Functional differences between two CRF-related diuretic hormone receptors in Drosophila. J Exp Biol (2009) 212:3142–7 10.1242/Jeb.033175 [DOI] [PubMed] [Google Scholar]
- 130.Clark TM, Hayes TK, Beyenbach KW. Dose-dependent effects of CRF-like diuretic peptide on transcellular and paracellular transport pathways. Am J Physiol (1998) 274:F834–40 [DOI] [PubMed] [Google Scholar]
- 131.Jagge CL, Pietrantonio PV. Diuretic hormone 44 receptor in Malpighian tubules of the mosquito Aedes aegypti: evidence for transcriptional regulation paralleling urination. Insect Mol Biol (2008) 17:413–26 10.1111/J.1365-2583.2008.00817.X [DOI] [PubMed] [Google Scholar]
- 132.Zandawala M. Calcitonin-like diuretic hormones in insects. Insect Biochem Mol Biol (2012) 42:816–25 10.1016/J.Ibmb.2012.06.006 [DOI] [PubMed] [Google Scholar]
- 133.Coast GM, Garside CS, Webster SG, Schegg KM, Schooley DA. Mosquito natriuretic peptide identified as a calcitonin-like diuretic hormone in Anopheles gambiae (Giles). J Exp Biol (2005) 208:3281–91 10.1242/Jeb.01760 [DOI] [PubMed] [Google Scholar]
- 134.Kwon H, Lu HL, Longnecker MT, Pietrantonio PV. Role in diuresis of a calcitonin receptor (GPRCAL1) expressed in a distal-proximal gradient in renal organs of the mosquito Aedes aegypti (L.). PLoS One (2012) 7:e50374. 10.1371/journal.pone.0050374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant Methuselah. Science (1998) 282:943–6 10.1126/Science.282.5390.943 [DOI] [PubMed] [Google Scholar]
- 136.West AP, Llamas LL, Snow PM, Benzer S, Bjorkman PJ. Crystal structure of the ectodomain of Methuselah, a Drosophila G protein-coupled receptor associated with extended lifespan. Proc Natl Acad Sci U S A (2001) 98:3744–9 10.1073/Pnas.051625298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cvejic S, Zhu Z, Felice SJ, Berman Y, Huang XY. The endogenous ligand stunted of the GPCR Methuselah extends lifespan in Drosophila. Nat Cell Biol (2004) 6:540–6 10.1038/Ncb1133 [DOI] [PubMed] [Google Scholar]
- 138.Ja WW, Carvalho GB, Madrigal M, Roberts RW, Benzer S. The Drosophila G protein-coupled receptor, Methuselah, exhibits a promiscuous response to peptides. Protein Sci (2009) 18:2203–8 10.1002/Pro.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sugita S, Ichtchenko K, Khvotchev M, Südhof TC. α-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors – G-protein coupling not required for triggering exocytosis. J Biol Chem (1998) 273:32715–24 10.1074/Jbc.273.49.32715 [DOI] [PubMed] [Google Scholar]
- 140.Li YL, Hoxha V, Lama C, Dinh BH, Vo CN, Dauwalder B. The hector G-protein coupled receptor is required in a subset of fruitless neurons for male courtship behavior. PLoS One (2011) 6:e28269. 10.1371/journal.pone.0028269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baggerman G, Boonen K, Verleyen P, De Loof A, Schoofs L. Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J Mass Spectrom (2005) 40:250–60 10.1002/Jms.744 [DOI] [PubMed] [Google Scholar]
- 142.Overend G, Cabrero P, Guo AX, Sebastian S, Cundall M, Armstrong H, et al. The receptor guanylate cyclase Gyc76C and a peptide ligand, NPLP1-VQQ, modulate the innate immune IMD pathway in response to salt stress. Peptides (2012) 34:209–18 10.1016/J.Peptides.2011.08.019 [DOI] [PubMed] [Google Scholar]
- 143.Chang JC, Yang RB, Adams ME, Lu KH. Receptor guanylyl cyclases in Inka cells targeted by eclosion hormone. Proc Natl Acad Sci U S A (2009) 106:13371–6 10.1073/Pnas.0812593106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol (2006) 51:1–24 10.1146/annurev.ento.51.110104.151011 [DOI] [PubMed] [Google Scholar]
- 145.Fernandez-Almonacid R, Rosen OM. Structure and ligand specificity of the Drosophila melanogaster insulin receptor. Mol Cell Biol (1987) 7:2718–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petruzzelli L, Herrera R, Garcia-Arenas R, Rosen OM. Acquisition of insulin-dependent protein tyrosine kinase-activity during Drosophila embryogenesis. J Biol Chem (1985) 260:6072–5 [PubMed] [Google Scholar]
- 147.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol (2001) 11:213–21 10.1016/S0960-9822(01)00068-9 [DOI] [PubMed] [Google Scholar]
- 148.Antonova Y, Arik AJ, Moore W, Riehle MR, Brown MR. Insulin-like peptides: structure, signaling, and function. In: Gilbert LI, editor. Insect Endocrinology. New York, NY: Elsevier Academic Press; (2012). p. 63–92 [Google Scholar]
- 149.Wen Z, Gulia M, Clark KD, Dhara A, Crim JW, Strand MR, et al. Two insulin-like peptide family members from the mosquito Aedes aegypti exhibit differential biological and receptor binding activities. Mol Cell Endocrinol (2010) 328:47–55 10.1016/j.mce.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sajid W, Kulahin N, Schluckebier G, Ribel U, Henderson HR, Tatar M, et al. Structural and biological properties of the Drosophila insulin-like peptide 5 show evolutionary conservation. J Biol Chem (2011) 286:661–73 10.1074/Jbc.M110.156018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, et al. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A (2008) 105:5716–21 10.1073/Pnas.0800478105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gulia-Nuss M, Eum JH, Strand MR, Brown MR. Ovary ecdysteroidogenic hormone activates egg maturation in the mosquito Georgecraigius atropalpus after adult eclosion or a blood meal. J Exp Biol (2012) 215:3758–67 10.1242/Jeb.074617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gulia-Nuss M, Robertson AE, Brown MR, Strand MR. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS One (2011) 6:e20401. 10.1371/journal.pone.0020401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Castillo J, Brown MR, Strand MR. Blood feeding and insulin-like peptide 3 stimulate proliferation of hemocytes in the mosquito Aedes aegypti. PLoS Pathog (2011) 7:e1002274. 10.1371/Journal.Ppat.1002274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gouzi JY, Moressis A, Walker JA, Apostolopoulou AA, Palmer RH, Bernards A, et al. The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet (2011) 7:e1002281. 10.1371/Journal.Pgen.1002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, Brush G, et al. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS One (2011) 6:e22636. 10.1371/journal.pone.0022636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lorén CE, Scully A, Grabbe C, Edeen PT, Thomas J, McKeown M, et al. Identification and characterization of DAlk: a novel Drosophila melanogaster RTK which drives ERK activation in vivo. Genes Cells (2001) 6:531–44 10.1046/J.1365-2443.2001.00440.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J (2009) 420:345–61 10.1042/Bj20090387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang HL, Eriksson T, Vernersson E, Vigny M, Hallberg B, Palmer RH. The ligand jelly belly (Jeb) activates the Drosophila Alk RTK to drive PC12 cell differentiation, but is unable to activate the mouse ALK RTK. J Exp Zool B Mol Dev Evol (2007) 308(3):269–82 10.1002/Jez.B.21146 [DOI] [PubMed] [Google Scholar]
- 160.Ahier A, Rondard P, Gouignard N, Khayath N, Huang S, Trolet J, et al. A new family of receptor tyrosine kinases with a venus flytrap binding domain in insects and other invertebrates activated by amino acids. PLoS One (2009) 4:e5651. 10.1371/journal.pone.0005651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vicogne J, Pin JP, Lardans V, Capron M, Noël C, Dissous C. An unusual receptor tyrosine kinase of Schistosoma mansoni contains a venus flytrap module. Mol Biochem Parasitol (2003) 126:51–62 10.1016/S0166-6851(02)00249-9 [DOI] [PubMed] [Google Scholar]
- 162.McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, et al. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell (2007) 13:857–71 10.1016/J.Devcel.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science (2009) 326:1403–5 10.1126/Science.1176450 [DOI] [PubMed] [Google Scholar]
- 164.Smith W, Rybczynski R. Prothoracicotropic hormone. In: Gilbert LI, editor. Insect Endocrinology. New York, NY: Elsevier Academic Press; (2012). p. 1–62 [Google Scholar]
- 165.Telang A, Peterson B, Frame L, Baker E, Brown MR. Analysis of molecular markers for metamorphic competency and their response to starvation or feeding in the mosquito, Aedes aegypti (Diptera: Culicidae). J Insect Physiol (2010) 56:1925–34 10.1016/j.jinsphys.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Whisenton LR, Kelly TJ, Bollenbacher WE. Multiple forms of cerebral peptides with steroidogenic functions in pupal and adult brains of the yellow fever mosquito, Aedes aegypti. Mol Cell Endocrinol (1987) 50:3–14 10.1016/0303-7207(87)90071-2 [DOI] [PubMed] [Google Scholar]
- 167.Zhang Q, Denlinger DL. Molecular structure of the prothoracicotropic hormone gene in the northern house mosquito, Culex pipiens, and its expression analysis in association with diapause and blood feeding. Insect Mol Biol (2011) 20:201–13 10.1111/j.1365-2583.2010.01058.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jenkins SP, Brown MR, Lea AO. Inactive prothoracic glands in larvae and pupae of Aedes aegypti: ecdysteroid release by tissues in the thorax and abdomen. Insect Biochem Mol Biol (1992) 22:553–9 10.1016/0965-1748(92)90032-A [DOI] [Google Scholar]
- 169.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development (2011) 138:1045–55 10.1242/Dev.056671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shilo BZ. Regulating the dynamics of EGF receptor signaling in space and time. Development (2005) 132:4017–27 10.1242/Dev.02006 [DOI] [PubMed] [Google Scholar]
- 171.Lycett G, Blass C, Louis C. Developmental variation in epidermal growth factor receptor size and localization in the malaria mosquito, Anopheles gambiae. Insect Mol Biol (2001) 10:619–28 10.1046/J.0962-1075.2001.00302.X [DOI] [PubMed] [Google Scholar]
- 172.Bond D, Foley E. Autocrine platelet-derived growth factor-vascular endothelial growth factor receptor-related (Pvr) pathway activity controls intestinal stem cell proliferation in the adult Drosophila midgut. J Biol Chem (2012) 287:27359–70 10.1074/Jbc.M112.378018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cho NK, Keyes L, Johnson E, Heller J, Ryner L, Karim F, et al. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell (2002) 108:865–76 10.1016/S0092-8674(02)00676-1 [DOI] [PubMed] [Google Scholar]
- 174.Kadam S, McMahon A, Tzou P, Stathopoulos A. FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development (2009) 136:739–47 10.1242/Dev.027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hahn M, Bishop JM. Expression pattern of Drosophila ret suggests a common ancestral origin between the metamorphosis precursors in insect endoderm and the vertebrate enteric neurons. Proc Natl Acad Sci U S A (2001) 98:1053–8 10.1073/Pnas.021558598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vidal M, Wells S, Ryan A, Cagan R. ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res (2005) 65:3538–41 10.1158/0008-5472.Can-04-4561 [DOI] [PubMed] [Google Scholar]
- 177.Yoda A, Oishi I, Minami Y. Expression and function of the Ror-family receptor tyrosine kinases during development: lessons from genetic analyses of nematodes, mice, and humans. J Recept Signal Transduct Res (2003) 23(1):1–15 10.1081/RRS-120018757 [DOI] [PubMed] [Google Scholar]
- 178.Boyle M, Nighorn A, Thomas JB. Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development (2006) 133:1845–54 10.1242/Dev.02353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hinck AP. Structural studies of the TGF-βs and their receptors – insights into evolution of the TGF-β superfamily. FEBS Lett (2012) 586:1860–70 10.1016/j.febslet.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 180.Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol Biol (2009) 9:28. 10.1186/1471-2148-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Massagué J. TGF-β signaling in development and disease. FEBS Lett (2012) 586:1833. 10.1016/j.febslet.2012.05.030 [DOI] [PubMed] [Google Scholar]
- 182.Gesualdi SC, Haerry TE. Distinct signaling of Drosophila activin/TGF-β family members. Fly (Austin) (2007) 1:212–21 [DOI] [PubMed] [Google Scholar]
- 183.Lieber MJ, Luckhart S. Transforming growth factor-βs and related gene products in mosquito vectors of human malaria parasites: signaling architecture for immunological crosstalk. Mol Immunol (2004) 41:965–77 10.1016/J.Molimm.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 184.Gibbens YY, Warren JT, Gilbert LI, O’Connor MB. Neuroendocrine regulation of Drosophila metamorphosis requires TGF-β/activin signaling. Development (2011) 138:2693–703 10.1242/Dev.063412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gregory SJ, Kaiser UB. Regulation of gonadotropins by inhibin and activin. Semin Reprod Med (2004) 22:253–67 10.1055/S-2004-831901 [DOI] [PubMed] [Google Scholar]
- 186.Brown ML, Kimura F, Bonomi LM, Ungerleider NA, Schneyer AL. Differential synthesis and action of TGF-β superfamily ligands in mouse and rat islets. Islets (2011) 3:367–75 10.4161/Isl.3.6.18013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Surachetpong W, Singh N, Cheung KW, Luckhart S. MAPK ERK signaling regulates the TGF-β1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog (2009) 5:e1000366. 10.1371/journal.ppat.1000366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vodovotz Y, Zamora R, Lieber MJ, Luckhart S. Cross-talk between nitric oxide and transforming growth factor-β1 in malaria. Curr Mol Med (2004) 4:787–97 10.2174/1566524043359999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Akerberg H, Fallmar H, Sjodin P, Boukharta L, Gutierrez-de-Teran H, Lundell I, et al. Mutagenesis of human neuropeptide Y/peptide YY receptor Y2 reveals additional differences to Y1 in interactions with highly conserved ligand positions. Regul Pept (2010) 163:120–9 10.1016/J.Regpep.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 190.Merten N, Beck-Sickinger AG. Molecular ligand-receptor interaction of the NPY/PP peptide family. In: Zukowsak Z, Feuerstein GZ, editors. NPY Family of Peptides in Neurobiology, Cardiovascular and Metabolic Disorders: from Genes to Therapeutics. Basel: Birkhäuser Verlag; (2005). p. 35–62 [Google Scholar]
- 191.Walker P, Munoz M, Martinez R, Peitsch MC. Acidic residues in extracellular loops of the human Y1 neuropeptide Y receptor are essential for ligand binding. J Biol Chem (1994) 269:2863–9 [PubMed] [Google Scholar]
- 192.Sautel M, Martinez R, Munoz M, Peitsch MC, Beck-Sickinger AG, Walker P. Role of a hydrophobic pocket of the human Y1 neuropeptide Y receptor in ligand binding. Mol Cell Endocrinol (1995) 112:215–22 10.1016/0303-7207(95)03603-5 [DOI] [PubMed] [Google Scholar]
- 193.Sautel M, Rudolf K, Wittneben H, Herzog H, Martinez R, Munoz M, et al. Neuropeptide Y and the nonpeptide antagonist BIBP 3226 share an overlapping binding site at the human Y1 receptor. Mol Pharmacol (1996) 50:285–92 [PubMed] [Google Scholar]
- 194.Rosenkilde C, Cazzamali G, Williamson M, Hauser F, Søndergaard L, DeLotto R, et al. Molecular cloning, functional expression, and gene silencing of two Drosophila receptors for the Drosophila neuropeptide pyrokinin-2. Biochem Biophys Res Commun (2003) 309:485–94 10.1016/J.Bbrc.2003.08.022 [DOI] [PubMed] [Google Scholar]
- 195.Strotmann R, Schröck K, Böselt I, Stäubert C, Russ A, Schöneberg T. Evolution of GPCR: change and continuity. Mol Cell Endocrinol (2011) 331:170–8 10.1016/J.Mce.2010.07.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.