Abstract
During a decade of proof-of-principle analysis in model organisms, protein networks have been used to further the study of molecular evolution, to gain insight into the robustness of cells to perturbation, and for assignment of new protein functions. Following these analyses, and with the recent rise of protein interaction measurements in mammals, protein networks are increasingly serving as tools to unravel the molecular basis of disease. We review promising applications of protein networks to disease in four major areas: identifying new disease genes; the study of their network properties; identifying disease-related subnetworks; and network-based disease classification. Applications in infectious disease, personalized medicine, and pharmacology are also forthcoming as the available protein network information improves in quality and coverage.
With the completion of the Human Genome Project, focus has shifted from cataloging the “parts list” of genes and proteins to mapping the networks of interactions that take place among them. Understanding this network is important because proteins do not function in isolation, but rather interact with one another and with DNA, RNA, and small molecules to form molecular machines. These machines are modular, involve both static and dynamic assemblies of macromolecules, and transmit as well as respond to intra- and extracellular signals (Hartwell et al. 1999).
Just as genome sequencing was first demonstrated in model organisms, analysis of protein networks has progressed initially and most rapidly in the yeast Saccharomyces cerevisiae. Due to its ease of genetic manipulation, yeast has been an ideal test bed for efforts to increase the throughput and scale of protein interaction measurements, with the ultimate goal of obtaining complete coverage of all interactions encoded by an organism. Although it is unlikely that the current network coverage of yeast is saturating, yeast nonetheless has available the greatest number of networks, and many of the largest generated for any organism. These data include yeast two-hybrid (Y2H) interaction networks (Uetz et al. 2000; Ito et al. 2001), maps of immunoprecipitated protein complexes (Gavin et al. 2006; Krogan et al. 2006), a large-scale network of protein–DNA interactions (Harbison et al. 2004), a kinase–substrate interaction map (Ptacek et al. 2005), and numerous large synthetic–lethal networks (Tong et al. 2001, 2004; Schuldiner et al. 2005; Pan et al. 2006; Collins et al. 2007). These yeast networks have provided a superb resource for development of new algorithmic approaches and conceptual frameworks for analyzing molecular interactions.
In just the past two to three years, large biomolecular interaction networks have also become available for humans. Two studies have applied the Y2H system to test for interactions among large sets of human proteins (Rual et al. 2005; Stelzl et al. 2005). These studies used full-length human open reading frames (ORFs) (Rual et al. 2005) or a combination of ORF-based clones and cDNA libraries (Stelzl et al. 2005) to clone human genes within Y2H bait and prey constructs. Using a prey pooling strategy, an “interaction space” of ∼8100 × 8100 protein pairs was tested by Rual et al. (2005) versus 4500 × 5500 by Stelzl et al. (2005), yielding over 5000 novel interactions across the two data sets. A proof-of-concept study has applied coimmunoprecipitation, followed by mass spectrometry, to characterize the interactors of 338 proteins that were selected based on their putative involvement in disease (Ewing et al. 2007). In addition to experimentation, several efforts have attempted to build large networks of human protein interactions curated from prior studies in the literature (Ramani et al. 2005; Gandhi et al. 2006) (for review, see Mathivanan et al. 2006). The effort by Gandhi et al. summarized in the HPRD database (www.hprd.org), currently catalogs over 38,000 human protein interactions.
With the increase in availability of human protein interaction data, the focus of bioinformatics development has shifted from understanding networks encoded by model species to understanding the networks underlying human disease (Kann 2007). Many of these newer studies are directly inspired by earlier developments in yeast network analysis, while others are “uniquely human.” In the remainder of this review, we briefly describe some foundations of network analysis that have emerged from studies in yeast, and then address more recent developments in network analysis of human disease. These latter developments fall into four categories: the study of network properties of human disease genes; the use of protein networks to implicate additional genes in disease; the identification of disease-related subnetworks; and the network-based classification of case-control studies.
Network analysis in yeast: A brief tour
Although numerous methods have been applied to analyze gene and protein networks in yeast, they are best understood according to their ultimate goals of analysis. A first set of methods (Jansen et al. 2003; von Mering et al. 2003; Bader et al. 2004; Lee et al. 2004; Myers and Troyanskaya 2007) was developed to clean the noise from raw interaction measurements, leading to higher-confidence networks with quantitative measures of accuracy associated with each interaction. Typically, these methods integrate many sources of evidence for protein interaction, using them as features for a classifier of interactions/noninteractions. The classifier is trained on a gold-standard set of true and false interactions and assigns to each possible interaction a confidence score. Given a high-confidence weighted network, a second set of methods was developed to predict new annotations for proteins, such as protein function, localization, and functional orthology, based on the protein's network neighbors and other network information (Letovsky and Kasif 2003; Vazquez et al. 2003; Espadaler et al. 2005; Leone and Pagnani 2005; Bandyopadhyay et al. 2006; Lee et al. 2006).
The remaining efforts in yeast fall into two categories, which might be called “synthetic” and “divisive,” respectively. Synthetic methods attempt to synthesize global properties of biology through analysis of molecular interaction networks. Many analyses in this category have examined how the number of interactions per protein (the “degree” of each protein) is distributed over all proteins in the network. In yeast protein–protein interaction networks, the degree was found to follow a power law (Yook et al. 2004) or a truncated power-law distribution (Huang et al. 2007). In complex systems, power laws are often interpreted as signatures of hierarchy and robustness (see Box 1; for review, see Barabasi and Oltvai 2004). Some of these authors also showed that proteins with high degree are likely to be essential for growth (Jeong et al. 2001). Later, Said et al. (2004) and Shachar et al. (2007) showed that protein degree correlates with other phenotypes in addition to essentiality. Said et al. (2004) investigated proteins required for the response to genotoxic agents, showing that these lay in the “middle” between nonessential and essential proteins with respect to their average degree, network distance, size of connected components, and clustering coefficient (Box 1). Similar conclusions were reported by Shachar et al. (2007) for genes involved in maintenance of telomere length.
Box 1.
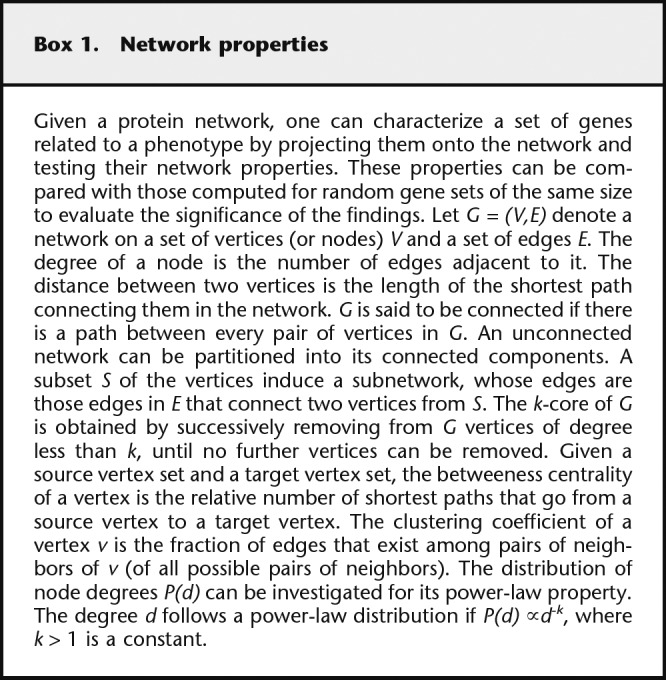
Network properties
In contrast to the above synthetic methods, divisive methods attempt to decompose or partition networks into smaller building blocks (Alon 2006). Uri Alon and colleagues (Milo et al. 2002) searched a yeast transcriptional interaction network (linking transcription factors to the genes they regulate) for instances of each possible subnetwork topology containing three or four proteins. They showed that certain subnetworks, which they called network “motifs,” were highly enriched compared with randomized networks (an example is the “feed-forward circuit,” in which one transcription factor regulates another along with the second factor's regulated genes; another is “autoregulation,” in which a transcription factor directly regulates its own gene). Some have argued that each type of network motif encodes a specific biological function: For instance, feed-forward loops may correspond to sign-sensitive accelerators, which speed up the response in an off-to-on direction, but not in an on-to-off direction (Mangan and Alon 2003).
Beyond enriched motifs, methods have been devised to identify all sorts of network structures, such as densely connected network regions (for review, see Brohee and van Helden 2006), which are suggestive of protein complexes, and paths of interactions connecting membrane proteins to transcription factors (Steffen et al. 2002; Scott et al. 2006), which are suggestive of signaling pathways. In cases in which interaction data are available across several species, there is now a burgeoning area of algorithm development aimed at detecting subnetwork structures that are evolutionarily conserved across multiple networks (for review, see Sharan and Ideker 2006). Finally, there is a rich yeast literature on methods for integrating gene-expression data with molecular networks and pathways, with the goal of identifying network “hot spots” or “expression-activated modules” (Zien et al. 2000; Ge et al. 2001; Ideker et al. 2001, 2002; Hanisch et al. 2002). Expression-activated modules are sets of proteins enriched for both interaction and coexpression across several conditions; they provide an important means of distilling the thousands of interactions present in a typical molecular network to arrive at a smaller number of discrete modules of activity.
Network analysis in human: Properties of disease genes
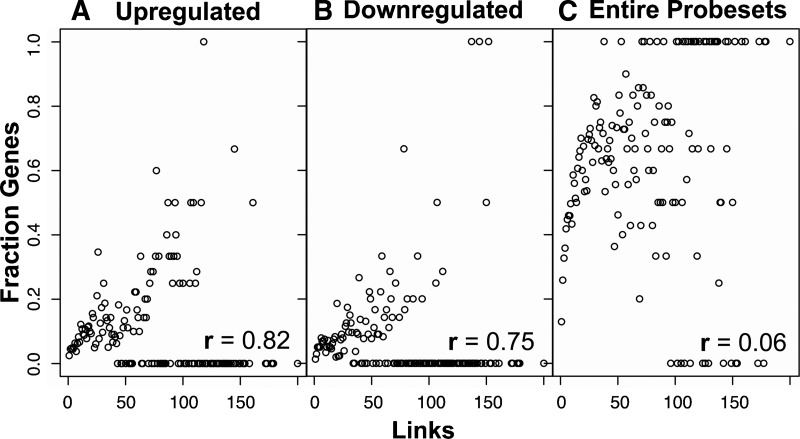
Inspired by the findings that essential yeast proteins tend to have high network degrees, several groups have now tailored such analyses to focus on phenotypes related to human disease. Wachi et al. (2005) studied genes that are differentially expressed in lung squamous cancer tissues (Fig. 1). They tested the degree distribution and centrality of the set of differentially expressed genes in a human PPI network, obtained by combining curated interactions with interactions that were transferred from model organisms (so-called “interologs”) (Matthews et al. 2001). As a centrality measure, they used the fraction of differentially expressed genes that were present in a k-core of the network (see Box 1). Up-regulated genes in the cancerous tissues tended to be highly connected and central. The authors explained this finding by suggesting that up-regulated genes are essential for proliferation of the cancerous tissue and, hence, share the topological characteristics of essential genes.
Figure 1.
Differentially expressed cancer genes tend toward higher network connectivity. Human proteins of each network degree (X-axis) were analyzed to compute their fraction of genes up-regulated (A) or down-regulated (B) in the microarray profiles of five lung cancer tissue biopsies. Both up- and down-regulated genes show significant positive correlation to protein degree, in contrast to the set of all genes on the microarray (C). Reproduced from Wachi et al. (2005) and reprinted with permission from Oxford University Press © 2005.
Jonsson and Bates (2006) investigated the network position of 346 genes that had been implicated in a comprehensive census of all human cancer genes (Futreal et al. 2004). They showed that these proteins tended to have, on average, twice as many interaction partners as noncancer proteins. In a clustering of the network (into overlapping subnetworks), the cancer proteins tended to reside in larger clusters. Moreover, such proteins tended to participate in more clusters than noncancer proteins.
Goh et al. (2007) created a network of human disease/human gene associations, in which each genetic disease is connected to the genes known to cause it, as documented in the database of Online Mendelian Inheritance in Man (Hamosh et al. 2005). They found that disease genes exhibit an increased tendency for their protein products to interact with one another, tend to be coexpressed in specific tissues, and display coherent functions with respect to all three branches of the Gene Ontology hierarchy (Harris et al. 2004). They also found that the vast majority of disease genes are nonessential and, in contrast to the above studies, have no tendency toward higher degree in the human protein–protein interaction network. In a related work, some of the same authors performed a similar analysis of a human gene/drug network, composed of gene-drug associations known to the USA Food and Drug Administration (Yildirim et al. 2007).
Combining these network-based disease studies with the original analyses of network properties in yeast, the overriding conclusion is that genes associated with a particular phenotype or function, including the progression of disease, are not randomly positioned in the network. Rather, they tend to exhibit high connectivity, cluster together, and occur in central network locations. In yeast (Said et al. 2004; Shachar et al. 2007), one sees that their network property values, such as overall degree or average distance to one another, tend to lie between those of the essential genes and those of the nonessential ones. In humans, some groups report an elevated degree for disease genes (Wachi et al. 2005; Jonsson and Bates 2006), while others do not (Goh et al. 2007). Further work will be needed to resolve this discrepancy and carefully examine the different studies for possible sources of bias. For instance, one possible explanation is that the former two studies focused on cancer genes in particular, while Goh et al. (2007) investigated disease in general. A potential source of bias, especially in literature-curated networks, is that disease-causing proteins may have higher degrees simply because they are better studied.
Prediction of disease-causing genes
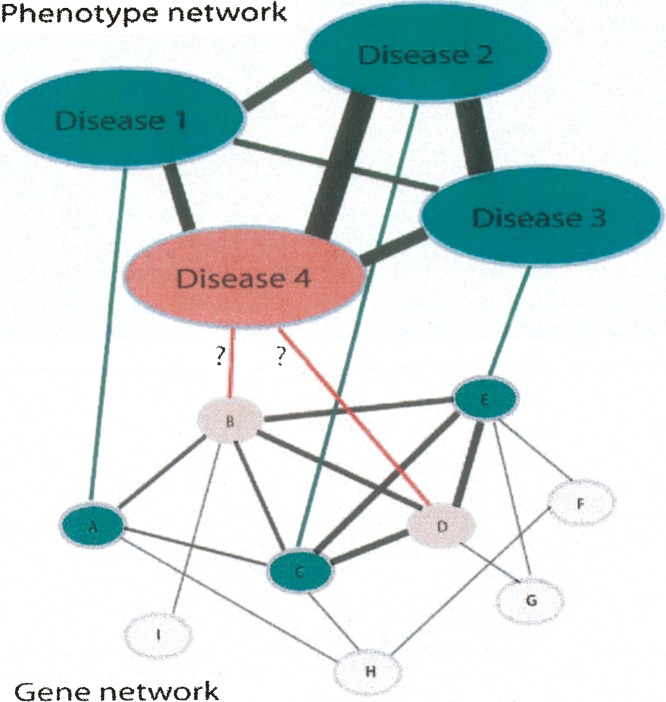
A second area in which biomolecular interaction networks have informed the study of human disease is in prediction of new disease-associated genes. The key assumption in these studies is that a network-neighbor of a disease-causing gene is likely to cause either the same or a similar disease (Goh et al. 2007; Oti and Brunner 2007). This concept is illustrated in Figure 2.
Figure 2.
A gene-phenotype network. Shown is a combined gene–gene, gene–phenotype, and phenotype–phenotype interaction network. In this hypothetical example, diseases 1, 2, and 3 have known causative genes (genes A, C, and E, respectively), and are all phenotypically related to disease 4, which lacks an identified causative gene. If the known causative genes are functionally closely related, as in this case, then candidate genes (genes B and D) can be hypothesized for disease 4 due to their close functional relationships to the known genes of the phenotypically related diseases. Black lines of varying thickness indicate the degree of phenotypic and functional similarity between diseases and genes, respectively. Reproduced from Oti and Brunner (2007) and reprinted with permission from Blackwell Publishing Ltd. © 2007 (www.blackwell-synergy.com).
Oti et al. (2006) aimed to predict disease-causing genes for genetically heterogeneous diseases in which some of the causative genes had been identified, while for other genetic factors, only locus information was available. This scenario is typical of large gene association studies, in which several significant loci are usually found (containing polymorphic markers associated with disease), but the specific causative gene in each locus is not easily identified. For a given disease, Oti et al. (2006) predicted new disease genes as those that fell within one of the significant loci and had a protein interaction with a gene already well known to cause disease. They showed that predictions using this method are 10-fold enriched in true disease-causing genes, as compared with a random selection of genes at the same locus.
Franke et al. (2006) devised the Prioritizer algorithm based on similar principles. Their algorithm ranks a set of candidate disease-causing genes in multiple susceptibility loci for further sequence or association analysis. To this end, they constructed a functional human gene network based on known molecular interactions as well as computationally predicted functional relations. The network was used to rank the candidate genes on the basis of their interactions, assuming that the causative genes for any one disorder will be involved in only a few distinct biological pathways. This assumption implies that genes from different susceptibility loci would cluster, resulting in shorter network distances (see Box 1) between disease genes than the random expectation.
Finally, Lage et al. (2007) extended the above works by using information on several related diseases to tackle the prediction task. Specifically, they devised a phenotype similarity score and used it to look for protein complexes whose genes were associated with similar phenotypes. The search for protein complexes was conducted on a network of protein–protein interactions, including reported interactions and interactions that were transferred from model organisms. Each candidate protein was ranked by the phenotype similarity score of the diseases associated with the protein and its direct network neighbors. The biological interpretation of a high-scoring candidate was that this protein was likely to be involved in the molecular pathology of a disorder of interest, since it is part of a high-confidence candidate complex in which some proteins are known to be involved in highly similar (or identical) disorders.
Thus, the idea that proteins close to one another in a network cause similar diseases is becoming an increasingly important factor in the hunt for disease genes. Different approaches tackle the prediction problem using different kinds of integrated data, but all of them involve superimposing a set of candidate genes alongside a set of known disease genes on a physical or functional network. “De-novo” approaches that do not depend on prior knowledge of disease genes are yet to be developed.
Identification of disease-related subnetworks
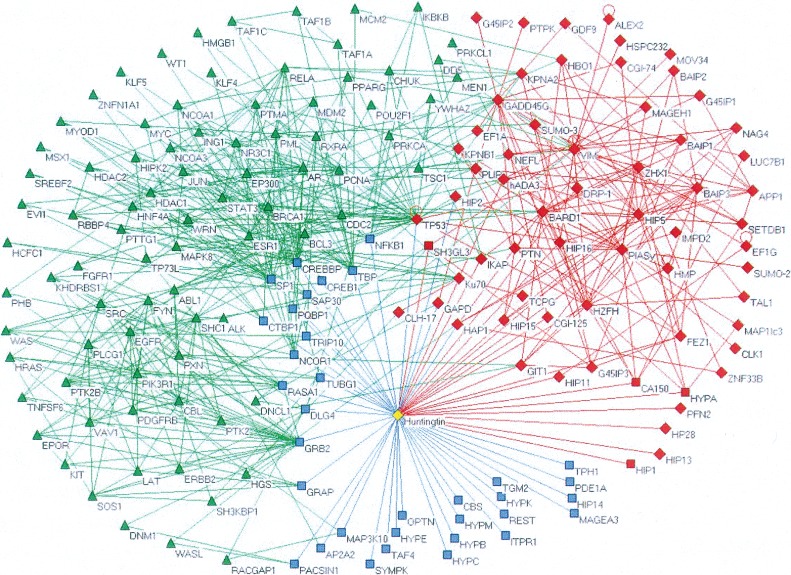
In addition to predicting individual disease proteins, a biomolecular network can also be used to predict disease-related subnetworks. Subnetworks are significant because, in contrast to individual proteins, they provide concrete hypotheses as to the molecular complexes, signaling pathways, and other mechanisms that impact the disease outcome. As one example, Goehler et al. (2004) constructed a PPI subnetwork around HTT, mutations that cause Huntington disease (Fig. 3). Each direct interactor of HTT was tested for its ability to enhance HTT aggregation, which is linked to disease progression (Sanchez et al. 2003). This screen identified a new enhancer of HTT aggregation, GIT1, and additional tests verified its role in disease pathogenesis.
Figure 3.
A protein interaction network for Huntington disease. (Red diamonds) Y2H interactors of huntingtin (HTT) newly identified by the Goehler et al. (2004) study. (Blue squares) Previously published interactors. (Green triangles) Interactors culled from human protein interaction databases (HRPD) (Gandhi et al. 2006), MINT (Chatr-aryamontri et al. 2007), and BIND (Bader et al. 2003). (Red squares) HTT interactors that were both newly identified and previously reported. Reproduced from Goehler et al. (2004) and reprinted with permission from Elsevier Ltd. © 2004.
Calvano et al. (2005) assembled an endotoxin inflammatory response network by integrating functional interactions curated from the literature with gene-expression information. Here, gene-expression profiling was used to identify genes responsive to endotoxin administration. The response network enabled the identification of new endotoxin-responsive modules and revealed a reprioritization of the transcriptional regulatory program in leukocytes in response to endotoxin, suppressing mitochondrial energy production and protein synthesis machinery. More generally, the theme of interpreting expression profiles as hot spots of activity within a protein network has potential application to many disease studies; moving forward, it might benefit substantially by drawing on the earlier methods developed in yeast to identify expression-activated network modules (see above).
While the above approach specifies a subnetwork by overlaying expression profiles as states on a functional network, expression profiles have also been used to define the network itself, which is then integrated with other types of data. For instance, Ghazalpour et al. (2006) constructed a gene coexpression network using microarray profiles gathered from the livers of a panel of mice, in which genes were linked if the correlation between their expression profiles exceeded a certain threshold. This network was then integrated with genetic marker data from the same individuals, indicating which genetic loci had association with 22 different physiological traits such as body weight. Various subnetwork “modules” of the coexpression network were found to be enriched for genes in loci with strong associations to a physiological trait, yielding a matrix of module/trait associations.
Lim et al. (2006) used Y2H screens to construct a PPI network around 23 proteins involved in inherited ataxias. The resulting network was expanded by literature-curated interactions and interologs and contained a total of ∼7000 interactions among ∼3500 proteins. The network was found to be highly connected, and the mean distance between the ataxia-causing proteins in the network was much lower than in a network nucleated around 30 disease proteins sampled independent of phenotypes. The authors demonstrated the network's utility in uncovering novel ataxia-causing genes and genetic modifiers for ataxia.
In a conceptually similar study (Pujana et al. 2007), a breast cancer-related network was constructed starting with four known breast cancer-associated genes: BRCA1, BRCA2, ATM, and CHEK2. To form the network, proteins were linked based on coexpression, phenotypic similarity, and genetic or physical interactions among orthologs of the proteins in other species. The network neighbors of the starting four disease genes implicated additional factors important for breast cancer progression, including a new gene, HMMR, which was shown by the authors to be associated with a higher risk of breast cancer in two case-control studies.
Common to all of the above studies is the understanding that integrating disease genes with physical or functional networks can lead to the identification of additional disease-related genes and generate subnetworks that offer mechanistic hypotheses about the causes of disease. The interactions within such subnetworks are often suggestive of functional signaling cascades, metabolic pathways, or molecular complexes that are either causes or effects of the disease phenotype. They help to explain the influence of the many genetic and environmental factors influencing a disease in the context of a smaller number of discrete modules.
Network-based classification of case-control studies
A final emerging application of molecular network analysis is the use of networks to improve the task of disease classification. Classification has long been an important technique for identifying biomarkers able to separate “cases” from “controls.” In disease research in particular, cases versus controls are used to separate individuals who have a disease versus those who do not, to predict individuals likely to have severe disease outcomes versus those who may be treated less aggressively, or to distinguish between different diseases that might otherwise appear superficially similar (Quackenbush 2006). In terms of biomarker identification, large-scale profiling experiments (mRNA, protein, and small-molecular) provide quantitative measurements for thousands of molecules; however, a major challenge has been to develop methods able to sift through the enormous number of potential markers to identify the particular set having the highest predictive ability (Sotiriou and Piccart 2007). One solution to this problem may be to integrate the search for biomarkers with a network-level map of the cell, which focuses the search around sets of markers functioning in the same pathway.
As one example, Tuck et al. (2006) examined the utility of local features of a transcriptional interaction network in classifying disease states. As the features for classification, they considered both single transcriptional interactions and sets of interactions incident to the same regulated gene. For a given sample, a transcriptional interaction feature was said to be “active” (i.e., activity = 1) if the corresponding transcription factor and gene were coexpressed in the sample. The activity of a set of interactions was defined as the sum of their activities. These features were successfully used for classifying patients to disease states. The authors noted that the network-based classification compared favorably with gene-expression–based classification. Moreover, they observed that genes that optimally classify samples (based on their set of incident interactions) are close to one another in the network.
Ma et al. (2007) used this same notion, i.e., that informative genes fall into the same network neighborhoods, to identify genes predictive of Alzheimer's disease. Their method, termed “combining gene expression and protein interaction data” (CGI), uses the protein interaction network to define a Markov Random Field, in which the score of association between a gene's expression level and the disease class depends on the scores of its network neighbors according to a diffusion kernel. CGI was reported to perform better than when gene-expression data were used as the sole source of information.
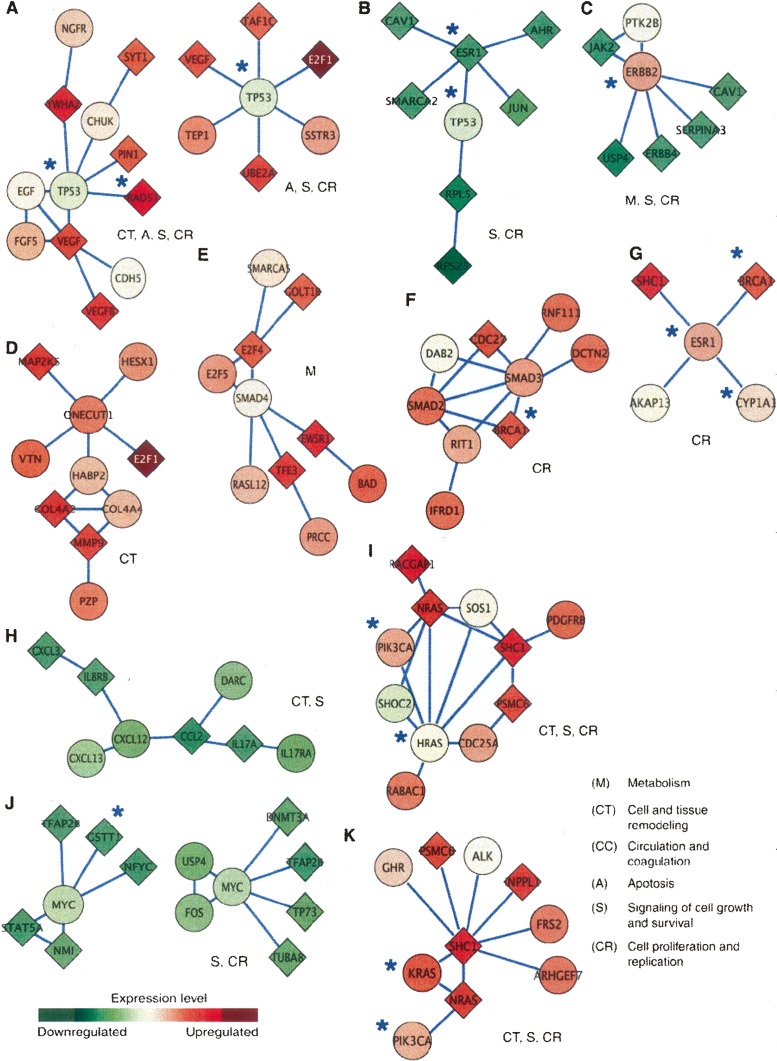
Chuang et al. (2007) applied a network-based classifier to the prognosis of breast cancer metastasis. According to their method, the gene-expression profiles of metastatic and nonmetastatic patients are superimposed on a human protein–protein interaction network. Subnetworks are identified whose expression levels correlate with metastasis, where the expression level of a subnetwork is defined as a function of the expression levels of its member genes (Fig. 4). They found that subnetwork markers were more reproducible than individual marker genes selected without network information, and that they achieved higher accuracy in the classification of metastatic versus nonmetastatic tumors.
Figure 4.
Discriminative subnetworks enriched with hallmarks of cancer. Vertices and edges represent human proteins and protein interactions, respectively. The color of each node scales with the change in expression of the corresponding gene for metastatic (red) versus nonmetastatic (green) cancer. The shape of each node indicates whether its gene is significantly differentially expressed (diamond; P <0.05 from a two-tailed t-test) or not (circle). The predominant cellular functions are indicated next to each module, and known breast cancer susceptibility genes are marked by a blue asterisk. Reproduced from Chuang et al. (2007) with permission from Macmillan Publishers Ltd. © 2007.
Efroni et al. (2007) performed a related study, in which they sought to identify pathways associated with cancer gene-expression data sets. Expression data were used to score known pathways (taken from curated databases) for the level of activity and consistency of their interactions in a given sample. These scores were then used as features for classifying samples to disease states, subsequently deriving small collections of pathways that distinguish the phenotypes with high accuracy. The main difference versus Chuang et al. (2007) is that curated pathways were used rather than subnetworks dynamically extracted from a protein network.
Thus, protein networks are proving to be a powerful source of information for disease classification. Typically, one superimposes gene-expression data onto the network to identify links, or more composite subnetwork structures, whose aggregate expression discriminates between disease states. These discriminating subnetworks are more reproducible than single genes and can improve the prediction accuracy.
The future of networks and disease
One means of charting the road ahead is to recognize emerging studies in yeast that are just beginning to be adapted to the more complex networks of human disease. One such area is protein network evolutionary comparison. In yeast, there is now a sizable literature reporting methods to align, contrast, and compare protein networks across species spanning a wide range of evolutionary distances (see above and Sharan and Ideker 2006). In humans, network comparative methods are receiving significant attention in the study of infectious disease. An increasing number of protein interaction networks are becoming available for microbial pathogens, including Helicobacter pylori (Rain et al. 2001), Campylobacter jejuni (Parrish et al. 2007), and Plasmodium falciparum (LaCount et al. 2005). Comparison to the human protein network is providing useful insights by identifying which protein interactions are common between pathogen and host and, more importantly from the standpoint of drug development, which interactions might be wired differently. For instance, Suthram et al. (2005) analyzed an interaction map of the malarial pathogen P. falciparum and showed that it was significantly divergent from the interaction maps of other eukaryotes.
Network-level analyses of viral pathogens are also underway (Flajolet et al. 2000; McCraith et al. 2000; von Schwedler et al. 2003; Uetz et al. 2006; Calderwood et al. 2007). Projects to map protein networks among viral proteins, or between viral and host proteins, are appealing because of the greatly reduced number of proteins and protein interactions that must be tested—i.e., even the largest viral genomes encode no more than 1000 proteins (Raoult et al. 2004), versus the more than 20,000 that must be considered in higher eukaryotes. Comparison of these networks to each other and with the human protein network may be able to elucidate key mechanisms of infection: Toward this goal, Uetz et al. (2006) used interactions between herpesviral and human proteins to connect the viral interactome into the known human protein interaction network and to simulate infection.
Another important area to watch will be the application of protein interaction networks to interpret the effects of genetic and environmental perturbations on human populations. In yeast, genetic perturbations have been profiled by expression profiling of gene knockout strains (Hughes et al. 2000; Workman et al. 2006; Hu et al. 2007) or by analysis of expression quantitative trait loci (eQTLs) (Brem and Kruglyak 2005), which identifies associations between polymorphic genomic markers and gene-expression changes. In both of these cases, perturbed genes (either explicitly by gene deletion or implicitly through population genetics) are linked to genes downstream whose expression levels are affected by perturbation. For integrative analysis of these data, a number of network-based approaches have been proposed that map the molecular interaction pathways leading from each causal gene to its affected genes (Yeang et al. 2004, 2005; Ourfali et al. 2007; Shachar et al. 2007). These approaches typically model a signaling pathway as a path of interactions through a protein–protein or protein–DNA interaction network, in which each interaction is associated with a direction of information flow and a regulatory influence (activating “+” or repressing “−”). The most plausible pathway explanations for each cause-and-effect relationship are inferred from the data, along with the likely direction and regulatory influence of each pathway interaction. Recently, two studies have taken a network-based approach to eQTL analysis in particular, relying on the idea that the causal gene should be well connected to the affected genes in the network (Tu et al. 2006; Suthram et al. 2007).
In humans, network analysis may offer a powerful means of mapping the molecular mechanisms underlying the genetic and environmental perturbations at the heart of disease. During the past few years, a substantial body of cause-and-effect pertubation data have been generated in humans, including a number of eQTL studies (Dixon et al. 2007; Goring et al. 2007; Stranger et al. 2007). As for yeast, interpretation of human eQTLs could greatly benefit from integration with protein–protein and transcriptional interactions. However, despite the availability of human protein–protein interaction networks, large-scale measurements of human transcriptional interactions are still underway, mainly using systematic chromatin immunoprecipitation experiments in human cell lines and in-vitro technologies such as the protein-binding microarray (PBM) (Berger et al. 2006). As human transcriptional networks become available, the approaches demonstrated in yeast may also provide a powerful means for identifying human disease genes and their associated transcriptional regulatory pathways.
Yet another direction in which network-based analysis might inform human disease is in pharmacology, i.e., drug discovery and targeting. Pioneering work in yeast (Parsons et al. 2004, 2006) has demonstrated the utility of combining molecular interaction networks with measurements of chemical–genetic interactions (in which the combination of a chemical agent with a gene knockout leads to cell death). These studies are based on the observation that the chemical–genetic interaction profile of an agent is similar to the genetic interaction profile of genes in its target pathway. Moreover, gene deletions that result in hypersensitivity to a specific drug can identify pathways that buffer the cell against the toxic effects of the drug, providing clues about its mode of action.
All of these and other applications of molecular networks to disease will continue to face technological, biological, and algorithmic challenges. Human network data remain sparse, and many important types of networks, such as networks of regulatory and synthetic–lethal or chemical–genetic interactions, are still forthcoming. Issues of data collection and interpretation are complicated by the large size of the proteome in human and its diversity of cells and tissues. In addition, existing computational frameworks are ill-suited to cope with the ongoing explosion in network-level measurements and information.
Nonetheless, elucidating the mechanisms of human disease remains a holy grail of bioinformatics. Most previous studies in this regard have analyzed single genes and the changes they exhibit in the diseased state. The recent availability of human molecular interaction networks has revolutionized this view by demonstrating the importance not only of the proteins themselves, but of their inter-relationships. If the recent progress in the field is any indicator, exploiting these networks is also destined to revolutionize our view of fundamental human biology as well as disease progression, diagnosis, and treatment.
Acknowledgments
T.I. is a David and Lucille Packard Fellow and is supported by NIH grants ES14811 and GM070743. R.S. is supported by an Alon fellowship and by the Israel Science Foundation (grant no. 385/06).
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.071852.107.
References
- Alon U. Introduction to systems biology: Design principles of biological circuits. Chapman and Hall; London, UK: 2006. [Google Scholar]
- Bader G.D., Betel D., Hogue C.W. BIND: The Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Sharan R., Ideker T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 2006;16:428–435. doi: 10.1101/gr.4526006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.L., Oltvai Z.N. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., 3rd, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R.B., Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohee S., van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood M.A., Venkatesan K., Xing L., Chase M.R., Vazquez A., Holthaus A.M., Ewence A.E., Li N., Hirozane-Kishikawa T., Hill D.E., et al. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano S.E., Xiao W., Richards D.R., Felciano R.M., Baker H.V., Cho R.J., Chen R.O., Brownstein B.H., Cobb J.P., Tschoeke S.K., et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A., Ceol A., Palazzi L.M., Nardelli G., Schneider M.V., Castagnoli L., Cesareni G. MINT: The Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H.Y., Lee E., Liu Y.T., Lee D., Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Efroni S., Schaefer C.F., Buetow K.H. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS ONE. 2007;2:e425. doi: 10.1371/journal.pone.0000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espadaler J., Aragues R., Eswar N., Marti-Renom M.A., Querol E., Aviles F.X., Sali A., Oliva B. Detecting remotely related proteins by their interactions and sequence similarity. Proc. Natl. Acad. Sci. 2005;102:7151–7156. doi: 10.1073/pnas.0500831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M.D., O'Connor L., Li M., et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspe G., Tiollais P., Transy C., Legrain P. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–379. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Franke L., Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi T.K., Zhong J., Mathivanan S., Karthick L., Chandrika K.N., Mohan S.S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B., et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ge H., Liu Z., Church G.M., Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A., Doss S., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K.S., Knoblich M., Haenig C., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabasi A.L. The human disease network. Proc. Natl. Acad. Sci. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch D., Zien A., Zimmer R., Lengauer T. Co-clustering of biological networks and gene expression data. Bioinformatics. 2002;(Suppl. 1) 18:S145–S154. doi: 10.1093/bioinformatics/18.suppl_1.s145. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Hu Z., Killion P.J., Iyer V.R. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- Huang H., Jedynak B.M., Bader J.S. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 2007;3:e214. doi: 10.1371/journal.pcbi.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Ideker T., Ozier O., Schwikowski B., Siegel A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;(Suppl. 1) 18:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jonsson P.F., Bates P.A. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M.G. Protein interactions and disease: Computational approaches to uncover the etiology of diseases. Brief. Bioinform. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- LaCount D.J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J.R., Schoenfeld L.W., Ota I., Sahasrabudhe S., Kurschner C., et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- Lage K., Karlberg E.O., Storling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tumer Z., Pociot F., Tommerup N., et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Lee I., Date S.V., Adai A.T., Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Lee H., Tu Z., Deng M., Sun F., Chen T. Diffusion kernel-based logistic regression models for protein function prediction. OMICS: A Journal of Integrative Biology. 2006;10:40–55. doi: 10.1089/omi.2006.10.40. [DOI] [PubMed] [Google Scholar]
- Leone M., Pagnani A. Predicting protein functions with message passing algorithms. Bioinformatics. 2005;21:239–247. doi: 10.1093/bioinformatics/bth491. [DOI] [PubMed] [Google Scholar]
- Letovsky S., Kasif S. Predicting protein function from protein/protein interaction data: A probabilistic approach. Bioinformatics. 2003;(Suppl. 1) 19:i197–i204. doi: 10.1093/bioinformatics/btg1026. [DOI] [PubMed] [Google Scholar]
- Lim J., Hao T., Shaw C., Patel A.J., Szabo G., Rual J.F., Fisk C.J., Li N., Smolyar A., Hill D.E., et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Ma X., Lee H., Wang L., Sun F. CGI: A new approach for prioritizing genes by combining gene expression and protein-protein interaction data. Bioinformatics. 2007;23:215–221. doi: 10.1093/bioinformatics/btl569. [DOI] [PubMed] [Google Scholar]
- Mangan S., Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S., Periaswamy B., Gandhi T.K., Kandasamy K., Suresh S., Mohmood R., Ramachandra Y.L., Pandey A. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;(Suppl. 5) 7:S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L.R., Vaglio P., Reboul J., Ge H., Davis B.P., Garrels J., Vincent S., Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs.”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S., Holtzman T., Moss B., Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Myers C.L., Troyanskaya O.G. Context-sensitive data integration and prediction of biological networks. Bioinformatics. 2007;23:2322–2330. doi: 10.1093/bioinformatics/btm332. [DOI] [PubMed] [Google Scholar]
- Oti M., Brunner H.G. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- Oti M., Snel B., Huynen M.A., Brunner H.G. Predicting disease genes using protein-protein interactions. J. Med. Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourfali O., Shlomi T., Ideker T., Ruppin E., Sharan R. SPINE: A framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics. 2007;23:i359–i366. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Parrish J.R., Yu J., Liu G., Hines J.A., Chan J.E., Mangiola B.A., Zhang H., Pacifico S., Fotouhi F., DiRita V.J., et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons A.B., Brost R.L., Ding H., Li Z., Zhang C., Sheikh B., Brown G.W., Kane P.M., Hughes T.R., Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Parsons A.B., Lopez A., Givoni I.E., Williams D.E., Gray C.A., Porter J., Chua G., Sopko R., Brost R.L., Ho C.H., et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Pujana M.A., Han J.D., Starita L.M., Stevens K.N., Tewari M., Ahn J.S., Rennert G., Moreno V., Kirchhoff T., Gold B., et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray analysis and tumor classification. N. Engl. J. Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Ramani A.K., Bunescu R.C., Mooney R.J., Marcotte E.M. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Said M.R., Begley T.J., Oppenheim A.V., Lauffenburger D.A., Samson L.D. Global network analysis of phenotypic effects: Protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2004;101:18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez I., Mahlke C., Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Scott J., Ideker T., Karp R.M., Sharan R. Efficient algorithms for detecting signaling pathways in protein interaction networks. J. Comput. Biol. 2006;13:133–144. doi: 10.1089/cmb.2006.13.133. [DOI] [PubMed] [Google Scholar]
- Shachar R., Unger L., Kupiec M., Ruppin R., Sharan R. Proc. RECOMB satellite workshop on systems biology. Columbia University; NY: 2007. A systems-level approach to mapping the telomere-length maintenance gene circuitry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R., Ideker T. Modeling cellular machinery through biological network comparison. Nat. Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- Sotiriou C., Piccart M.J. Taking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care? Nat. Rev. Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- Steffen M., Petti A., Aach J., D'Haeseleer P., Church G. Automated modelling of signal transduction networks. BMC Bioinformatics. 2002;3:34. doi: 10.1186/1471-2105/3/34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthram S., Sittler T., Ideker T. The Plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108–112. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthram S., Beyer A., Ideker T. Proc. RECOMB satellite workshop on systems biology. Columbia University; NY: 2007. eQED: An efficient method for interpreting eQTL associations using protein networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tu Z., Wang L., Arbeitman M.N., Chen T., Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics. 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- Tuck D.P., Kluger H.M., Kluger Y. Characterizing disease states from topological properties of transcriptional regulatory networks. BMC Bioinformatics. 2006;7:236. doi: 10.1186/1471-2105-7-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Uetz P., Dong Y.A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S.V., Roupelieva M., Rose D., Fossum E., et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- Vazquez A., Flammini A., Maritan A., Vespignani A. Global protein function prediction from protein-protein interaction networks. Nat. Biotechnol. 2003;21:697–700. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U.K., Stuchell M., Muller B., Ward D.M., Chung H.Y., Morita E., Wang H.E., Davis T., He G.P., Cimbora D.M., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Wachi S., Yoneda K., Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C.T., Mak H.C., McCuine S., Tagne J.B., Agarwal M., Ozier O., Begley T.J., Samson L.D., Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang C.H., Ideker T., Jaakkola T. Physical network models. J. Comput. Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- Yeang C.H., Mak H.C., McCuine S., Workman C., Jaakkola T., Ideker T. Validation and refinement of gene-regulatory pathways on a network of physical interactions. Genome Biol. 2005;6:R62. doi: 10.1186/gb-2005-6-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M.A., Goh K.I., Cusick M.E., Barabasi A.L., Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Yook S.H., Oltvai Z.N., Barabasi A.L. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- Zien A., Kuffner R., Zimmer R., Lengauer T. Analysis of gene expression data with pathway scores. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:407–417. [PubMed] [Google Scholar]