SUMMARY
Natural killer (NK) cells play an essential role in the defense against influenza virus, one of the deadliest respiratory viruses known today. The NKp46 receptor, expressed by NK cells, is critical for controlling influenza infections, as influenza-virus-infected cells are eliminated through the recognition of the viral hemagglutinin (HA) protein by NKp46. Here, we describe an immune-evasion mechanism of influenza viruses that is mediated by the neuraminidase (NA) protein. By using various NA blockers, we show that NA removes sialic acid residues from NKp46 and that this leads to reduced recognition of HA. Furthermore, we provide in vivo and in vitro evidence for the existence of this NA-mediated, NKp46-dependent immune-evasion mechanism and demonstrate that NA inhibitors, which are commonly used for the treatment of influenza infections, are useful not only as blockers of virus budding but also as boosters of NKp46 recognition.
INTRODUCTION
Influenza viruses are responsible for most respiratory infections, affecting all age groups (Thompson et al., 2004). In influenza-virus-infected cells, the assembly and budding of progeny viruses is the final and critical step in the life cycle of the virus (Rossman and Lamb, 2011). This step requires the coordinated localization of the viral hemagglutinin (HA) and neuraminidase (NA) proteins to lipid raft domains (Nayak et al., 2004; Wang et al., 2007), which allows the virus to acquire these glycoproteins by simply budding through the host cell membrane (Rifkin and Quigley, 1974). To complete the viral budding, the viral NA enzyme cleaves sialic acid residues that attach the progeny virus to the infected cells (Nayak et al., 2004).
HA is recognized by natural killer (NK) cells through their cytotoxic receptors NKp44 and NKp46, either on the infected cells or when the virus adheres to the cells, and this recognition leads to sialic-acid-dependent, NKp44/NKp46-mediated killing (Achdout et al., 2010; Arnon et al., 2004; Ho et al., 2008; Mandelboim et al., 2001). In vivo studies confirmed the significance of the NKp46-HA interaction by showing that in the absence of NCR1 (NKp46 mouse orthologous protein), all NCR1-deficient mice (Ncr1gfp/gfp) succumbed to A/Puerto Rico/8/34 influenza-virus infection, whereas most of the wild-type (WT) mice survived (Gazit et al., 2006; Glasner et al., 2012).
Here we demonstrate both in vitro and in vivo that influenza viruses use their NA proteins to counter NKp46/NCR1 recognition, and that NA inhibitors, which are widely used to treat influenza infections (Jefferson et al., 2009), not only affect virus budding but also boost NKp46/NCR1 activity.
RESULTS
Assessment of NCR1 Activity against Influenza Virus
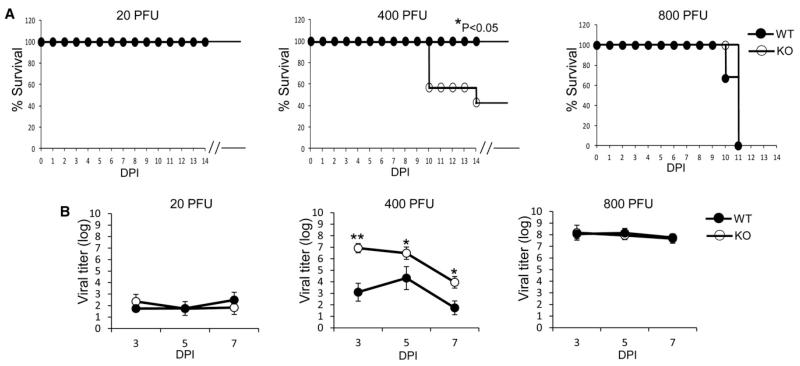
We previously showed that in the absence of NCR1, the NCR1 knockout mice (Ncr1gfp/gfp) are more sensitive to infection as compared with their WT or heterozygous littermates (Gazit et al., 2006; Glasner et al., 2012). To test whether the increased sensitivity of the Ncr1gfp/gfp mice would be observed at various doses of influenza virus, we infected Ncr1+/+ (WT) and Ncr1gfp/gfp (KO) mice with various plaque-forming units (PFU) of the A/Puerto Rico/8/34 influenza virus. In mice infected with a low dose (20 PFU) of the A/Puerto Rico/8/34 influenza virus, a very mild disease developed, as all of the Ncr1gfp/gfp and Ncr1+/+ mice survived the infection (Figure 1A). When a higher dose was applied (400 PFU), a more severe disease developed, and differences were seen in the survival of the Ncr1+/+ mice compared with the Ncr1gfp/gfp mice (Figure 1A, middle). When 800 PFU were administered, all mice died due to the infection (Figure 1A).
Figure 1. The Absence of NCR1 Affects Influenza Infection at a Certain Dose.
(A and B) Survival rate (A) and viral titers in the lungs (B, log scale) were evaluated in Ncr1+/+ mice (WT) and Ncr1gfp/gfp mice (KO) following infection with the A/Puerto Rico/8/34 influenza virus at various PFU (indicated in the figure). Virus titers in the lungs were determined at various DPIs (days postinfection). The figure shows one representative experiment out of two performed. Statistically significant differences are indicated (*p < 0.05, **p < 0.01). The break in the x axis indicates that all mice that survived the infection stayed healthy. In (B), mean values and SD are shown. In (A) and (B), the x axis indicates DPI.
We also analyzed the virus titers in the infected lungs. When 20 PFU were administered, low virus titers were detected in the lungs, and in mice infected with 800 PFU, high virus titers were detected at all time points irrespective of whether NCR1 was present (Figure 1B). In contrast and in agreement with the survival data (Figure 1A), in mice infected with 400 PFU, differences in the virus titers were detected between the Ncr1gfp/gfp and Ncr1+/+ mice at all time points tested (Figure 1B).
Because we observed that the differences in lethality and virus titers in the absence of NCR1 were only noted at a particular dose of infection and that at slightly higher viral titers the absence of NCR1 had no effect, and because viruses often develop various means to counterattack important antiviral immune mechanisms, we hypothesized that influenza viruses might have developed mechanisms to interfere with NKp46/NCR1-HA recognition and that following the neutralization of these mechanisms, NKp46/NCR1 would be able to better control influenza infections.
NA Impairs NKp46/NCR1 Recognition of Influenza Viruses
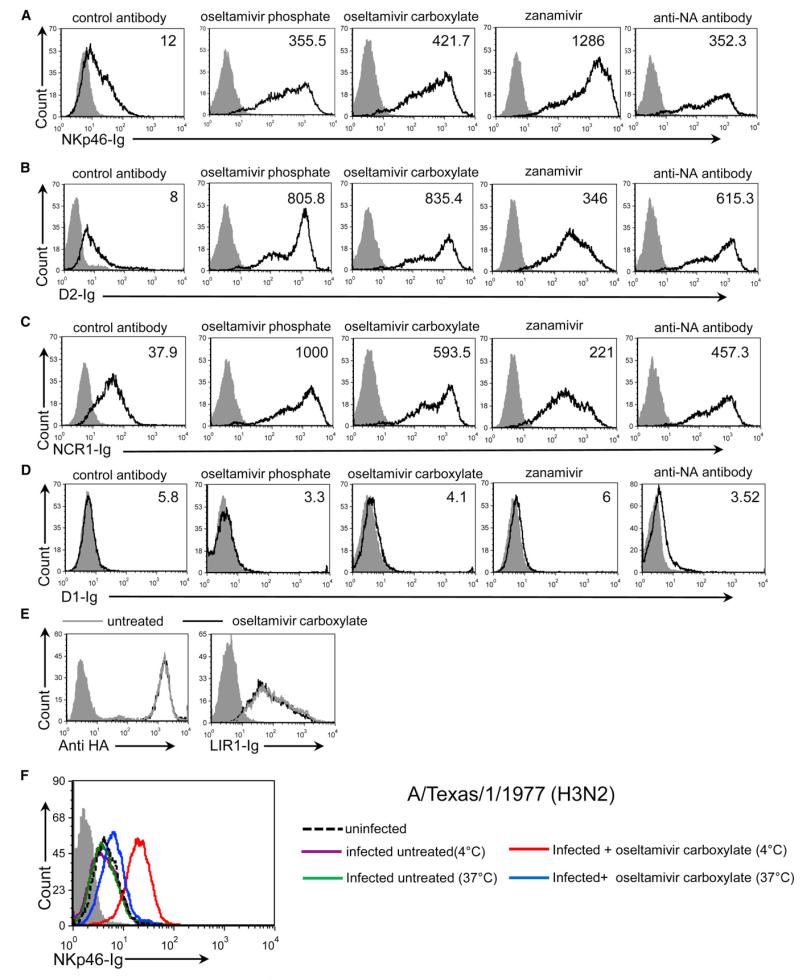
We previously showed that sialic acid residues attached to the human NKp46/NCR1 are essential for their interactions with HA (Achdout et al., 2010; Arnon et al., 2004; Glasner et al., 2012). Because the viral NA glycoprotein removes sialic acid residues, we wondered whether NA counters the HA recognition of NKp46/NCR1 by removing their sialic acid residues. To address this issue, we examined the binding of NKp46-immunoglobulin (NKp46-Ig) to A/Puerto Rico/8/34-infected Jeg3 cells following treatment with the NA inhibitor oseltamivir phosphate. As reported previously (Achdout et al., 2010), in the absence of influenza, little or no NKp46-Ig binding was observed (data not shown). However, upon infection, Jeg3 cell expression of both HA and NA was detected and NKp46-Ig staining was noticed (Figure 2A, left histogram and data not shown). Importantly, the binding of NKp46-Ig was significantly enhanced following inhibition of NA activity with oseltamivir phosphate, oseltamivir carboxylate, or zanamivir, or after blocking with anti-NA monoclonal antibody (mAb; Figure 2A).
Figure 2. Increased Binding of NKp46/NCR1 to Infected Cells following Blocking of NA Activity.
(A–D) FACS staining of infected cells. Staining was performed with NKp46-Ig (A), D2-Ig (B), NCR1-Ig (C), and D1-Ig (D). In each part of the figure, the left panels show the staining of the infected cells treated with control antibody (12E7), and the other panels show the staining of infected cells treated with oseltamivir phosphate, oseltamivir carboxylate, zanamivir, or anti-NA mAb (NA2-1C1). The various treatments are indicated above the figure. The empty black histograms depict staining with the indicated fusion protein. The median fluorescent intensity (MFI) of each staining is indicated in the histograms. See also Figure S1. (E) FACS analysis of infected cells stained with anti-HA mAb (left histogram) or LIR1-Ig (right histogram). In each panel, the empty gray histogram depicts the staining of the untreated cells with the indicated fusion protein, and the empty black histogram depicts the staining following oseltamivir carboxylate treatment. (F) FACS analysis of A/Texas/1/1977-infected EL-4 cells stained with NKp46-Ig at 4°C and at 37°C with and without oseltamivir carboxylate treatment. In all figure parts the staining is presented after gating of the live-cell population only, and the filled gray histograms depict staining with the secondary mAb only. The figure shows representative staining, and four independent experiments were performed.
The NKp46 receptor contains two extracellular C2-type Ig-like domains: a membrane-distal domain (D1) and a membrane-proximal domain (D2). The latter was shown to contain the sialic acid residue (Thr 225), which is essential for HA-NKp46 recognition (Arnon et al., 2004). Thus, to further corroborate our results, we generated truncated versions of NKp46 protein that contained the membrane-distal domain (D1-Ig) or the membrane-proximal domain including the stalk region (D2-Ig) of NKp46 fused to human IgG1. Using these fusion proteins, we observed increased binding of D2-Ig to influenza-virus-infected cells that were treated with NA inhibitors (Figure 2B). A similar effect was observed when the cells were stained with NCR1-Ig (Figure 2C), but no binding was observed when D1-Ig was used (Figure 2D). To control our experiments, we stained both treated and untreated influenza-virus-infected cells with an anti-influenza virus type A HA mAb and with a fusion protein that contains the extracellular domain of the NK inhibitory receptor LIR1 (LIR1-Ig). We observed that untreated and treated cells were similarly recognized by anti-HA mAb and LIR1-Ig (Figure 2E).
The A/Puerto Rico/8/34 virus (H1N1) is a human virus that has been adapted for mouse studies. This virus is able to bind both Neu5NAcα(2,3) and Neu5NAcα(2,6), sialic acid residues that are found on NKp46 (Mendelson et al., 2010). Because most human viruses preferentially interact with Neu5NAcα(2,6) sialic acid residues, it was important to test whether similar results would be obtained with human viruses. To that end, we tested NKp46-Ig binding to the influenza A/Texas/1/1977 (H3N2) virus. We used this virus to infect mouse EL4 cells because we observed that in some tumor cells that express an unknown tumor ligand for NKp46 (such as EL4), binding of NKp46-Ig is not observed following infection (Figure 2F and data not shown). We reasoned that this is because the expression of the unknown tumor ligand for NKp46 is already quite high, and thus upon infection with influenza, the binding of NKp46 to HA cannot be detected. We further thought that upon oseltamivir carboxylate treatment, we might be able to observe the HA-NKp46 interactions.
To test the above hypotheses, EL-4 cells were infected with the A/Texas/1/1977 (H3N2) influenza virus, treated or not with oseltamivir carboxylate, and incubated with NKp46-Ig at 4°C and 37°C. Importantly, upon oseltamivir carboxylate treatment, the binding of NKp46 to the A/Texas/1/1977-infected cells was significantly increased (Figure 2F). Interestingly, the increased binding of NKp46 to A/Texas/1/1977-infected cells (Figure 2F) and A/Puerto Rico/8/34-infected cells (Figures 2A-2E) following NA inhibition was observed at 4°C, even though this temperature decreases the enzymatic activity of NA. We expected, however, that at 37°C the NA activity would be more prominent, and indeed when the A/Texas/1/1977-infected cells were treated with oseltamivir carboxylate and incubated at 37°C, the increased NKp46-Ig binding was markedly reduced compared with incubation at 4°C (Figure 2F), probably because the oseltamivir carboxylate treatment (at this dose) was insufficient to inhibit the fully active NA enzyme.
To further demonstrate that anti-NA drugs boost the NKp46-Ig recognition of infected cells, we treated influenza-virus-infected cells with two concentrations of oseltamivir phosphate or oseltamivir carboxylate. At a concentration of 10 μg/ml, both drugs enhanced the NKp46-Ig binding (Figure S1), whereas at a concentration of 10 ng/ml, increased NKp46-Ig binding was observed only with oseltamivir carboxylate (which is more active than oseltamivir phosphate; Lindemann et al., 2010).
NA Impairs the Direct Interaction between HA and NKp46/NCR1 by Removing their Sialic Acid Residues
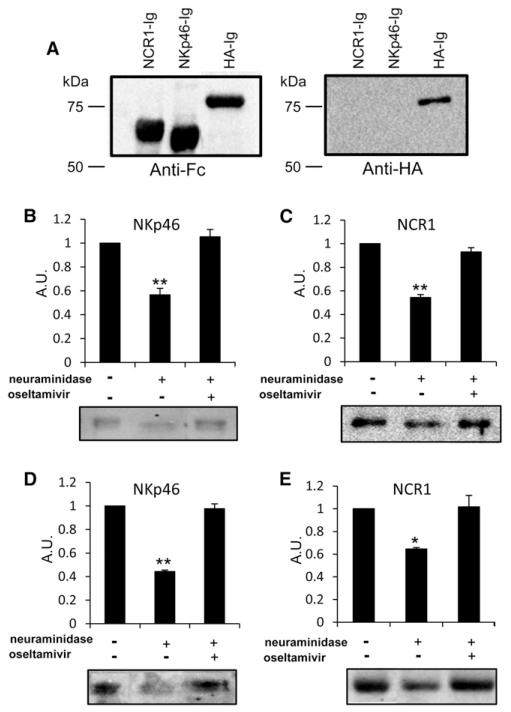
We next tested whether NA treatment impairs the interaction between NKp46/NCR1 and HA by reducing the sialic acid content of these two receptors. For this, we generated an additional fusion protein in which the extracellular portion of the HA protein (A/Puerto Rico/8/34) was fused to the human IgG1 Fc domain (HA-Ig). We verified by western blotting that all fusion proteins were highly purified, had the correct size, and were not degraded by the use of anti-human Fc antibodies (Figure 3A, left panel) or anti-HA antibody (Figure 3A, right panel). NKp46-Ig and NCR1-Ig were then treated with NA in the presence or absence of oseltamivir carboxylate and run on SDS-PAGE gels. We previously demonstrated that α2,6-linked sialic acid residues are important for NKp46-HA recognition (Arnon et al., 2004). We therefore performed western blotting using Sambucus nigra agglutinin (SNA) lectin, which preferentially binds these residues. As can be seen in Figures 3B and 3C, the NA treatment significantly reduced the binding of SNA lectin to NKp46-Ig and NCR1-Ig as compared with the untreated proteins. Inhibition of NA activity by oseltamivir carboxylate restored the SNA lectin binding to both proteins (Figures 3B and 3C).
Figure 3. The Removal of Sialic Acid Residues from NKp46/NCR1 Reduces HA Recognition.
(A) NCR1-Ig, NKp46-Ig, and HA-Ig were western blotted with biotinylated anti-human IgG1 Fc antibodies (left panel) and an anti-influenza virus type A (H1) mAb (right panel).
(B–E) NKp46-Ig (B and D) and NCR1-Ig (C and E) proteins were incubated with and without NA beads, in the presence or absence of oseltamivir carboxylate. The proteins were western blotted with SNA lectin (B and C) or HA-Ig (D and E). The figure shows the quantification (in arbitrary units) of the relative intensity of the binding. Representative results from three independent experiments are shown. Shown are mean values and SD. Statistically significant differences are indicated (*p < 0.05, **p < 0.01).
NA treatment of almost any protein will result in reduced sialic acid content. However, most sialylated proteins we have tested are unable to interact with HA (Arnon et al., 2001; Mandelboim et al., 2001). Therefore, it was important to demonstrate that the removal of sialic acid residues from NKp46/NCR1 will directly affect HA recognition. To demonstrate this, the proteins were treated with NA as above, in the presence or absence of oseltamivir carboxylate, run on SDS-PAGE gels, and western blotted with biotinylated HA-Ig. Importantly, we observed that the direct interaction between HA and NKp46 (Figure 3D) or NCR1 (Figure 3E) was impaired upon NA treatment and restored when NA inhibitors were added.
NA Impairs NKp46/NCR1 Activity In Vitro and In Vivo
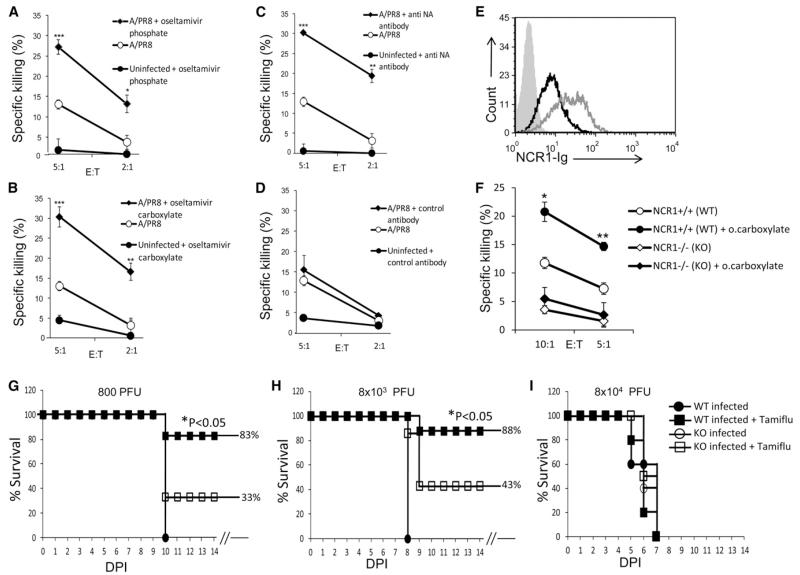
To test whether blocking of NA activity would directly affect the killing of influenza-virus-infected cells, we performed NK cytotoxicity assays using the Jeg3 cell line, which is negative for known NK ligands (Achdout et al., 2010) and human NK cells. In the absence of infection, only minimal killing of Jeg3 cells was observed, and following influenza infection, NK killing was detected (Figures 4A-4D). Importantly, when infected cells were pretreated with oseltamivir phosphate (Figure 4A), oseltamivir carboxylate (Figure 4B), or an anti-NA mAb (Figure 4C), a significant increase in NK cytotoxicity was observed. In contrast, preincubation of the infected cells with a control mAb (12E7) had no effect (Figure 4D).
Figure 4. Blocking of NA Boosts NKp46/NCR1 Activity.
(A–D) Infected Jeg3 cells that were not treated or treated with various NA inhibitors (oseltamivir phosphate [A], oseltamivir carboxylate [B], anti-NAmAb [C]) and control mAb (D) were tested in killing assays. The effector-to-target ratio (E:T) is indicated in the x axis. Shown are mean values and SD derived from triplicates. Statistically significant differences are indicated (*p < 0.05, **p < 0.01 or ***p < 0.005). The figure shows the results of one representative experiment out of three performed.
(E) Infected EL4 cells treated (gray empty histogram) or not (black empty histogram) with oseltamivir carboxylate were stained with NCR1-Ig. The filled gray histogram depicts staining with the secondary mAb only.
(F) Infected EL4 cells treated or not with oseltamivir carboxylate were tested in killing assays against mouse NK cells isolated from Ncr1+/+ (WT) and Ncr1gfp/gfp mice (depicted in the figure as NCR1−/− [KO] mice). The E:T is indicated in the x axis. Shown are mean values and SD derived from triplicates. Statistically significant differences are indicated (*p < 0.05, **p < 0.01). The experiment was repeated twice.
(G–I) Percent survival of Ncr1+/+ and Ncr1gfp/gfp mice infected with 800 PFU (G), 8 × 103 PFU (H), or 8 × 104 PFU (I) of A/Puerto Rico/8/34 influenza virus, treated with oseltamivir phosphate (Tamiflu) or mock treated. The experiment was repeated twice. Statistically significant differences are indicated (*p < 0.05). The x axis indicates DPI. The break in the x axis indicates that all mice that survived the infection stayed healthy.
We also tested whether oseltamivir carboxylate treatment would affect the NCR1-Ig binding and mouse NK killing of EL4 cells infected with A/Puerto Rico/8/34 influenza virus (we used A/Puerto Rico/8/34 because it was used later in the in vivo assays). As seen for the NKp46-Ig staining (Figure 2F), upon infection, little or no change in the binding of NCR1-Ig to the infected cells was observed (Figure 4E and data not shown). However, upon oseltamivir carboxylate treatment, the binding of NCR1-Ig was markedly enhanced (Figure 4E). We next performed NK cell cytotoxicity assays using mouse NK cells derived from Ncr1+/+ (WT) and Ncr1gfp/gfp (KO) mice. When NK cells were derived from the Ncr1gfp/gfp mice, only minimal killing of the infected cells was observed and the level of killing was not affected by oseltamivir carboxylate treatment (Figure 4F). In contrast, when NK cells were derived from Ncr1+/+ mice, killing of the infected cells was observed and, importantly, it was enhanced following oseltamivir carboxylate treatment (Figure 4F).
In our final set of experiments, we evaluated the NA effect in vivo and infected Ncr1+/+ (WT) and Ncr1gfp/gfp (KO) mice with various doses of A/Puerto Rico/8/34 virus. At 800 PFU (a dose that causes equal lethality in both Ncr1gfp/gfp and Ncr1+/+ mice [Figure 1]), ~10 days postinfection (DPI) all untreated mice (both Ncr1+/+ and Ncr1gfp/gfp) died due to the infection (Figure 4G). Importantly, when the mice were treated with oseltamivir carboxylate (Tamiflu), 83% of the Ncr1+/+mice survived the infection, whereas only 33% of the Ncr1gfp/gfp mice remained alive (Figure 4G). Similar differences between the Ncr1gfp/gfp and Ncr1+/+mice were observed even when a 10-fold higher viral dose (8 × 103 PFU) was administered (Figure 4H). When 100-fold more virus (8 × 104 PFU) was administered, the NKp46 effect was no longer seen, the disease progression was extremely rapid and all mice died at 6 or 7 DPI (Figure 4I).
DISCUSSION
We started this investigation because we were surprised to see that the absence of NCR1 affected mouse mortality and virus titers in the lungs only at a certain dose of the influenza virus. We hypothesized that the influenza virus developed mechanisms to counter the NKp46/NCR1 recognition of HA and that this might impair NK-mediated killing, similarly to the effect seen in HSV-1 infections, in which the viral evasion from complement-mediated neutralization masks the effect of knocking down the complement component C3 (Lubinski et al., 1998). Indeed, we have demonstrated here that NA removes sialic acid residues from NKp46/NCR1 and that this results in reduced recognition of HA. Furthermore, we showed that when the NA activity is blocked in vivo, the NCR1 effect is observed over a range of virus titers. We thus provide in vivo evidence that NA inhibitors exert their effect not only by inhibiting the release of virus progeny from the infected cells but also by enhancing NKp46-mediated elimination of infected cells. Interestingly, it was recently demonstrated in vitro that NA inhibitors have additional immune-related properties, because they facilitate the ability of innate immune proteins such as ficolins to inhibit viral infectivity (Verma et al., 2012). Taken together, these results suggest that NA inhibitors may have several beneficial effects in the defense against influenza infection.
The main influenza virus strain used in this research was A/Puerto Rico/8/34 (H1N1). Because of the special characteristics of this virus strain, it was important to verify our findings with another human virus strain. Importantly, similar findings were obtained when the human influenza virus A/Texas/1/1977 (H3N2) was used, indicating that NKp46 interacts with several HAs and that the NA-based immune-evasion mechanism is a general mechanism that is shared by different influenza viruses and NA subtypes.
NA inhibitors, such as oseltamivir (Tamiflu), are widely used to treat influenza infections. Unfortunately, the virus evades the activity of those drugs by mutating the NA active site (Dolin, 2011). Thus, our results not only provide an insight into influenza virus immune-evasion mechanisms, they may also provide the information needed to develop more efficient drugs that will better target NA and at the same time more efficiently boost NKp46-mediated killing.
EXPERIMENTAL PROCEDURES
Cells and Viruses
The cell lines used in this study were the human choriocarcinoma cell line Jeg3 and the mouse lymphoma cell line EL4. The influenza viruses A/Puerto Rico/8/34 (H1N1) and A/Texas/1/1977 (H3N2) were grown in eggs as previously described (Achdout et al., 2003). Viral titers were determined by using quantitative RT-PCR (qRT-PCR), a method used for the detection of influenza virus in patients (Hindiyeh et al., 2005).
Fusion Proteins and Compounds
NKp46-Ig, D2-Ig, D1-Ig, NCR1-Ig, and LIR1-Ig fusion proteins were produced as previously described (Arnon et al., 2004). For generation of the HA-Ig fusion protein, the extracellular domain of the A/Puerto Rico/8/34-HA was cloned in frame with human IgG1 Fc to a mammalian expression vector containing the Fc portion of human IgG1.The HA-Ig protein used in the western blot experiments was biotinylated (21331; Thermo Scientific). The NA inhibitors used were oseltamivir phosphate (Tamiflu; F. Hoffmann-La Roche), oseltamivir carboxylate (sc-212484; Santa Cruz), and zanamivir (sc-208495; Santa Cruz).
Fluorescence-Activated Cell Sorting Staining
For all fluorescence-activated cell sorting (FACS) staining, cells were incubated for 1 hr or overnight with the different influenza virus strains at a ratio of 500 PFU/103 cells at 37°C. The cells were then washed and incubated on ice (unless indicated otherwise) with the appropriate fusion protein (5 μg/well) for 2 hr. For NA inhibition assays, the cells were incubated for 1 hr or overnight with the different influenza virus strains at a ratio of 500 PFU/103 cells at 37°C, washed, and incubated for 1 hr on ice with 5 μg/ml of anti-NA mAb or with 10 ng/ml to 10 μg/ml of NA inhibitors, and then stained with the appropriate fusion proteins.
Western Blotting
NKp46-Ig and NCR1-Ig were incubated for 2 hr with NA beads (Sigma) at a ratio of 2 μl beads/1 μg protein. For NA inhibition, NA beads were pretreated with 50 μl of oseltamivir carboxylate (10 ng/ml to 10 μg/ml). Samples (3 μg) were run on 10% SDS-PAGE gels in reducing conditions and blotted with 20 μg/ml of biotinylated SNA lectin (Vector Laboratories) or with biotinylated HA-Ig (4 μg/ml) and then incubated with Avidin-HRP (Bio Legend). The nontreated NKp46-Ig or NCR1-Ig proteins were used as control. NKp46-Ig, NCR1-Ig, and HA-Ig were also blotted with biotinylated anti-human Fc antibody (Jackson ImmunoResearch) and anti-influenza virus type A (H1) mAb.
Cytotoxicity Assay
The cytotoxic activity of human NK cells against various targets was assessed in 5 hr 35S release assays as previously described (Mandelboim et al., 1996). For NA inhibition, cells were incubated overnight with the different influenza virus strains at a ratio of 500 PFU/103 cells at 37°C and treated with 0.5 μg of anti-NA mAb or with 1–10 μg/ml of NA inhibitors. Mouse NK cells were isolated from WT and Ncr1gfp/gfp mice 18 hr following i.p. injection of 200 μg poly(I):poly(C) (P1530; Sigma).
Infection of Mice and Oseltamivir Treatment
Ncr1+/+ (WT) and Ncr1gfp/gfp (KO) female mice (4–6 weeks old) were infected intranasally with various PFU of the A/Puerto Rico/8/34 virus. Each group contained at least eight mice. For Tamiflu treatment, mice were treated orally with 50 μl of 10 mg/ml oseltamivir phosphate (Tamiflu) diluted in double deionized water (DDW) or with DDW only as control. The first treatment was given 3 hr after infection. The Tamiflu treatment was then given twice a day at 12 hr intervals for 3 days, and later at 6 DPI.
Statistical Methods
Student’s t test and log rank test (for the survival experiments) were used to determine significant differences.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Yewdell for kindly providing the anti-HA and anti-NA antibodies. This study was supported by a Croatia Israel Research Grant, a MOST-DKFZ Research Grant, an ICRF Professorship Grant, an ERC Advanced Grant, and grants from the Israeli Science Foundation, the Israeli ICORE, and the Association for International Cancer Research (all to O.M.). O.M. is a Crown Professor of Molecular Immunology.
REFERENCES
- Achdout H, Arnon TI, Markel G, Gonen-Gross T, Katz G, Lieberman N, Gazit R, Joseph A, Kedar E, Mandelboim O. Enhanced recognition of human NK receptors after influenza virus infection. J. Immunol. 2003;171:915–923. doi: 10.4049/jimmunol.171.2.915. [DOI] [PubMed] [Google Scholar]
- Achdout H, Meningher T, Hirsh S, Glasner A, Bar-On Y, Gur C, Porgador A, Mendelson M, Mandelboim M, Mandelboim O. Killing of avian and Swine influenza virus by natural killer cells. J. Virol. 2010;84:3993–4001. doi: 10.1128/JVI.02289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Arnon TI, Achdout H, Lieberman N, Gazit R, Gonen-Gross T, Katz G, Bar-Ilan A, Bloushtain N, Lev M, Joseph A, et al. The mechanisms controlling the recognition of tumor- and virus-infected cells by NKp46. Blood. 2004;103:664–672. doi: 10.1182/blood-2003-05-1716. [DOI] [PubMed] [Google Scholar]
- Dolin R. Resistance to neuraminidase inhibitors. Clin. Infect. Dis. 2011;52:438–439. doi: 10.1093/cid/ciq184. [DOI] [PubMed] [Google Scholar]
- Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- Glasner A, Zurunic A, Meningher T, Lenac Rovis T, Tsukerman P, Bar-On Y, Yamin R, Meyers AF, Mandeboim M, Jonjic S, Mandelboim O. Elucidating the mechanisms of influenza virus recognition by Ncr1. PLoS One. 2012;7:e36837. doi: 10.1371/journal.pone.0036837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindiyeh M, Levy V, Azar R, Varsano N, Regev L, Shalev Y, Grossman Z, Mendelson E. Evaluation of a multiplex real-time reverse transcriptase PCR assay for detection and differentiation of influenza viruses A and B during the 2001-2002 influenza season in Israel. J. Clin. Microbiol. 2005;43:589–595. doi: 10.1128/JCM.43.2.589-595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Hershkovitz O, Peiris M, Zilka A, Bar-Ilan A, Nal B, Chu K, Kudelko M, Kam YW, Achdout H, et al. H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. J. Virol. 2008;82:2028–2032. doi: 10.1128/JVI.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T, Jones M, Doshi P, Del Mar C. Neuraminidase inhibitors for preventing and treating influenza in healthy adults: systematic review and meta-analysis. BMJ. 2009;339:b5106. doi: 10.1136/bmj.b5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann L, Jacobsen H, Schuhbauer D, Knoflach F, Gatti S, Wettstein JG, Loetscher H, Chu T, Ebeling M, Paulson JC, et al. In vitro pharmacological selectivity profile of oseltamivir prodrug (Tamiflu) and active metabolite. Eur. J. Pharmacol. 2010;628:6–10. doi: 10.1016/j.ejphar.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski JM, Wang L, Soulika AM, Burger R, Wetsel RA, Colten H, Cohen GH, Eisenberg RJ, Lambris JD, Friedman HM. Herpes simplex virus type 1 glycoprotein gC mediates immune evasion in vivo. J. Virol. 1998;72:8257–8263. doi: 10.1128/jvi.72.10.8257-8263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O, Reyburn HT, Valés-Gómez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- Mendelson M, Tekoah Y, Zilka A, Gershoni-Yahalom O, Gazit R, Achdout H, Bovin NV, Meningher T, Mandelboim M, Mandelboim O, et al. NKp46 O-glycan sequences that are involved in the interaction with hemagglutinin type 1 of influenza virus. J. Virol. 2010;84:3789–3797. doi: 10.1128/JVI.01815-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Hui EK, Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin DB, Quigley JP. Virus-induced modification of cellular membranes related to viral structure. Annu. Rev. Microbiol. 1974;28:325–351. doi: 10.1146/annurev.mi.28.100174.001545. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- Verma A, White M, Vathipadiekal V, Tripathi S, Mbianda J, Ieong M, Qi L, Taubenberger JK, Takahashi K, Jensenius JC, et al. Human H-ficolin inhibits replication of seasonal and pandemic influenza A viruses. J. Immunol. 2012;189:2478–2487. doi: 10.4049/jimmunol.1103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.