Abstract
Hundreds of genes are proposed to contribute to nociception and pain perception. Historically, most studies of pain-related genes have examined them in isolation or alongside a handful of other genes. More recently the use of systems biology techniques has enabled us to study genes in the context of the biological pathways and networks in which they operate.
Here we describe a Web-based resource, available at http://www.PainNetworks.org. It integrates interaction data from various public databases with information on known pain genes taken from several sources (eg, The Pain Genes Database) and allows the user to examine a gene (or set of genes) of interest alongside known interaction partners. This information is displayed by the resource in the form of a network.
The user can enrich these networks by using data from pain-focused gene expression studies to highlight genes that change expression in a given experiment or pairs of genes showing correlated expression patterns across different experiments. Genes in the networks are annotated in several ways including biological function and drug binding.
The Web site can be used to find out more about a gene of interest by looking at the function of its interaction partners. It can also be used to interpret the results of a functional genomics experiment by revealing putative novel pain-related genes that have similar expression patterns to known pain-related genes and by ranking genes according to their network connections with known pain genes.
We expect this resource to grow over time and become a valuable asset to the pain community.
Keywords: Systems biology, Protein–protein interactions, Protein interaction network, Web-based resource, Pain genes, Microarrays
1. Introduction
A variety of human and animal model studies have revealed a large number of genes associated with altered pain sensibility. Such genes, derived from human and animal studies, have been deposited in databases such as Online Mendelian Inheritance in Man (OMIM) [1] and the Pain Genes Database [3]. However these genes are often studied in isolation or alongside a handful of other genes.
Systems biology has developed approaches for combining diverse data such as protein interactions/associations with gene expression data from functional genomics studies, to allow us to study these genes in a broader context [2] and to computationally predict functional associations. These approaches are likely to generate a more in-depth understanding of the molecular basis of pain and potentially find novel candidates (genes or signalling pathways) susceptible to pharmacological intervention for the treatment of pain conditions. Here we describe a Web-based resource that enables the user to visualise known and putative pain genes in the context of an interaction network, available at http://www.PainNetworks.org.
2. Methods
In order to build the networks presented on the PainNetworks Web site, protein–protein interaction/association data were taken from a number of public resources. These data are combined to build interaction networks, in which the nodes are genes and the edges connect genes having at least one experimentally validated interaction/association in one of the public resources. The resource also contains information on known pain-associated genes from several sources: OMIM, which contains genes that alter pain sensitivity in man; the Algynomics Pain Panel, an experimental assay designed to measure genetic variation in a number of pain-related genes; and the Pain Genes Database, which contains genes shown to alter pain-like behaviour in rodents. This means that known and putative pain-related genes can be highlighted in the networks.
Networks can be human-, mouse-, or rat-centric. Orthology-based tools are used to transfer gene annotation and interaction data between species. Clustering algorithms can be applied to look for modules in networks that contain a large number of highly connected genes. To enrich the networks, microarray datasets produced using animal models of pain were analysed to provide information on differentially expressed genes and genes showing similar patterns of differential expression across different datasets. Networks can be annotated with such genes. Further annotation data were extracted from a number of other databases, including drug-binding data from DrugBank and functional annotation taken from Gene Ontology (GO). The Web site itself was developed using Python, and Cytoscape Web was used to display and edit the networks.
3. Results
3.1. Building pain networks
In its most basic usage, the user submits a query gene, or genes, into the search bar, and the site will display an interaction network including the query gene(s), and their known interactors as nodes. Interaction information includes data from a variety of public sources that store literature-curated interactions, protein–protein interactions derived from high-throughput screens (such as yeast-2-hybrid) and co-complex membership (determined, for example, through tandem affinity purification-mass spectrometry (TAP-MS)). Verified physical/direct interactions are represented in the networks by solid lines, whereas co-complex memberships are represented by broken lines.
The default settings only encompass experimentally verified protein interactions/associations, but users can also add predicted interactions to the network if they wish. Furthermore, interactions can also be inherited between species, allowing a comparison of putative pain-related networks between organisms.
3.2. Gene expression data
The user can also enrich the networks using data from a number of pain-focused gene expression studies, both in terms of genes that change in expression in a given experiment and genes showing correlated patterns of expression in a number of different experiments. Datasets used can be those available on the Web site or added by the user. The Web site currently contains several pain-related datasets (detailed on the Web site and in the full article, available online).
The user is also able to input data from their own experiment and view it alongside these included datasets, without the need to send any of their data over the web. They can also browse the current datasets, and download the data in a variety of formats. We also invite users to submit their own data to the Web site and to allow others to view it.
3.3. Network clustering
Additional features of the site include the ability to cluster the network, essential for interpretation when networks get beyond a certain size. Two graph-clustering algorithms are available. The user can also look for over-representation of GO categories in different network clusters.
3.4. Filtering the network with tissue-specific expression data
The site allows the user to further refine the networks using tissue-specific expression data, taken from a compendia of mice expression data produced using a variety of techniques (http://www.proteinatlas.org/) [5]. This can be achieved via a checkbox and drop-down menu on the Web site that allows the user to restrict his search to a given tissue.
3.5. Annotating the network
It is possible to use annotation data from various sources to highlight components of the network (genes or groups of genes) with specific properties. The user can also download the network in a variety of formats, alongside annotations, such as GO annotation, what drugs are available that bind to the protein encoded by the gene, and a list of experiments in which the gene shows differential expression.
3.6. Potential uses
Fig. 1 presents a number of potential uses for the site. It can be used to filter lists of genes returned from microarray studies, find out more about a gene of interest, find potential interactors for a less well-known gene, find putative pain-associated genes and look for potential novel drug targets. More details of the potential uses for the site are given in the online e-pain article. Example uses are described and the performance of the site in reproducing known interactions in canonical pathways is reported. Information on the methods used for ranking lists of genes according to interactions with known pain genes and network topology is also given.
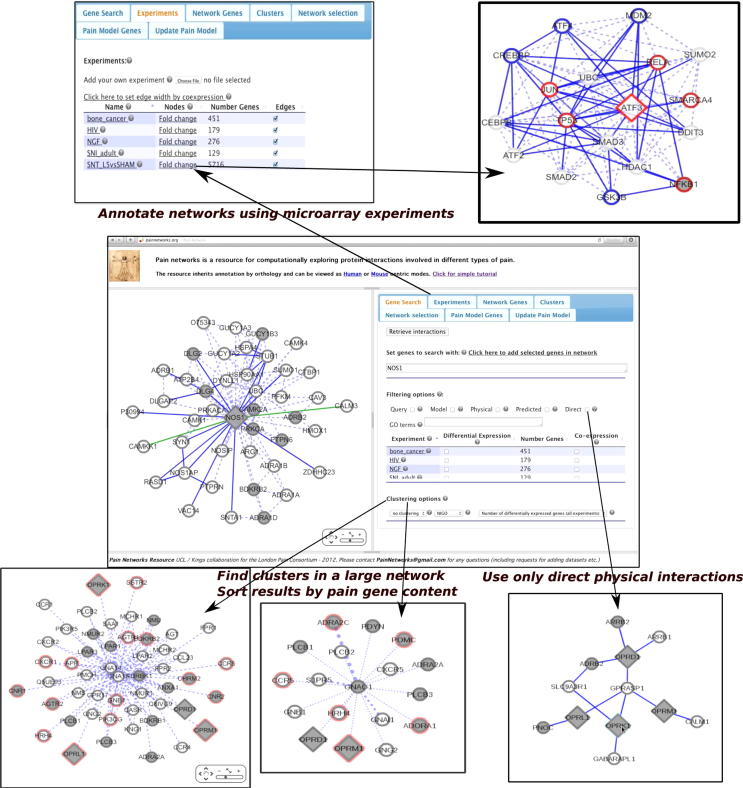
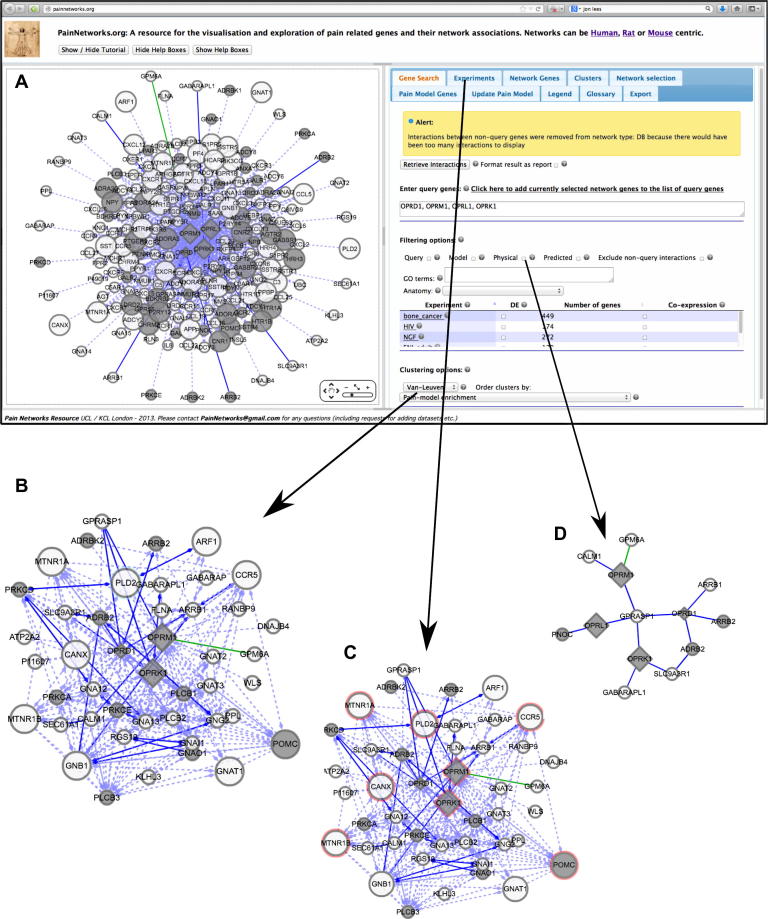
Fig. 1.
Example uses of the site. The central panel, from which the red arrows protrude, shows the homepage of the site. The red circle highlights the search bar, where the user enters their gene(s) of interest, and any other search parameters (such as the type of interactions they want to bring back). They then click search and the site will bring back a network involving their search gene(s) and the proteins interacting with this gene(s) (ie, interactors), as well as any interactions between these interactors. Three examples are shown: searching with ATF3 (cyclic AMP-dependent transcription factor), with the nodes in the resulting network coloured based on microarray data (top), opioid receptors, with the resultant network clustered into smaller networks (bottom left), and angiotensin receptors, with results filtered so that only direct physical interactions are displayed (bottom right). Figs. Fig. 2, Fig. 3, Fig. 4 of the full article show these networks in more detail.
Tutorials are available from the site itself, from the panel on the left hand side of the homepage. Screencasts showing potential usage of the site are available from http://www.youtube.com/channel/UCfl06Zr51Sy3BgN9eca_w0A/videos?view=0. Full details of the different features of the site are available from the reference manual, accessible from http://www.PainNetworks.org/tutorials/RefMan.pdf.
3.7. Site updates
The site will be updated periodically to ensure that new pain genes added to OMIM [1] and the Pain Genes Database [3] are included, as well as any new interactions, and annotations. Currently, updates are scheduled to coincide with new releases of Ensembl.
3.8. Conclusions
We have provided a resource for the pain community, allowing researchers to view protein networks based on their gene(s) of interest and all the proteins interacting with this gene(s). This network is enriched with various information including gene expression data and functional involvement in pain. We expect this resource to grow over time and become a valuable asset to the pain community.
References
[1] Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res 2009;37:D793–6.
[2] Antunes-Martins A, Perkins JR, Lees J, Hildebrandt T, Orengo C, Bennett DLH. Systems biology approaches to finding novel pain mediators. Wiley Interdiscip Rev Syst Biol Med 2013;5:11–35.
[3] Lacroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies. Pain 2007;131:3.e1–4.
[4] Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 2009;27:199–204.
[5] Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248–50.
1. Introduction
Persistent pain has a complex pathophysiological basis in which multiple mechanisms contribute to its initiation and maintenance [17], [23], [44], [65]. The risk of developing this condition is dependent on the interaction between environmental and genetic factors. Multiple techniques are currently being employed to investigate the molecular basis of pain: transcriptional profiling of experimental models of persistent pain in rodent and human [14], [20], [33], [40], the study of inbred rodent strains and genetically modified animals to understand the genetic basis of acute and chronic pain [1], [22], [34], [66], and, more recently, human genetics, describing both high impact variants [31], [64] and the use of gene association studies in experimental pain models and patient cohorts [15], [56]. Systems biology (defined as “the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources …” [6]), provides a means to organise, integrate, and maximise the utility of these large disparate pain-related data sets [3].
Gene expression profiling experiments using microarray technology represent one means to study pain in an unbiased, high-throughput manner [14], [29], [33], [40]. These functional genomics studies provide a large amount of information on biological systems, including processes involved in pain. However, they generate large gene lists. By considering the relationship between these putative genes and those responsible for pain processes, a biologist can focus on genes interacting with pain-related genes and therefore likely to be pain related.
Protein interactions can be detected using a range of technologies [48], and these data have been collected and made available by several public resources [4], [13], [18], [27], [52], [57]. By combining gene expression data with protein networks, built from this interaction/association data, we can identify parts of the network activated under particular conditions and detect differentially expressed genes likely to be involved in pain due to connections with known pain genes.
Network-based approaches have been successfully applied to many human diseases, including cancer [10], [47], [59], diabetes [38], asthma [25], Parkinson’s [45], Alzheimer’s [30], [55], and systemic inflammation [11]. Several resources exist that enable a user to build networks and examine their gene(s) of interest in the context of their network associations [35], [58], [63].
The PainNetworks resource displays pain-specific gene networks built by combining publicly available protein interaction data with microarray-derived gene expression data from a wide range of pain-related experiments. Users can query with a single gene or list of genes generated by their own transcriptional profiling experiments. The resulting network shows interactions between the query genes and other nonquery genes, including pain-related genes, for example, from the Pain Genes Database [34]. Clustering of the networks can be performed to detect modules of related genes; filtering can be used to select genes from a specific tissue type or anatomical structure of interest. Annotation can also be obtained for network genes and interactions.
Here we describe the Web site and underlying methodology. We also demonstrate the power of the networks to reproduce known associations in some pain-related biological pathways of interest and we give examples of using the site to find out more about genes and experiments of interest.
2. Methods
2.1. Network construction
In order to build the networks presented on the site, protein–protein interaction data were taken from a number of databases, as described below. These interactions were then combined in an unweighted manner to build a network, that is, the network will have an edge between 2 nodes (representing genes/proteins) provided at least one public resource reports an experimental validation for this interaction/association. PainNetworks then allows the user to view a part of this network, using a query gene or set of genes, and to further filter this network using various data sources.
2.2. Interaction/association data
Protein interactions were taken from 6 public databases (MINT, IntAct, BioGRID, DIP, HPRD, and Reactome [4], [13], [18], [27], [52], [57]) and combined to produce a network.
These resources contain information on interactions that have been experimentally detected using a variety of experimental techniques (for example, yeast 2-hybrid, tandem affinity purification). Further details on these techniques are provided in [48]. Only interactions that have a PubMed ID reference are included in this resource; the relevant PubMed ID for an interaction and its source database can be viewed by clicking on an edge in a network.
Proteins were mapped to their genes, allowing the data from different databases to be combined using a common identifier. This allowed us to resolve ambiguous mapping between databases. Direct protein–protein interactions (such as interactions curated from the literature or found using yeast 2-hybrid screens) are displayed in the network as solid lines. Indirect associations (such as associations inferred through co-complex membership) are displayed as broken lines.
The focus of PainNetworks is on experimentally defined interactions. However, we also provide the user with the ability to add predicted interactions from the STRING resource [58], v 9.0. Only high-confidence predictions (ie, with a STRING score >800) are included. More details on the STRING score can be found at the resource itself, http://www.string-db.org, and in [43]. We also provide predicted protein interaction data derived from homology-inferred inheritance of interactions between orthologues (HIPPO-C, HIPPO-DB; see below for details). As with many features of the Web site, this option is not enabled by default, but can be selected by the user and may be valuable for poorly annotated, less-well-studied genes for which few interactions are known, or for genes that cannot be assayed easily using experimental methods.
Interaction data are currently available for humans, rats, and mice. Therefore, the site has 3 potential species-centric views. Each view constructs the network using the interaction data available for that species. In addition, direct physical interactions detected in another species are transferred to that network, as described below.
2.3. Transferring annotation between species
The known pain genes deposited in the Pain Genes Database are from mice [34]. The protein–protein interaction (PPI) and Online Mendelian Inheritance in Man (OMIM) data are largely from humans, though some PPI data come from other species. The pain-related gene expression microarray data are produced using rat models, and the tissue-specific expression data are from humans. To integrate the data so they can be viewed together on the Web site, annotations must be transferred to one of the species, and therefore it is necessary to map genes between the species. PainNetworks currently allow the data to be transferred to mice, rats, or humans. Thus, there are 3 different possible views of the site: human-centric, mouse-centric, or rat-centric.
The Ensembl Compara method for identifying orthologues is used to map genes between species [62]. In order to transfer PPIs between species, a method similar to the interlogs procedure [41] is used; we name this procedure, “Homologous Inheritance of Protein–Protein Interactions by Orthology – Conservative” (HIPPO-C). Given an interaction between a pair of genes in one species, if both of these genes have unambiguous orthologues in another species, this interaction can be inherited. This method has been shown to work well in previous studies [51]. Inherited interactions will be visualised as a green edge in the network, as opposed to blue for interactions not inherited from other species. For example, when looking at the mouse interaction network (ie, using the mouse-centric view), inherited interactions found using human-based methodology would be coloured green; interactions discovered using mouse-based methodology would be coloured blue.
A second method for protein interaction inheritance is also included, named HIPPO-DB (Homologous Inheritance of Protein–Protein interactions by Orthology – Domain Based). This method uses a combination of CATH and Pfam domain classifications [19], [37], [50] to identify the likely subset of domain interactions underpinning a given protein interaction. This method provides increased confidence in protein interactions and indicates the domain combination that may be underpinning the interaction.
Predicted gene associations taken from the STRING database [58] are available for the human-centric view, and are not transferred between species. Table 1 shows the total numbers of genes and interactions in the respective networks.
Table 1.
Overview of the different types of data contained in PainNetworks.
| Direct interactions | All physical interactions | PGD genes | Algynomics chip v2.0 | OMIM pain | OMIM absence of pain | Predicted interactions | |
|---|---|---|---|---|---|---|---|
| Human (genes) | 10,297 | 15,927 | 360 | 542 | 8 | 10 | 17,404 |
| Human (interactions) | 40,832 | 230,500 | 2884 | 4213 | 2 | 9 | 414,786 |
| Mouse (genes) | 2104 | 5815 | 195 | 0 | 5 | 4 | 16,101 |
| Mouse (interactions) | 2716 | 12,190 | 130 | 0 | 0 | 1 | 320,625 |
| Rat (genes) | 545 | 1948 | 118 | 0 | 6 | 4 | 12,248 |
| Rat (interactions) | 438 | 2530 | 51 | 0 | 1 | 0 | 142,034 |
PGD, Pain Genes Database; OMIM, Online Mendelian Inheritance in Man.
2.4. Pain-related gene lists
The user can input a set of query genes in order to produce a network. However, the Web site also contains lists of pain-related genes obtained from public resources: 1) The OMIM database, which is an online database of diseases and related genes (containing traditional “Mendelian” diseases and more complex polygenic diseases). 2) The Pain Genes Database (PGD), maintained by Jeffrey Mogil’s Pain Genetics Lab. This resource contains genes that, according to a published study, lead to a change in pain-related behaviour when knocked out in mice [34]. 3) The Algynomics Pain Research Panel. This is a custom microarray that allows the user to assay single nucleotide polymorphisms in a number of genes that may have some involvement in pain-related processes: http://www.algynomics.com/pain-research-panel.html. Table 1 shows the number of pain-related genes present in the various interaction networks for the different organisms, and the number of interactions between them.
PainNetworks will be updated regularly to capture any new microarray, RNA-seq experiments, additional genes placed in the PGD, OMIM, or other published pain-based resources, and for future versions of the Algynomics Pain Research Panel. The user can use genes from these resources to annotate their network, or to identify clusters in the network with an over-representation of pain-related genes.
Note that there is not an exact match between the number of pain-related genes in PainNetworks and PGD or OMIM. This is due to ambiguities between databases with regard to gene identifiers: a given gene in one resource might not have an equivalent entry in another, or might have more than one.
Currently, PainNetworks contains, for humans, 10,297 genes with Ensembl IDs, with 40,832 direct interactions between them. The respective numbers for mice are 2104 and 2716, and for rats, 545 and 438. Most efforts to map interactions and associations between genes/proteins focussed on humans; this explains the discrepancy in numbers between the different organisms.
2.5. Expression data and analysis
The current datasets used by the Web site consist of gene expression data taken from microarray analyses of 4 animal pain models, performed by researchers in the London Pain Consortium and their collaborators, using rat as the model organism. Three of these are published [16], [40].
Details on experimental design have either been published or are provided on the Web site and are summarized in Table 2. Further details of all currently available experiments can be viewed by clicking on the experiments tab. Lists of genes and fold changes can be downloaded for each experiment in a variety of formats, or browsed online. More datasets will be added in time, and we invite members of the pain community to contact us to upload datasets to the site. Users can also view their own expression datasets through the site locally (client side), without having to send any data via the Web. Instructions on how to do this are given on the site, along with an example file.
Table 2.
Details of the pain-related microarray datasets currently contained in PainNetworks.
| Name | Experimental details | Species | Platform | Contributor | Reference |
|---|---|---|---|---|---|
| HIV | Rats received concomitant delivery of ddC (Zalcitabine) and HIV-gp120 to the sciatic nerve. Gene expression in L5-DRG at 15 days was compared to vehicle-receiving controls | Rat | Affymetrix Rat Genome 230 2.0 array | Klio Maratou/Andrew Rice | [40] |
| SNT-L5 | Spinal nerve transection (SNT) was performed on the rat L5 spinal nerve. Gene expression in the L5-DRG (dorsal root ganglion) at 15 days was compared to that of the L5-DRG for rats that received sham surgery | Rat | Affymetrix Rat Genome 230 2.0 array | Klio Maratou/Andrew Rice | [40] |
| NGF | Rats received recombinant nerve growth factor (NGF) for seven days through a pump placed subcutaneously beneath the scapulae. Gene expression in the L4 and L5-DRG was compared to vehicle receiving controls | Rat | Affymetrix Rat Genome 230 2.0 array | Stephen McMahon | Unpublished |
| SNI | Spared nerved injury (SNI) was performed on tibial and peroneal branches of the sciatic nerve of rats. Gene expression in the L4 and L5-DRG was compared to sham surgery receiving control animals | Rat | Affymetrix Rat Genome 230 2.0 array | David Vega-Avelaira/Maria Fitzgerald | [61] |
All microarray-derived gene expression datasets were produced using Affymetrix GeneChip technology. The resultant data were preprocessed using the RMA package [9], [26]. Differential gene expression was calculated using limma (Linear Models for Microarray Data) [54], which produced a P-value for each gene probed by the array. False discovery rate was estimated from these P-values using the Benjamini–Hochberg method [7]. Fold changes in gene expression were computed between case samples (ie, animals in which pain-related behaviour was induced) and control samples (ie, the corresponding control for the model). A false discovery rate of 0.1 was used to decide if a gene was differentially expressed between case and control samples.
2.6. Filtering to obtain context specific networks
Networks can be filtered using various methods to make the networks more context-specific. For example, a list of Gene Ontology [5] (GO) category names can be entered, and only genes annotated with one or more of these terms would be retained in the network. In this way, networks can be filtered to only display genes involved in specific biological processes (eg, GO:0006954, inflammatory response), molecular functions (eg, GO:0005244, voltage-gated ion channel activity), or expressed in given cellular components (eg, GO:0045202, synapse).
The user can also filter the network to retain genes differentially expressed in one or more of the gene expression microarray experiments. They may also filter the network so that it retains interactions where both genes show significant differential expression (in the same direction) in user-specified experiments. Such filtering can greatly reduce the size of the interaction network, making it easier to examine and interpret.
Networks can also be filtered using tissue-specific expression data, in order to produce networks that only contain genes expressed in a given tissue/anatomical structure. These data are taken from the Human Protein Atlas Project [60]. This project combines Antibody-Based Proteomic and tissue-microarray based techniques in order to profile protein expression across various cells and tissues. For example, the user is able to filter the network so that it only contains genes expressed in a given cell or tissue, such as peripheral nerve tissue.
2.7. Clustering the network
Since there are large amounts of experimental data available for many genes, it is very easy to generate large networks (ie, >100 nodes), which are difficult to interpret. Therefore, PainNetworks allows the user to cluster the network in order to break it up into smaller groups of related genes. The advantages of clustering are 2-fold: to help visualisation and to find modules enriched in specific functions. We use established graph-clustering algorithms to find clusters in the network. The aim of these algorithms is to find groups in the network with a high number of interactions between group members, but a small number of interactions between members of the group and the other genes in the network outside the group.
Two graph-clustering algorithms are implemented: Louvain clustering [8] and Links clustering [2]. Louvain looks for distinct clusters of genes in the network, not allowing a given gene to belong to more than one network. Links clustering groups the edges of the network, that is, the interactions. This second approach has the potential advantage that it allows communities to be found with overlapping nodes; the same gene can belong to different clusters in the network. This fits in well with the functional pleiotropy of many genes.
Once clusters have been built, they can be ranked according to their size or the number of pain-related genes they contain. Enriched GO categories for each cluster are also displayed. Enrichment is performed using a method similar to that employed by the DAVID functional enrichment tool [21]: for each GO category, the proportion of category members in the cluster is compared to the proportion of category members outside of the cluster using Fisher’s exact test. Categories are deemed significant if they have a P-value of <0.05 following Bonferroni correction for multiple testing, and if the proportion of category members inside the cluster is at least 3 times higher than the proportion outside the cluster.
2.8. Additional gene annotation data
Additional functional annotation can be obtained for the genes in the network by clicking on the relevant node. This will bring up a box containing details of the GO categories to which the gene belongs, drugs that can target the protein encoded by the gene (taken from DrugBank [28]), and any potential domains the protein contains (InterPro [24], Pfam [50]). Clicking on the network selection tab allows the user to highlight the genes in the network that have drug-binding annotations, predicted coiled-coil regions (ncoils), predicted transmembrane regions (TMHMM [32]), and signal peptide sequence features (SignalP [49]).
2.9. Pain gene enrichment score
The ratio of interactions that a gene has with known pain genes, compared to all interactors, multiplied by 100, is deemed the Pain Gene Enrichment (PGE) score. It is viewable in the table of network genes found in the “Network Genes” tab of the Web site. In order to contextualise this score, a “percentile” value is given in the same table. This value represents the percentage of genes in the resource that have a higher PGE score than a given gene. For example, for a gene with a PGE score of 90: if there were a total of 10,000 genes in the resource and 1000 of these genes had a PGE-score higher than 90, the percentile for this gene would be 10.
2.10. Ranking a set of query genes by their probability to be pain associated
As well as providing a PGE score, we provide a ranking for query lists of genes, for example, lists of differentially expressed genes identified by a microarray experiment. This ranking is generated by the GeneMANIA algorithm, a publicly available method that is well established and shown to be effective in analysing protein network topologies to determine the likelihood that a query gene is functionally associated with a set of genes in the network known to be involved in a particular biological process. We use the default GO biological process-weighted default kernel combination, and apply label affinity propagation as described in [63] to derive scores for ranking. The ranking can be viewed as an extra column in the Network Genes tab of the site.
2.11. Implementation
The Web site was implemented using Python and CSS. Cytoscape Web [39], a Flash-based plug-in, available from http://cytoscapeweb.cytoscape.org/, was used to visualise the networks.
In addition to customising the content of their network, the user can move the protein components around on the screen interactively, allowing them to focus on whatever part of the network they wish to examine further.
A detailed reference manual, video tutorials, and several interactive search tutorials and are also available from the site.
3. Results
3.1. Overview of the site
Here we present a publicly available resource, PainNetworks. The resource combines data from various sources in order to allow the user to view pain-related biological networks. These networks can be used for various purposes: to learn more about a gene of interest, to further analyse a group of genes that have a putative role in pain, and to look for novel drug targets. A further benefit is the ability to submit gene lists obtained by high-throughput studies in order to use the networks to identify a more focused subset of genes likely to be associated with pain-related processes.
Therefore, the purpose of the site ranges from educational through hypothesis generation to the interpretation of large datasets obtained in high-throughput functional genomics studies.
In this section, we will demonstrate the power of the resource to recognise known pathways implicated in pain and we will describe some example usage scenarios for the site. We have tried to make the site as intuitive as possible, however, there is a large amount of functionality and a number of ways to view the networks and export the data. Although we will describe some key features of the site here, more extensive details are provided in the reference manual: http://www.PainNetworks.org/tutorials/RefMan.pdf. This manual describes how to use all of the features of the site, illustrated with screenshots. There are also video tutorials, available at http://www.youtube.com/channel/UCfl06Zr51Sy3BgN9eca_w0A/videos?view=0, which present potential usage scenarios. It is also possible to search the site using example queries presented on the site itself, easily accessible from the panel to the left of the network panel on the site’s homepage.
A nonexhaustive summary of potential uses for the site is also presented in Table 3. Below we demonstrate the performance of the site in reproducing known pathways. We subsequently show how the resource could be used to identify novel genes that are likely to be involved in pain as they have many interactions/associations with known pain genes. Finally, we describe some potential uses of the site, in obtaining more information for putative pain gene(s).
Table 3.
Example usages scenarios for the site.
| Use | Gene (s)/experiment (s) involved | Description | Link |
|---|---|---|---|
| Find out more about a gene of interest | NRG1 (Neuregulin 1) | Enter NRG1 into the search bar, examine the resultant network | http://www.youtube.com/watch?v=5iYnZW-YLag |
| Use the results of a gene expression microarray experiment to highlight differentially expressed genes in a network | ATF3 (Cyclic AMP-dependent transcription factor) | Enter ATF3 and filter results by direct physical interactions. The results of a pain-related microarray gene expression experiment can then be used to highlight nodes in the network | http://www.youtube.com/watch?v=A6e7oyK58CM |
| Look at predicted associations for a given gene for which no known physical interactions exist | VGF (VGF nerve growth factor inducible) | VGF is used as a query gene, but no interactions are found using default search parameters. However, the user can include predicted association in their query – this will return a network of VGF and its predicted associations | http://www.youtube.com/watch?v=Z0jx7IuahJg |
| Make a large network from a group of related genes and cluster in order to look for subsets of highly connected genes | OPRD1, OPRK1 OPRM1 and ORL1 (opioid receptors) | These genes are used to query the site and generate the network. This resulting network is then clustered, these clusters are then sorted by the number of pain-related genes they contain | http://www.youtube.com/watch?v=4RdFMBogeio |
| Search for experimental interactions between a large list of genes identified as differentially expressed from a microarray gene expression experiment | SNT (spinal nerve transection model): L5 vs Sham | In this search the top six genes from the SNT experiment (largest fold changes amongst the significantly differentially expressed genes) were used as query genes. The resultant network shows the interactions between these genes. Genes are: VIP, ARG1, CSRP3, POSTN, TSLP, CLEC7A | http://www.youtube.com/watch?v=1Y9a8RaIKEs |
| Look for interaction partners for a given gene that are druggable using approved drugs | SCN10A (sodium channel, voltage-gated, type X, alpha subunit) | Network produced using SCN10A as a query genes. Annotation used to show that many of the interactors are druggable, and that many are predicted to be membrane bound proteins | http://www.youtube.com/watch?v=1Mta9QpLO14 |
3.2. Demonstrating the performance of the PainNetworks site in reproducing known pathways reported in the literature
To demonstrate the value of PainNetworks in providing information on genes involved in pain, we show the performance of the resource in reproducing information on known biological pathways reported as pathway figures in the literature. We do this by reporting the proportion of interactions between the pathway genes, shown in published pathway figures that are also highlighted on the PainNetworks Web site.
Given that the resource uses experimental data on protein interactions (both direct and indirect), the PainNetworks Web site is unlikely to match these diagrams perfectly. Pathway diagrams taken from the literature are necessarily reductive models of the real processing that occurs within the cell, biased towards the system being considered. It is therefore possible that some of the proteins in the pathway are also involved in other processes [36], [53]. Furthermore, with regards to any missing connections in the networks constructed by PainNetworks, the site uses protein interaction data stored in public databases, and these resources do not currently capture all the protein interactions reported in the literature. However, they are the most comprehensive public source of such data.
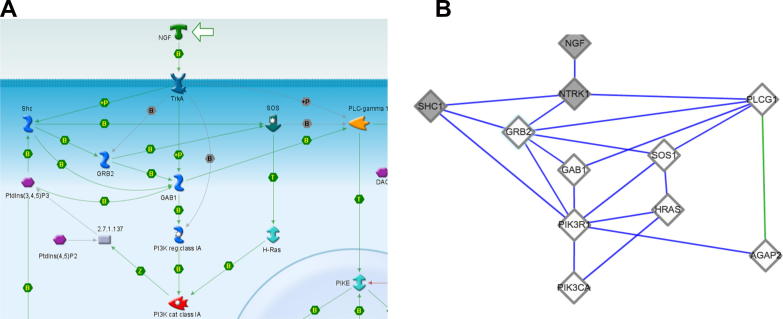
Despite these caveats, we explored how well PainNetworks could reproduce two such canonical pathway diagrams, shown in Fig. 1, Fig. 2, taken from http://pathwaymaps.com/maps/652/ and [46]. Fig. 1 shows the TrkA-NGF signalling pathway; PainNetworks built the network shown in this figure using the pathway members as query genes, filtering the network to include only physical interactions and only considering interactions between the query genes. As we see in Fig. 1, PainNetworks was able to recover 76% of the interactions in the original pathway. Moreover, PainNetworks displays some additional interactions between the query genes that are not shown in the canonical pathway figure (6 interactions, 5 of which involve the same gene, PIK3R1). Since these interactions are associated with experiments reported in one of the public databases integrated by PainNetworks, these links may suggest novel associations between genes in the network that had not previously been considered.
Fig. 1.
Using PainNetworks to reproduce the NGF-TrkA signalling pathway. (A) The original pathway diagram, adapted from http://pathwaymaps.com/maps/652/. (B) The pathway as reproduced by PainNetworks, created by querying the resource with the pathway members, restricting the results to only include experimentally validated direct physical interactions, and only showing interactions occurring between the query genes.
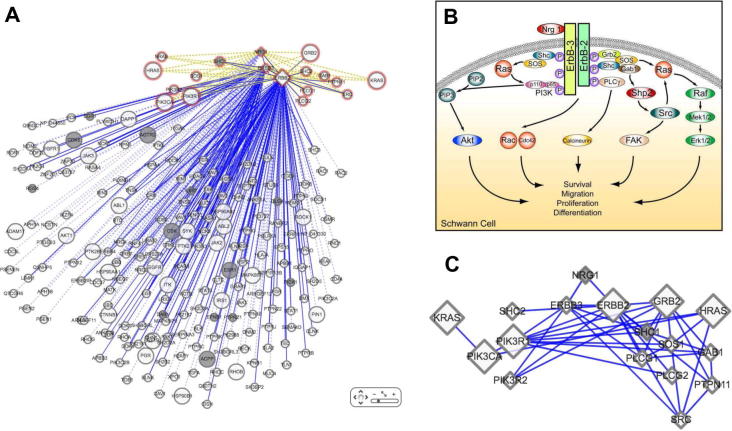
Fig. 2.
Using PainNetworks to reproduce the NRG1-ErbB signalling pathway. (A) The pathway as reproduced by PainNetworks, showing the interactions involving NRG1/ErbB2/ErbB3. The genes from the original pathway are shown in red. In order to create this network, PainNetworks was queried using NRG1, ErbB2, and ErbB3; results were filtered so that only interactions that include these genes were shown; interactions between the nonquery genes were removed to prevent the number of edges in the network from obscuring the image. (B) The original pathway, taken from [43]. Note that only a part of the pathway is reproduced, in order to stop the network becoming too large and difficult to interpret. (C) A second reproduction of the pathway. In this network, only direct interactions between the query genes are shown, and results have been limited to include only direct physical interactions.
Fig. 2 shows another example in which PainNetworks was used to reproduce the NRG1-ErbB signalling pathways, which has been implicated in pain [12]. A figure taken from a recent review [46] was used to make the comparison. PainNetworks was queried using all the members of the pathway (NRG1, ErbB2 and ErbB3): the resource is able to find all of the interactions within the canonical pathway diagram (Fig. 2C), plus many more interactions with other genes from outside the pathway (Fig. 2A). These may be good targets for putative novel pain genes implicated in this pathway.
3.3. Using the information in PainNetworks to suggest putative novel pain genes
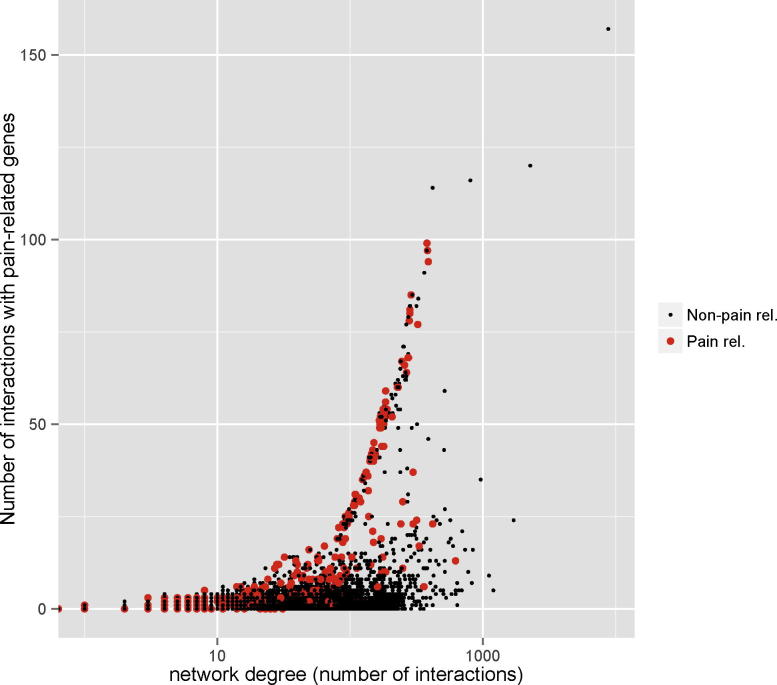
Fig. 3 shows the number of interactions for each gene in the resource with at least one known interaction (ie, network degree, x-axis), plotted against the number of these interactions that are pain related (y-axis).
Fig. 3.
Network degree of all genes in PainNetworks with at least one known interaction. The x-axis shows the number of interactions that each gene is involved in. The y-axis shows the number of these interactions that are with pain genes. Black points = non-pain-related genes, red points = pain genes. This figure was generated using direct and indirect physical interaction data but not predicted interactions.
The red points on the figure represent pain-related genes, taken from the pain genes database. It can be seen that, as the node degree increases (ie, for genes with higher numbers of interactions), the known pain genes have many more interactions with other known pain genes.
This is to be expected – we are showing that many known pain-related genes interact with other known pain-related genes. However, we also see that there are several non-pain-related genes that interact with a large number of pain-related genes. These genes are likely to be of interest to a pain researcher as one can hypothesise that they are potentially novel pain genes and target them for experimental validation. This figure is produced at the human level, that is, annotations and interactions have been transferred to humans, using the strategies described in Methods.
3.4. Using PainNetworks to find out more about genes of interest and their interactors
3.4.1. Opioid receptors
In Fig. 4, we show some network analyses of the 4 different human opioid receptors – the δ opioid receptor, κ opioid receptor, μ opioid receptor, and nociceptin receptor – known to be implicated in pain. Fig. 4A shows the results of a simple search of the Web site using the genes that encode these receptors: OPRD1, OPRK1, OPRM1, and ORL1, in the human-centric view. This query results in a very large network, too large to interpret visually. Therefore, only interactions that involve the query genes are shown in the network; interactions between the interactors of the query genes are not shown.
Fig. 4.
Looking for druggable proteins associated with opioid receptors. Building the network using all 4 opioid receptors and their interactors, as shown in panel (A), results in a large network. Different ways of reducing the network to make it more interpretable are presented and explained further in the text: (B) and (C) show the results of clustering the original network to find groups of genes that interact with each other more frequently than they interact with genes outside of the group. (C) Genes highlighted in red represent genes with known drug targets, according to DrugBank [27]. This annotation is obtained from the “Network selection” tab. (D) Shows the network with indirect interactions removed, leaving only direct physical interactions.
The grey nodes show pain-associated genes, taken from the PGD; the white nodes show the genes not reported in PGD. Three further analyses that refine the network are shown in the bottom half of the figure. Fig. 4B shows one of the clusters found by clustering the original network by the Louvain clustering, which partitions the genes into separate, nonoverlapping clusters. We see that three of the opioid receptors are placed into a cluster with beta-2-adrenergic receptor (ADRB2), pro-opiomelanocortin (POMC), and a number of other genes from the pain genes database.
Fig. 4B shows this same network cluster, but with druggable targets highlighted in red, generated using the “Network selection” tab. Fig. 4D shows the results of filtering the original network so that only direct physical interactions (not indirect interactions based on co-complex membership) are shown. Because the network produced by this method is small, the interactions between the nonopioid genes are displayed. Investigation of these networks can further elucidate pathways in which opioid compounds operate by revealing novel genes highly connected to these receptors. Some of these novel genes may be druggable proteins that might influence opioid receptor activity.
3.4.2. Angiotensin II receptor, type 2 gene analysis
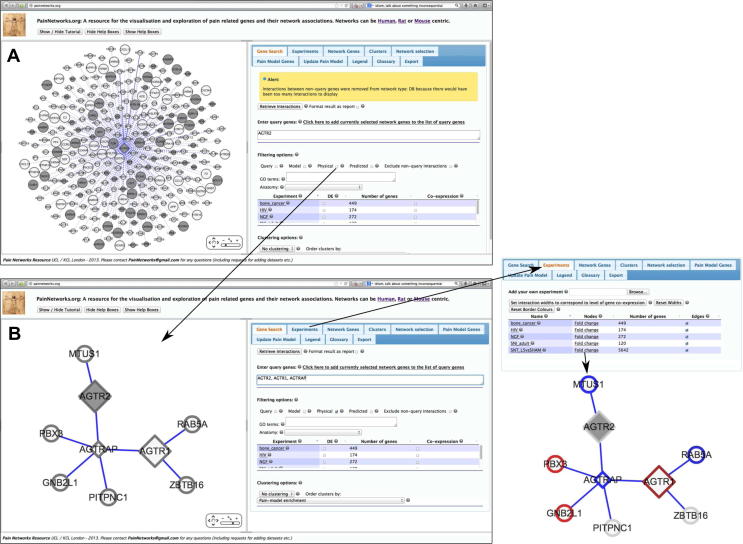
Fig. 5 shows how PainNetworks can be used to find out more about the angiotensin II receptor, type 2 gene (AGTR2). A recent phase II clinical trial showed that an inhibitor of this receptor (EMA 401) is effective in reducing pain reported by patients with postherpetic neuralgia [42]. Fig. 5A shows AGTR2 and its network interactions. A large number of the interactors of AGTR2 are pain-associated genes: 76 of its 284 interactors are in the pain-related gene list; this would give it a PGE score of 27. A PGE score of 27 is higher than the score associated with 97% of the genes in PainNetworks and within the range of PGE-scores obtained by other known pain genes.
Fig. 5.
Finding out more about angiotensin II type 2 receptors and their potential role in pain signalling. (A) Looking at direct and indirect interactors of angiotensin II receptor, type 2 gene (AGTR2). Note that the shaded circles represent pain-related genes. (B) The direct physical interactions of AGTR1, AGRT2, and AGTRAP. (C) The same network as shown in (B), but highlighting genes that are differentially expressed according to a gene expression microarray experiment of a model of neuropathic pain (spinal nerve transection). Red = increased expression following nerve injury; blue = decreased expression.
Fig. 5B shows the network formed by considering direct physical interactions involving AGTR2, along with related proteins AGTR1 (angiotensin II receptor, type 1) and AGTRAP (angiotensin II receptor-associated protein).
Genes that change in expression following spinal nerve transection (quantified using gene expression microarrays) are highlighted in Fig. 6. We notice that many of the genes in this network are differentially expressed, suggesting a potential role of this network in neuropathic pain. Thus, PainNetworks could be used as a starting point to further elucidate the mechanisms by which AGTR2 leads to pain relief.
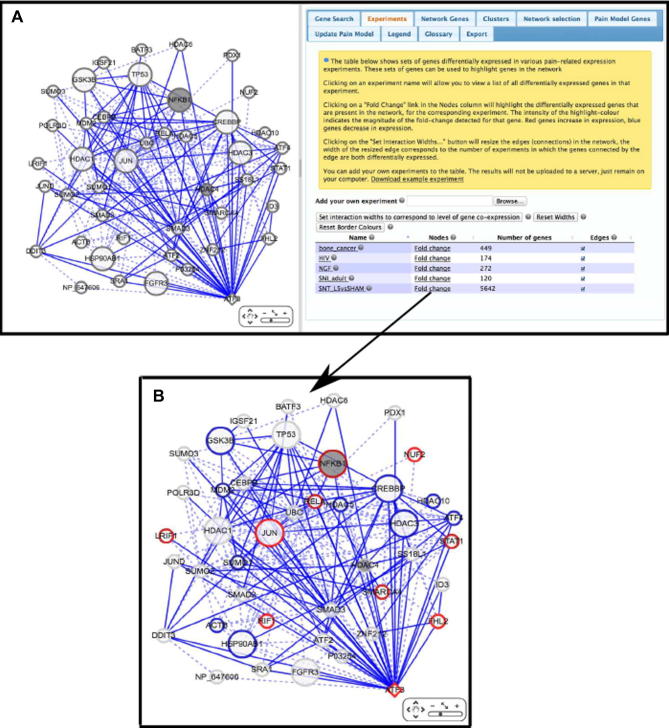
Fig. 6.
AMP-dependent transcription factor (ATF3) interactors that change in expression in a nerve injury model of pain. (A) The network returned by querying PainNetworks with ATF3, using default parameters. Pain-related genes, which in this case are obtained from the Pain Genes Database, are highlighted as grey in the network. (B) The genes that in L5 DRGs in the spinal nerve transection vs sham dataset are highlighted in the network by the addition of red or blue rings, for increased or decreased expression (respectively) in SNT compared to sham.
3.4.3. Cyclic AMP-dependent transcription factor
Cyclic AMP-dependent transcription factor (ATF3) has been shown to change in expression in a number of microarray experiments profiling gene expression in animal models of pain [33]. Fig. 6 shows ATF3 together with its network associations/interactions (A). Although ATF3 shows differential expression in a number of different neuropathic pain models, relatively few of its interaction partners are known to be pain associated. We see only 2 genes from the pain-associated gene list interacting with ATF3, NFKB1, and HDAC4. However, if we click on the “Fold Change” link for the spinal nerve transection, transected L5 nerve vs sham experiment, we see that many of ATF3’s network interaction partners are differentially expressed in this experiment (Fig. 6B). As we would expect, many of these differentially expressed interaction partners are also transcription factors. This suggests that spinal nerve transection may be leading to changes in gene expression through the actions of a network of transcription factors during the induction phase of neuropathic pain. This supports the hypothesis that ATF3 is indeed associated with the response to nerve injury, and these differentially expressed genes could represent further targets for experimental investigation.
3.5. Using PainNetworks to filter putative pain genes identified by a microarray experiment
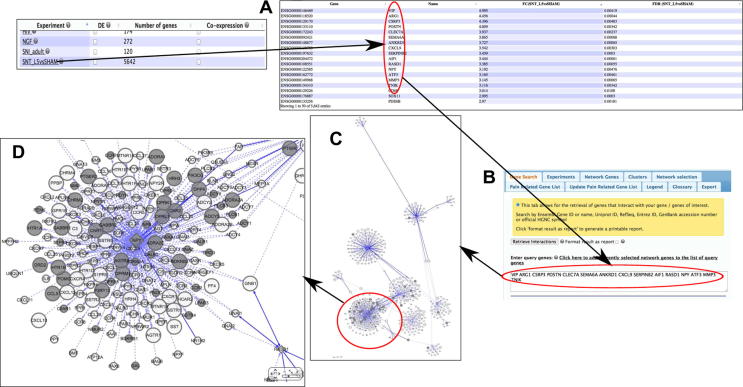
PainNetworks can also be used to analyse the results of a functional genomics experiment. Fig. 7 shows the network returned by querying the site using the differentially expressed genes with the largest fold changes from the spinal nerve transection experiment described above (Fig. 7A). We see that the network returned is quite large; however, by zooming in and inspecting different parts of the network, we see that there is a highly connected module that contains a large number of pain genes (Fig. 7D).
Fig. 7.
Using PainNetworks to analyse the results of a microarray experiment comparing L5-DRG following spinal nerve transection to L5 following sham surgery. (A) Clicking on the relevant experiment on the PainNetworks homepage returns a table of the differentially expressed genes from the experiment, alongside their log fold changes in expression. (B) The top 15 genes in terms of increased expression following spinal nerve transection are selected and used as query genes for the site. (C) The resulting network is displayed on the site, in the network panel. (D) The user can then zoom in on different areas in the network to look for relationships between the query genes and their interaction partners.
3.6. Conclusions
To conclude, we have developed a resource for the pain community that allows a pain scientist to view protein networks based on their gene(s) of interest. PainNetworks combines publicly available data on protein interactions, gene associations, gene expression data, and protein annotations. The value of the resource is increased considerably by including information on known pain genes given in the PGD of Mogil et al. [34], and by presenting information on putative gene functions reported in GO [5]. Networks provided by our resource allow a pain researcher to consider the function of their genes based on the functions of their interactors.
Our hope is that this novel view of the data will lead to hypothesis generation and a fuller understanding of the biological processes involved in pain. A particularly valuable aspect of the resource is the ability to interpret large gene datasets returned from high throughput functional genomics experiments. Users can obtain information on which genes in their list of putative novel targets are more likely to be pain associated because they have more interactions with known pain genes.
The resource will expand over time: we plan to add human genome-wide association study data, data from more transcriptional profiling experiments, and to increase the number of species covered. We expect that PainNetworks will become a valuable asset to the pain community.
Conflict of interest statement
None of the authors have financial or other relationships that might lead to a conflict of interest in the study.
Acknowledgements
We acknowledge the Wellcome Trust for a strategic award to the London Pain Consortium. D.L.H.B., J.R.P., S.B.M. and C.O. are members of the London Pain Consortium.
A.A.-M., S.B.M., D.L.H.B., J.L., and C.O. are part of the Europain Collaboration, which has received support from the Innovative Medicines Initiative Joint Undertaking, under grant agreement no 115007, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution. D.L.H.B. is a senior Wellcome clinical scientist (095698/z/11/z).
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
References
- 1.Abrahamsen B., Zhao J., Asante C.O., Cendan C.M., Marsh S., Martinez-Barbera J.P., Nassar M.A., Dickenson A.H., Wood J.N. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. doi: 10.1126/science.1156916. [DOI] [PubMed] [Google Scholar]
- 2.Ahn Y.-Y., Bagrow J.P., Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 3.Antunes-Martins A., Perkins J.R., Lees J., Hildebrandt T., Orengo C., Bennett D.L.H. Systems biology approaches to finding novel pain mediators. Wiley Interdiscip Rev Syst Biol Med. 2012 doi: 10.1002/wsbm.1192. [DOI] [PubMed] [Google Scholar]
- 4.Aranda B., Achuthan P., Alam-Faruque Y., Armean I., Bridge A., Derow C., Feuermann M., Ghanbarian A.T., Kerrien S., Khadake J., Kerssemakers J., Leroy C., Menden M., Michaut M., Montecchi-Palazzi L., Neuhauser S.N., Orchard S., Perreau V., Roechert B., van Eijk K., Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baitaluk M. System biology of gene regulation. Methods Mol Biol. 2009;569:55–87. doi: 10.1007/978-1-59745-524-4_4. [DOI] [PubMed] [Google Scholar]
- 7.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 8.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech: Theor Exp. 2008;10:P10008. [Google Scholar]
- 9.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 10.Bonifaci N., Berenguer A., Díez J., Reina O., Medina I., Dopazo J., Moreno V., Pujana M.A. Biological processes, properties and molecular wiring diagrams of candidate low-penetrance breast cancer susceptibility genes. BMC Med Genomics. 2008;1:62. doi: 10.1186/1755-8794-1-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvano S.E., Xiao W., Richards D.R., Felciano R.M., Baker H.V., Cho R.J., Chen R.O., Brownstein B.H., Cobb J.P., Tschoeke S.K., Miller-Graziano C., Moldawer L.L., Mindrinos M.N., Davis R.W., Tompkins R.G., Lowry S.F. Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 12.Calvo M., Zhu N., Tsantoulas C., Ma Z., Grist J., Loeb J.A., Bennett D.L.H. Neuregulin-ErbB signaling promotes microglial proliferation and chemotaxis contributing to microgliosis and pain after peripheral nerve injury. J Neurosci. 2010;30:5437–5450. doi: 10.1523/JNEUROSCI.5169-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceol A., Chatr Aryamontri A., Licata L., Peluso D., Briganti L., Perfetto L., Castagnoli L., Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costigan M., Befort K., Karchewski L., Griffin R.S., D’Urso D., Allchorne A., Sitarski J., Mannion J.W., Pratt R.E., Woolf C.J. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costigan M., Belfer I., Griffin R.S., Dai F., Barrett L.B., Coppola G., Wu T., Kiselycznyk C., Poddar M., Lu Y., Diatchenko L., Smith S., Cobos E.J., Zaykin D., Allchorne A., Gershon E., Livneh J., Shen P.-H., Nikolajsen L., Karppinen J., Männikkö M., Kelempisioti A., Goldman D., Maixner W., Geschwind D.H., Max M.B., Seltzer Z., Woolf C.J. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costigan M., Moss A., Latremoliere A., Johnston C., Verma-Gandhu M., Herbert T.A., Barrett L., Brenner G.J., Vardeh D., Woolf C.J., Fitzgerald M. T-cell infiltration and signaling in the adult dorsal spinal cord is a major contributor to neuropathic pain-like hypersensitivity. J Neurosci. 2009;29:14415–14422. doi: 10.1523/JNEUROSCI.4569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costigan M., Scholz J., Woolf C.J. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009;32:1–32. doi: 10.1146/annurev.neuro.051508.135531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., Jupe S., Kalatskaya I., Mahajan S., May B., Ndegwa N., Schmidt E., Shamovsky V., Yung C., Birney E., Hermjakob H., D’Eustachio P., Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuff A.L., Sillitoe I., Lewis T., Clegg A.B., Rentzsch R., Furnham N., Pellegrini-Calace M., Jones D., Thornton J., Orengo C.A. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawes J.M., Calvo M., Perkins J.R., Paterson K.J., Kiesewetter H., Hobbs C., Kaan T.K.Y., Orengo C., Bennett D.L.H., McMahon S.B. CXCL5 mediates UVB irradiation-induced pain. Sci Transl Med. 2011;3:90ra60. doi: 10.1126/scitranslmed.3002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 22.Foulkes T., Wood J.N. Pain genes. PLoS Genet. 2008;4:e1000086. doi: 10.1371/journal.pgen.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Hehn C.A., Baron R., Woolf C.J. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron. 2012;73:638–652. doi: 10.1016/j.neuron.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R.D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A.F., Rivoire C., Sangrador-Vegas A., Selengut J.D., Sigrist C.J.A., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P.D., Wu C.H., Yeats C., Yong S.-Y. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang S., Son S.-W., Kim S.C., Kim Y.J., Jeong H., Lee D. A protein interaction network associated with asthma. J Theor Biol. 2008;252:722–731. doi: 10.1016/j.jtbi.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D.S., Sebastian A., Rani S., Ray S., Harrys Kishore C.J., Kanth S., Ahmed M., Kashyap M.K., Mohmood R., Ramachandra Y.L., Krishna V., Rahiman B.A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., Pandey A. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V., Djoumbou Y., Eisner R., Guo A.C., Wishart D.S. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko J., Na D.S., Lee Y.H., Shin S.Y., Kim J.H., Hwang B.G., Min B.-I., Park D.S. cDNA microarray analysis of the differential gene expression in the neuropathic pain and electroacupuncture treatment models. J Biochem Mol Biol. 2002;35:420–427. doi: 10.5483/bmbrep.2002.35.4.420. [DOI] [PubMed] [Google Scholar]
- 30.Krauthammer M., Kaufmann C.A., Gilliam T.C., Rzhetsky A. Molecular triangulation: bridging linkage and molecular-network information for identifying candidate genes in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremeyer B., Lopera F., Cox J.J., Momin A., Rugiero F., Marsh S., Woods C.G., Jones N.G., Paterson K.J., Fricker F.R., Villegas A., Acosta N., Pineda-Trujillo N.G., Ramírez J.D., Zea J., Burley M.-W., Bedoya G., Bennett D.L.H., Wood J.N., Ruiz-Linares A. A gain-of-function mutation in TRPA1 causes familial episodic pain syndrome. Neuron. 2010;66:671–680. doi: 10.1016/j.neuron.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 33.LaCroix-Fralish M.L., Austin J.-S., Zheng F.Y., Levitin D.J., Mogil J.S. Patterns of pain: meta-analysis of microarray studies of pain. PAIN®. 2011;152:1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Lacroix-Fralish M.L., Ledoux J.B., Mogil J.S. The Pain Genes Database: an interactive web browser of pain-related transgenic knockout studies. PAIN®. 2007;131:3.e1–3.e4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 35.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Las Rivas J., Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lees J., Yeats C., Perkins J., Sillitoe I., Rentzsch R., Dessailly B.H., Orengo C. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012;40:D465–D471. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M., Liberzon A., Kong S.W., Lai W.R., Park P.J., Kohane I.S., Kasif S. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet. 2007;3:e96. doi: 10.1371/journal.pgen.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maratou K., Wallace V.C.J., Hasnie F.S., Okuse K., Hosseini R., Jina N., Blackbeard J., Pheby T., Orengo C., Dickenson A.H., McMahon S.B., Rice A.S.C. Comparison of dorsal root ganglion gene expression in rat models of traumatic and HIV-associated neuropathic pain. Eur J Pain. 2009;13:387–398. doi: 10.1016/j.ejpain.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthews L.R., Vaglio P., Reboul J., Ge H., Davis B.P., Garrels J., Vincent S., Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy TD, Desem N, Kitson G, Rice ASC, Dworkin BH, Bountra C, Anand P, Raff M. Clinical development of EMA401: an angiotensin II type 2 receptor antagonist as a potential new treatment for neuropathic pain. In: 4th World Congress on Pain, International Association for the Study of Pain (IASP) 2012. Available: http://www.iasp-pain.org/Milan/Program/Milan2012_Program.pdf.
- 43.Von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mogil J.S. Pain genetics: past, present and future. Trends Genet. 2012;28:258–266. doi: 10.1016/j.tig.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Moran L.B., Graeber M.B. Towards a pathway definition of Parkinson’s disease: a complex disorder with links to cancer, diabetes and inflammation. Neurogenetics. 2008;9:1–13. doi: 10.1007/s10048-007-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newbern J., Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. doi: 10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nibbe R.K., Markowitz S., Myeroff L., Ewing R., Chance M.R. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins J.R., Diboun I., Dessailly B.H., Lees J.G., Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 50.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E.L.L., Eddy S.R., Bateman A., Finn R.D. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojas A.M., Santamaria A., Malik R., Jensen T.S., Körner R., Morilla I., de Juan D., Krallinger M., Hansen D.A., Hoffmann R., Lees J., Reid A., Yeats C., Wehner A., Elowe S., Clegg A.B., Brunak S., Nigg E.A., Orengo C., Valencia A., Ranea J.A.G. Uncovering the molecular machinery of the human spindle–an integration of wet and dry systems biology. PLoS ONE. 2012;7:e31813. doi: 10.1371/journal.pone.0031813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer M.H., Lopes T.J.S., Mah N., Shoemaker J.E., Matsuoka Y., Fontaine J.-F., Louis-Jeune C., Eisfeld A.J., Neumann G., Perez-Iratxeta C., Kawaoka Y., Kitano H., Andrade-Navarro M.A. Adding protein context to the human protein-protein interaction network to reveal meaningful interactions. PLoS Comput Biol. 2013;9:e1002860. doi: 10.1371/journal.pcbi.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 55.Soler-López M., Zanzoni A., Lluís R., Stelzl U., Aloy P. Interactome mapping suggests new mechanistic details underlying Alzheimer’s disease. Genome Res. 2011;21:364–376. doi: 10.1101/gr.114280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorge R.E., Trang T., Dorfman R., Smith S.B., Beggs S., Ritchie J., Austin J.-S., Zaykin D.V., Vander Meulen H., Costigan M., Herbert T.A., Yarkoni-Abitbul M., Tichauer D., Livneh J., Gershon E., Zheng M., Tan K., John S.L., Slade G.D., Jordan J., Woolf C.J., Peltz G., Maixner W., Diatchenko L., Seltzer Z., Salter M.W., Mogil J.S. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med. 2012;18:595–599. doi: 10.1038/nm.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stark C., Breitkreutz B.-J., Chatr-Aryamontri A., Boucher L., Oughtred R., Livstone M.S., Nixon J., Van Auken K., Wang X., Shi X., Reguly T., Rust J.M., Winter A., Dolinski K., Tyers M. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J.L. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 60.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 61.Vega-Avelaira D., McKelvey R., Hathway G., Fitzgerald M. The emergence of adolescent onset pain hypersensitivity following neonatal nerve injury. Mol Pain. 2012;8:30. doi: 10.1186/1744-8069-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waxman S.G. Painful Na-channelopathies: an expanding universe. Trends Mol Med. 2013 doi: 10.1016/j.molmed.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Woolf C.J. Overcoming obstacles to developing new analgesics. Nat Med. 2010;16:1241–1247. doi: 10.1038/nm.2230. [DOI] [PubMed] [Google Scholar]
- 66.Zhao J., Lee M.-C., Momin A., Cendan C.-M., Shepherd S.T., Baker M.D., Asante C., Bee L., Bethry A., Perkins J.R., Nassar M.A., Abrahamsen B., Dickenson A., Cobb B.S., Merkenschlager M., Wood J.N. Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J Neurosci. 2010;30:10860–10871. doi: 10.1523/JNEUROSCI.1980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]