Abstract
Objective
Obesity-associated non-alcoholic fatty liver disease (NAFLD) may cause liver dysfunction and failure. In a previously reported genome-wide association meta-analysis, single nucleotide polymorphisms (SNPs) near PNPLA3, NCAN, GCKR, LYPLAL1 and PPP1R3B were associated with NAFLD and with distinctive serum lipid profiles. The present study examined the relevance of these variants to NAFLD in extreme obesity.
Methods
In 1,092 bariatric patients, the candidate SNPs were genotyped and association analyses with liver histology and serum lipids were performed.
Results
We replicated the association of hepatosteatosis with PNPLA3 rs738409[G] and with NCAN rs2228603[T]. We also replicated the association of rs2228603[T] with hepatic inflammation and fibrosis. Rs2228603[T] was associated with lower serum LDL, total cholesterol and triglycerides. After stratification by the presence or absence of NAFLD, these associations were present predominantly in the subgroup with NAFLD.
Conclusion
NCAN rs2228603[T] is a risk factor for liver inflammation and fibrosis, suggesting that this locus is responsible for progression from steatosis to steatohepatitis. In this bariatric cohort, rs2228603[T] was associated with low serum lipids only in patients with NAFLD. This supports a NAFLD model in which the liver may sequester triglycerides as a result of either increased triglyceride uptake and/or decreased lipolysis.
Keywords: Obesity, dyslipidemia, steatohepatitis, cirrhosis, steatosis
Obesity is a driving force for the metabolic syndrome, which is a combination of cardiovascular risk factors that include visceral obesity, hypertension, high serum triglycerides, low serum high-density lipoproprotein (HDL) cholesterol, and glucose intolerance or diabetes (1). In a substantial number of obese individuals with metabolic syndrome, fatty infiltration of the liver (hepatosteatosis) is observed. Non-alcoholic fatty liver disease (NAFLD) is often associated with inflammation (non-alcoholic steatohepatitis or NASH) which may progress to fibrosis and cirrhosis (2, 3). With the current epidemic of obesity, NAFLD and its progression is the leading cause of liver dysfunction and failure (4–7).
It is unknown why only a fraction of obese individuals develop hepatic steatosis and only a subset of those patients progress to NASH, fibrosis, and cirrhosis. Recently, genetic factors have been implicated. In a genome-wide association study by Hobbs and colleagues, a nonsynonymous variation I148M (rs738409) in the PNPLA3 gene was found to be associated with NAFLD (8). This variant has also been associated with increased serum AST and ALT levels and histological evidence of NASH (9, 10). More recently, the Genetics of Obesity-related Liver Disease (GOLD) Consortium, comprised of 7,176 subjects of European descent, performed a genome-wide association meta-analysis of NAFLD (11). In this study, three novel loci (NCAN, GCKR, and LYPLAL1) and the previously reported PNPLA3 locus were found to be associated with liver fat content as measured by both electron beam computer tomography as well as liver biopsy. Another locus, PPP1R3B was associated with liver content by electron beam computer tomography, but not with histologically-defined NAFLD. Interestingly, some of these variants were associated with distinct changes in serum lipid levels. In particular, the T allele of rs2228603 in NCAN was associated with increased liver fat and, seemingly paradoxically, lower serum triglyceride and low-density lipoprotein (LDL) cholesterol levels (11). These differences suggest that each gene locus may affect lipid metabolism and NAFLD through a distinct mechanism.
To further examine the role of these loci in NAFLD, we performed association studies of these variants in an independent cohort of severely obese patients who underwent bariatric surgery and from whom histologically well-characterized liver samples were obtained. Our aim was not simply to replicate the findings of the GOLD Consortium, but also to determine the relevance of these loci in extreme obesity, a condition in which both strong genetic and environmental factors contribute to the phenotype and its metabolic complications.
Methods
Study Population and Phenotyping
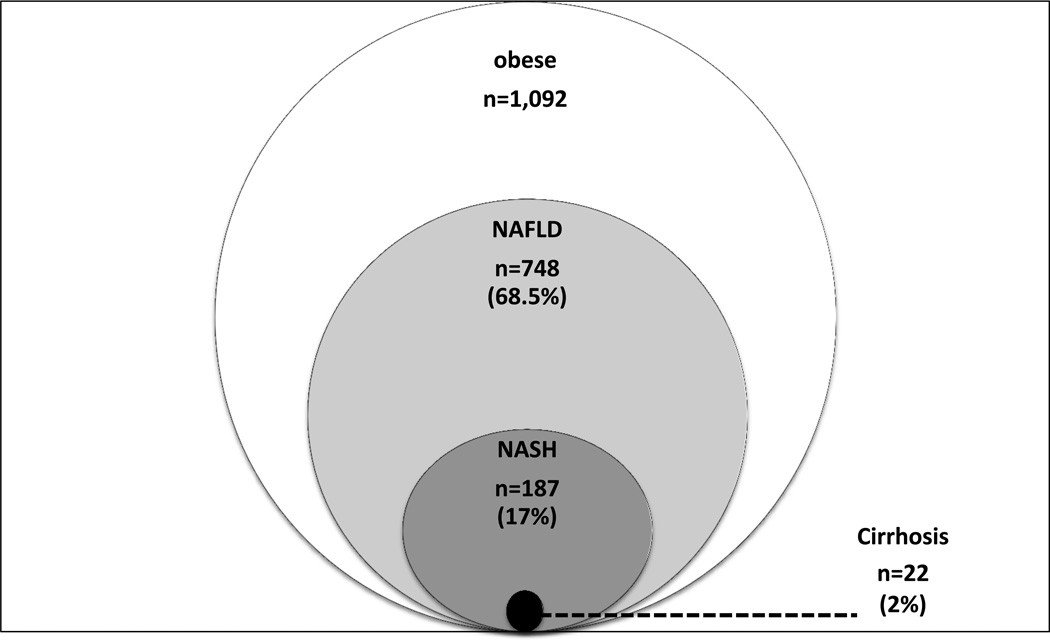
The participants were 1,092 bariatric surgery patients from the Geisinger Medical Center in Danville, PA with normal liver tissue or varying degrees of NAFLD, ranging from simple steatosis, to steatohepatitis, to cirrhosis (Figure 1) (these subjects were not included in the original GOLD Consortium). The protocol was approved by the Geisinger Clinic Institutional Review Board, and all subjects provided written informed consent. Prior to surgery, patients were extensively phenotyped, including comprehensive medical history and physical examination, anthropometry, fasting serum glucose and lipids, liver and kidney function, and medication usage (12).
Figure 1.
Spectrum of NAFLD represented in the bariatric cohort (n=1,092).
Intra-operative liver biopsy specimens were formalin fixed and stained with hematoxylin and eosin for routine histology and Masson’s trichrome for assessment of fibrosis (13). All specimens were read by experienced pathologists and graded according to standard NAFLD criteria (14). Steatosis was graded in severity from 0 (no steatosis) to 3 (severe steatosis). Scoring for lobular inflammation, hepatocyte ballooning, and perivenular fibrosis was dichotomized, with a score of 0 indicating absence of the feature and a score of 1 indicating any presence of the feature (ranging from mild to severe).
SNP Genotyping
Genomic DNA was isolated from peripheral blood leukocytes (15). The five hepatic steatosis-associated SNPs from the GOLD Consortium meta-analysis were genotyped using TaqMan® SNP Genotyping Assays (Life Technologies, Carlsbad, CA). These included rs12137855 (C/T) on chromosome 1 near LYPLAL1, rs780094 (C/T) on chromosome 2 in GCKR, rs4240624 (A/G) on chromosome 8 near PPP1R3B, rs2228603 (C/T) on chromosome 19 in NCAN, and rs738409 (G/C) on chromosome 22 in PNPLA3. PCR amplification was performed on the GeneAMP PCR System 9700 thermal cycler (Applied Biosystems) under the following conditions: 10 min at 95°C, then 40 cycles of 15 sec at 92°C and 1 min at 60°C, and allelic discrimination was performed on the ABI Prism 7900 HT Sequence Detection System (SDS) and SDS Software according to the manufacturer’s directions. All five SNPs passed genotyping quality control. The average genotype call rate was 96.7%. The genotype concordance rate of blind replicates was 99.2%. None of the SNP allele frequencies deviated significantly from the Hardy–Weinberg equilibrium.
Statistical Analysis
Statistical analyses were carried out using SAS version 9.2 (SAS Institute Inc., Cary, NC). For continuous traits (steatosis grade, serum total-, LDL- and HDL-cholesterol, and triglycerides), associations between SNPs and phenotypes were assessed by linear regression and dichotomous traits (lobular inflammation, ballooning, perivenular fibrosis and cirrhosis) were analyzed using logistic regression. Analyses were adjusted for age, sex, and lipid-lowering medication. Adjustment for lipid lowering therapy was performed using as covariates whether the subject was or was not on statin therapy and was or was not on fibrate therapy (the vast majority of subjects on lipid-lowering therapy were taking statins). Regression analyses tested for an additive SNP association between the number of copies (0, 1 or 2) of the NAFLD-associated allele and the trait of interest. Due to the relatively low minor allele frequency of the T allele of rs2228603 in NCAN and the G allele of rs4240624 in PPP1R3B (7.4% and 8.3% respectively), the rare homozygous and heterozygous genotype groups were combined into a single group and compared to the common homozygous genotype for analysis. Since a small number of SNPs were genotyped, each with a high posterior probability of being true positives, a two-sided p-value < 0.05 was used as the threshold for statistical significance.
Results
Like most bariatric surgery cohorts (16–18), the majority (80%) of the cohort was female (Table 1). The average age was 46 years and the mean pre-operative BMI was 50 kg/m2. Although BMI did not differ between patients with and without NAFLD (p=0.22), those with NAFLD had greater waist circumferences (p<0.0001), lower serum HDL-cholesterol levels (p<0.0001), and as shown in other studies (17, 19), higher serum triglycerides levels (p<0.0001). In addition, more patients in the NAFLD group had diabetes (p=0.0007). About one third of all subjects were on statin or fibrate therapy. However, the percentage of subjects on lipid-regulating medication was not significantly different between subjects with and without NAFLD or among any of the genotypes examined.
Table 1.
Characteristics of study population
| NAFLD + | NAFLD − | p-value* | |
|---|---|---|---|
| N | 748 | 344 | |
| Female, N (%) | 579 (77.4%) | 296.0 (86.0%) | 0.001 |
| Age, years (mean ± SD) | 47 ± 10.6 | 46 ± 11.8 | 0.36 |
| BMI, kg/m2 (mean ± SD) | 50.4 ± 8.9 | 49.6 ± 8.6 | 0.22 |
| Waist Circumference, in (mean ± SD) | 54.2 ± 6.7 | 52.4 ± 6.2 | <0.0001 |
| Diabetes, N (%) | 292 (39%) | 93 (27%) | 0.0007 |
| Total cholesterol, mg/dl (mean ± SD) | 191.7 ± 40.8 | 185.4 ± 40.8 | 0.006 |
| HDL-cholesterol, mg/dl (mean ± SD) | 46.3 ± 10.6 | 50.9 ± 12.1 | <0.0001 |
| LDL-cholesterol, mg/dl (mean ± SD) | 107.6 ± 34.5 | 106.4 ± 36.0 | 0.39 |
| Triglycerides, mg/dl (mean ± SD) | 197.4 ± 141.2 | 140.3 ± 69.4 | <0.0001 |
| Statin Therapy, N (%) | 228 (30%) | 117 (34%) | 0.07 |
| Fibrate Therapy, N (%) | 41 (5%) | 20 (6%) | 0.38 |
| Statin or Fibrate Therapy, N (%) | 250 (33%) | 122 (35%) | 0.14 |
BMI, Body mass index; NAFLD, Non-alcoholic fatty liver disease
Adjusted for age and sex (sex is only adjusted for age and age only adjusted for sex)
NAFLD was graded using the criteria of Brunt [12]. NAFLD − = grade 0; NAFLD + = grade ≥1.
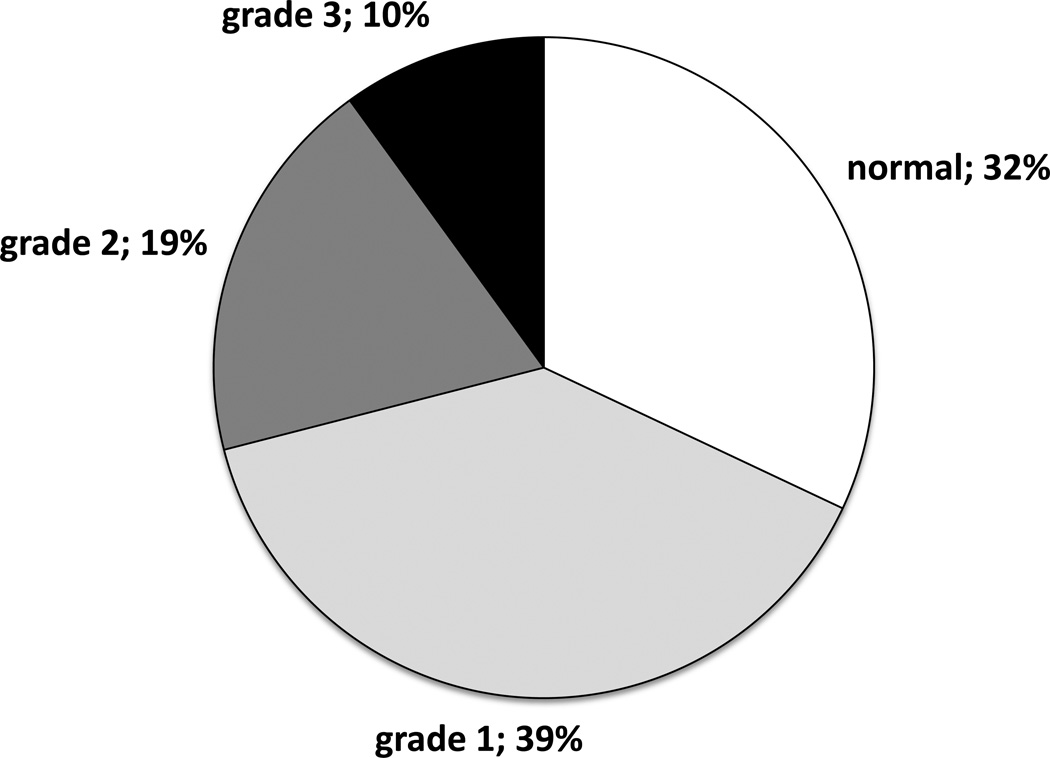
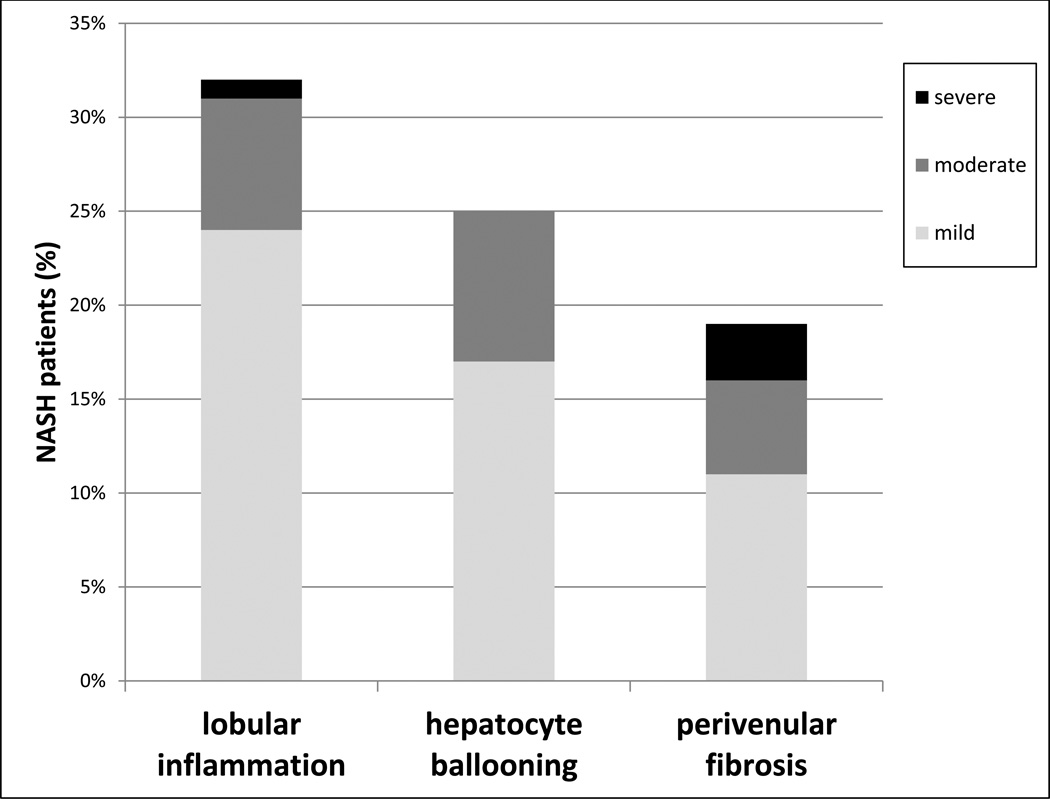
Of all 1,092 obese subjects, 32% had no evidence of hepatic steatosis, 39% had grade 1 steatosis, 19% had grade 2, and 10% had grade 3 (Figure 2). Of the 748 patients with evidence of hepatic steatosis, 187 (25%) had at least one histological feature of steatohepatitis. The biopsies from these 187 NASH patients revealed lobular inflammation (24% with mild, 7% with moderate, and 1% with severe), hepatocyte ballooning (17% with mild, 8% with moderate, and 0% with severe) and perivenular fibrosis (11% with mild, 5% with moderate, and 3% with severe) (Figure 3). Of the 187 NASH patients, 12% had cirrhosis.
Figure 2.
Proportions of bariatric surgery patients with varying degrees of hepatic steatosis (n=1,092). Grading was performed by experienced pathologists using the criteria of Brunt (1).
Figure 3.
Liver histology features (lobular inflammation, hepatocyte ballooning and perivenular fibrosis) in bariatric patients with NASH (n=187). Grading was performed by experienced pathologists using the criteria of Brunt (1).
Of the five SNPs associated with NAFLD in the GOLD Consortium meta-analysis, only rs780094[C] in GCKR was associated with higher weight (p=0.001) and BMI (p=0.001) in this bariatric cohort (Supplemental Table). SNPs rs780094[C] in GCKR and rs2228603[T] in NCAN were associated with increased waist circumference (p=0.02 and p=0.03, respectively) (Supplemental Table).
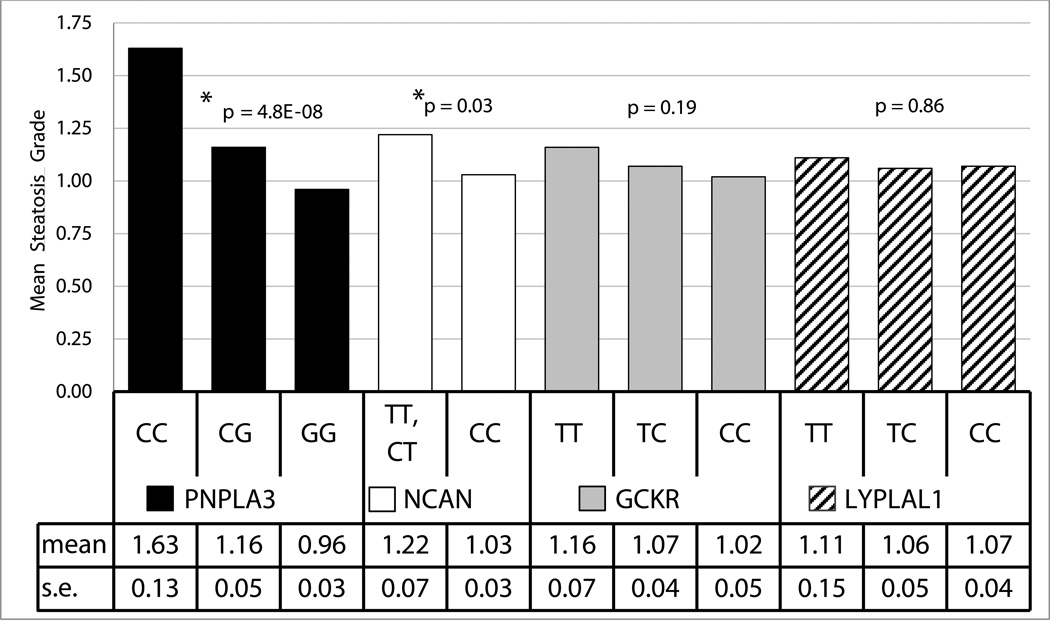
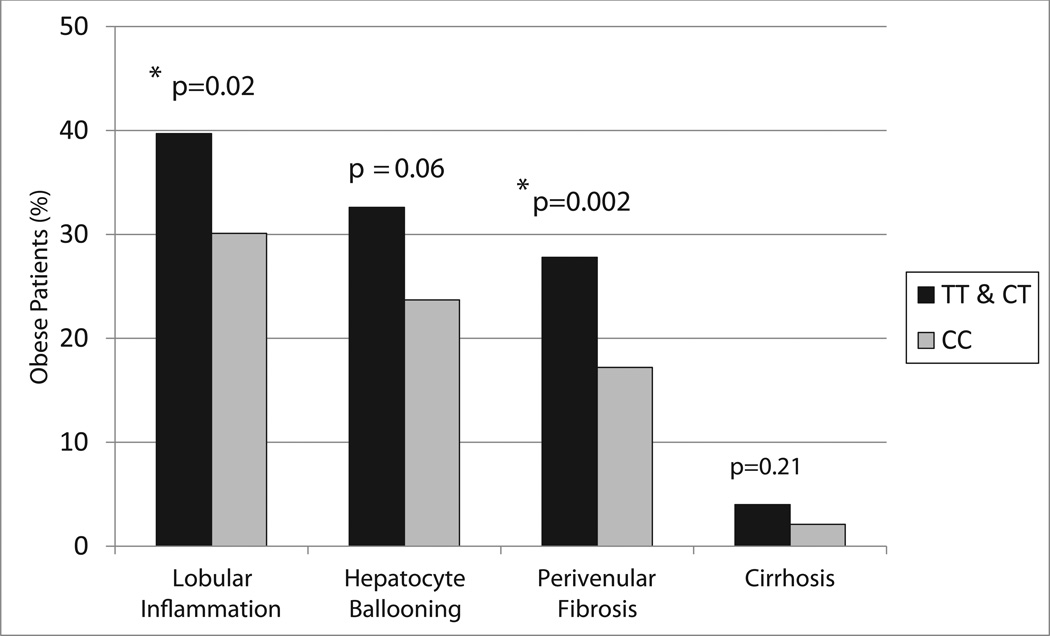
Except for rs1213785[C] near LYPLAL1, all of the SNPs were significantly associated with (or trended toward) increased steatosis in the same direction and with similar effect sizes as reported in the GOLD Consortium meta-analysis (Table 2). Consistent with the results from the GOLD Consortium (11), our data showed an association of PNPLA3 SNP rs738409[G] with both hepatic steatosis (p=4.8 ×10−8) and hepatocyte ballooning (p=0.006) (Figure 4, Table 2). Similarly, as previously reported (11), we found an association of the T allele of SNP rs2228603 in NCAN with liver steatosis (p=0.03) as well as with lobular inflammation (p=0.02) and perivenular fibrosis (p=0.002) (Figures 4 & 5, Table 2). Though not statistically significant, this allele also was associated with a trend toward increased hepatocyte ballooning (p=0.06) (Figure 5, Table 2). These findings show that in extreme obesity, PNPLA3 rs738409[G] and NCAN rs2228603[T] are associated with liver steatosis and an increased risk for progression from simple steatosis to NASH. By contrast, variants in LYPLAL1, GCKR and PPP1R3B did not appear to show increased risk for lobular inflammation, hepatocyte ballooning or perivenular fibrosis. Although there was only a trend toward statistical significance, the PPP1R3B SNP rs4240624[A] was the only variant associated with cirrhosis (p=0.07).
Table 2.
Association of SNPs with NAFLD characteristics*
| SNP | rs780094 | rs2228603 | rs1213785 | rs4240624 | rs738409 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect allele | T | T | C | A | G | ||||||||
| Other allele | C | C | T | G | C | ||||||||
| Effect allele Frequency | 40.9 | 7.4 | 73.5 | 8.3 | 22.0 | ||||||||
| Nearest gene | GCKR | NCAN | LYPLAL1 | PPP1R3B | PNPLA3 | ||||||||
| Genotypes | TT N=186 |
CT N=522 |
CC N=384 |
−T N=155 |
CC N=928 |
CC N=696 |
CT N=325 |
TT N=39 |
AA N=883 |
− G N=172 |
GG N=54 |
GC N=370 |
CC N=664 |
| Steatosis§ (mean) | 1.16 | 1.07 | 1.02 | 1.22 | 1.03 | 1.07 | 1.06 | 1.11 | 1.08 | 1.02 | 1.63 | 1.16 | 0.96 |
| SE | 0.07 | 0.04 | 0.05 | 0.07 | 0.03 | 0.04 | 0.05 | 0.15 | 0.03 | 0.07 | 0.13 | 0.05 | 0.03 |
| p-value | 0.19 | 0.03 | 0.86 | 0.49 | 4.80E-08 | ||||||||
| Lobular inflammation¥ (% > grade 0) | 34.4 | 30.9 | 31.1 | 39.7 | 30.1 | 31.4 | 32.2 | 32.4 | 31.4 | 32.7 | 32.7 | 34.8 | 29.6 |
| p-value | 0.68 | 0.02 | 0.98 | 0.68 | 0.11 | ||||||||
| Hepatocyte Ballooning¥ (% > grade 0) | 23.4 | 29.1 | 21.6 | 32.6 | 23.7 | 24.2 | 28.0 | 23.8 | 24.9 | 27.6 | 31.4 | 31.3 | 21.5 |
| p-value | 0.48 | 0.06 | 0.49 | 0.48 | 0.006 | ||||||||
| Perivenular fibrosis¥ (% > grade 0) | 20.0 | 18.8 | 18.8 | 27.8 | 17.2 | 19.2 | 18.0 | 21.6 | 19.1 | 17.3 | 21.2 | 22.0 | 17.1 |
| p-value | 0.98 | 0.002 | 0.74 | 0.65 | 0.06 | ||||||||
| Cirrhosis¥ (% > grade 0) | 2.8 | 2.5 | 1.6 | 4.0 | 2.1 | 2.5 | 1.9 | 2.7 | 2.0 | 4.2 | 1.9 | 3.3 | 1.7 |
| p-value | 0.47 | 0.21 | 0.61 | 0.07 | 0.21 | ||||||||
Age-, sex-, and lipid lowering medication-adjusted mean and standard error are presented for each genotype group.
Steatosis was examined histologically and graded in severity from 0 (no steatosis) to 3 (severe steatosis).
Dichotomized grade 0 vs. >0 for inflammation, ballooning, fibrosis and cirrhosis.
Figure 4.
Association between SNP genotype and liver steatosis in all bariatric patients (n=1,092).
Figure 5.
Association of NCAN rs2228603 genotype with features of NASH and cirrhosis in all bariatric patients (n=1,092).
As observed in the GOLD Consortium study, the data from this bariatric cohort revealed that some NAFLD-associated SNPs were associated with a distinct pattern of serum lipid levels (Table 3). These effects were most apparent in subjects with NAFLD. In NAFLD subjects, the NAFLD-associated G allele of rs738409 in PNPLA3 was associated with lower total cholesterol (p=0.03). Similarly, in NAFLD subjects, the NAFLD-associated T allele of rs2228603 in NCAN was associated with decreased serum total cholesterol (p=0.0002), LDL-cholesterol (p=0.009) and triglycerides (p=0.004). By contrast, as in the GOLD Consortium meta-analysis, the NAFLD-associated T allele of GCKR SNP rs780094 was associated with increased serum triglyceride levels; this effect was most evident in subjects with NAFLD (p=0.04). Also consistent with the GOLD Consortium findings, we did not find significant association between either LYPLAL1 SNP rs12137855 or PPP1R3B SNP rs4240624 and serum lipids.
Table 3.
Association of SNPs with fasting serum lipid levels*.
| SNP | rs780094 | rs2228603 | rs1213785 | rs4240624 | rs738409 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect allele | T | T | C | A | G | ||||||||
| Other allele | C | C | T | G | C | ||||||||
| Nearest gene | GCKR | NCAN | LYPLAL1 | PPP1R3B | PNPLA3 | ||||||||
| Patients With NAFLD | |||||||||||||
| Genotypes | TT N=129 |
CT N=352 |
CC N=247 |
− T N=114 |
CC N=583 |
CC N=449 |
CT N=207 |
TT N=27 |
AA N=575 |
− G N=107 |
GG N=47 |
GC N=244 |
CC N=411 |
| Total-C, mg/dL (mean) | 197.6 | 189.4 | 191.0 | 178.4 | 194.7 | 192.8 | 189.6 | 191.5 | 192.4 | 192.7 | 180.6 | 190.1 | 194.7 |
| SE | 3.9 | 2.1 | 2.6 | 3.6 | 1.7 | 1.9 | 2.7 | 6.9 | 1.7 | 3.8 | 5.9 | 2.5 | 2.0 |
| p-value* | 0.11 | 0.0002 | 0.58 | 0.96 | 0.03 | ||||||||
| LDL-C, mg/dL (mean) | 109.0 | 106.2 | 108.2 | 100.2 | 109.2 | 108.0 | 105.8 | 108.7 | 107.9 | 108.2 | 98.5 | 107.8 | 108.8 |
| SE | 3.5 | 1.8 | 2.2 | 3.1 | 1.5 | 1.7 | 2.3 | 5.8 | 1.5 | 3.2 | 5.3 | 2.2 | 1.7 |
| p-value* | 0.68 | 0.009 | 0.80 | 0.96 | 0.16 | ||||||||
| HDL-C, mg/dL (mean) | 46.8 | 45.9 | 46.0 | 45.3 | 46.3 | 46.6 | 44.9 | 48.3 | 46.2 | 47.4 | 46.1 | 45.8 | 46.5 |
| SE | 0.9 | 0.6 | 0.7 | 0.9 | 0.4 | 0.5 | 0.7 | 1.9 | 0.5 | 1.0 | 1.5 | 0.6 | 0.6 |
| p-value* | 0.13 | 0.47 | 0.73 | 0.41 | 0.38 | ||||||||
| Triglycerides§, mg/dL (mean) | 213.3 | 198.8 | 188.6 | 166.6 | 205.4 | 198.3 | 202.4 | 193.0 | 201.0 | 187.7 | 175.7 | 186.1 | 209.3 |
| SE | 11.6 | 7.6 | 9.8 | 8.2 | 6.3 | 6.1 | 11.4 | 36.7 | 6.5 | 11.9 | 14.7 | 6.3 | 8.2 |
| p-value | 0.04 | 0.004 | 0.64 | 0.78 | 0.08 | ||||||||
| Patients Without NAFLD | |||||||||||||
| Genotypes | TT N=49 |
CT N=156 |
CC N=122 |
− T N=31 |
CC N=295 |
CC N=208 |
CT N=103 |
TT N=8 |
AA N=262 |
− G N=55 |
GG N=5 |
GC N=103 |
CC N=218 |
| Total-C, mg/dL (mean) | 189.0 | 183.8 | 184.7 | 189.1 | 184.6 | 186.0 | 183.4 | 192.8 | 185.5 | 183.7 | 192.0 | 178.8 | 187.8 |
| SE | 6.2 | 3.2 | 3.7 | 6.1 | 2.4 | 2.9 | 3.6 | 14.6 | 2.6 | 5.1 | 26.4 | 3.7 | 2.8 |
| p-value* | 0.62 | 0.62 | 0.70 | 0.72 | 0.15 | ||||||||
| LDL-C, mg/dL (mean) | 106.1 | 106.2 | 105.9 | 106.5 | 106.0 | 107.2 | 104.3 | 111.5 | 106.3 | 106.3 | 111.6 | 100.4 | 108.5 |
| SE | 5.1 | 2.9 | 3.3 | 6.1 | 2.1 | 2.6 | 3.1 | 13.4 | 2.3 | 4.6 | 26.4 | 3.3 | 2.5 |
| p-value* | 0.90 | 0.92 | 0.56 | 0.89 | 0.15 | ||||||||
| HDL-C, mg/dL (mean) | 52.9 | 50.2 | 50.9 | 53.1 | 50.5 | 50.6 | 51.4 | 53.3 | 51.1 | 49.6 | 56.8 | 48.7 | 51.6 |
| SE | 1.6 | 0.9 | 1.1 | 2.0 | 0.7 | 0.9 | 1.1 | 5.4 | 0.8 | 1.4 | 4.9 | 1.1 | 0.8 |
| p-value* | 0.52 | 0.44 | 0.53 | 0.44 | 0.13 | ||||||||
| Triglycerides§, mg/dL (mean) | 145.5 | 138.3 | 139.7 | 147.6 | 140.4 | 140.0 | 140.5 | 139.6 | 140.5 | 139.3 | 116.8 | 148.1 | 138.3 |
| SE | 12.2 | 5.8 | 5.4 | 13.2 | 4.0 | 4.5 | 7.0 | 36.5 | 4.4 | 7.4 | 16.7 | 7.4 | 4.6 |
| p-value | 0.74 | 0.61 | 0.83 | 0.59 | 0.39 | ||||||||
| All Patients With and Without NAFLD | |||||||||||||
| Genotypes | TT N=186 |
CT N=522 |
CC N=384 |
− T N=155 |
CC N=928 |
CC N=696 |
CT N=325 |
TT N=39 |
AA N=883 |
− G N=172 |
GG N=54 |
GC N=370 |
CC N=664 |
| Total-C, mg/dL (mean) | 195.1 | 187.1 | 188.8 | 180.1 | 191.0 | 190.2 | 187.4 | 191.0 | 189.8 | 190.3 | 181.1 | 186.8 | 191.8 |
| SE | 3.3 | 1.8 | 2.1 | 3.1 | 1.4 | 1.6 | 2.2 | 6.0 | 1.4 | 3.0 | 5.7 | 2.1 | 1.6 |
| p-value* | 0.12 | 0.003 | 0.58 | 0.92 | 0.03 | ||||||||
| LDL-C, mg/dL (mean) | 108.1 | 105.6 | 107.3 | 101.2 | 107.9 | 107.4 | 105.2 | 108.6 | 107.0 | 108.0 | 99.9 | 105.8 | 108.2 |
| SE | 2.8 | 1.5 | 1.8 | 2.7 | 1.2 | 1.4 | 1.8 | 5.1 | 1.2 | 2.6 | 5.2 | 1.8 | 1.4 |
| p-value* | 0.75 | 0.02 | 0.61 | 0.85 | 0.12 | ||||||||
| HDL-C, mg/dL (mean) | 48.6 | 47.3 | 47.6 | 46.7 | 47.8 | 47.9 | 47.1 | 49.2 | 47.7 | 48.3 | 46.9 | 46.7 | 48.3 |
| SE | 0.8 | 0.5 | 0.5 | 0.9 | 0.4 | 0.4 | 0.6 | 1.9 | 0.4 | 0.8 | 1.5 | 0.5 | 0.5 |
| p-value* | 0.21 | 0.24 | 0.97 | 0.66 | 0.03 | ||||||||
| Triglycerides§, mg/dL (mean) | 193.5 | 178.5 | 172.1 | 162.4 | 182.7 | 179.0 | 181.0 | 180.6 | 181.1 | 171.3 | 167.3 | 173.7 | 184.2 |
| SE | 9.0 | 5.5 | 6.7 | 6.9 | 4.4 | 4.5 | 7.9 | 28.1 | 4.4 | 8.2 | 13.2 | 4.9 | 5.6 |
| p-value | 0.08 | 0.18 | 0.78 | 0.98 | 0.63 | ||||||||
Age-, sex-, and lipid lowering medication-adjusted mean and standard error are presented for each genotype group.
Log-transformed for analysis and back-transformed for presentation. C = cholesterol
Discussion
Using an independent sample of 1,092 well-phenotyped morbidly obese subjects who underwent bariatric surgery and in whom liver histology was determined, we found that despite similar BMI, subjects with NAFLD had a higher prevalence of diabetes, hyperlipidemia and greater waist circumference than those without NAFLD. These data support that NAFLD is associated with insulin resistance and other features of the metabolic syndrome largely independent of obesity. Association analyses of five NAFLD-associated genetic variants identified by the GOLD Consortium GWAS meta-analysis in our bariatric cohort revealed a nonsynonymous variant in PLPLA3 to be associated not only with the presence of steatosis but also with NASH, a more progressive and clinically ominous manifestation of NAFLD. These finding extend a previously reported studies to patients with extreme obesity. These findings are consistent with other studies in which the G allele of PLPLA3 rs738409 was not only associated with simple fat accumulation, but also with the degree of liver steatosis as evaluated by liver biopsy (23–25). In other studies, the same allele was associated with steatosis, portal inflammation, lobular inflammation, Mallory-Denk bodies, NAFLD activity score, and fibrosis (8, 10).
Furthermore, we replicated the GOLD Consortium’s association of the T allele of rs2228603 in NCAN with degree of liver steatosis as well as with signs of lobular inflammation and perivenular fibrosis, which are consistent with NASH, a more advanced stage of NAFLD. For GCKR rs780094 and PPP1R3B rs4240624, although not statistically significantly associated with NAFLD, the direction of effect and the effect size showed similar trends to the GOLD Consortium results. It is likely that the inability to achieve statistical significance is due to lack of power to discern the effect of variants of modest effect size. The absence of even a trend for association of LYPLAL1 rs1213785 with NAFLD may be due to differences among populations, a false negative finding, or a false positive finding in the initial GOLD Consortium study. Assuming a 24% population prevalence of NAFLD, our sample had 80% power to detect an OR of 1.48 for presence of steatosis for a variant with allele frequency of at least 0.05 at p<0.05. Using a higher population prevalence of 50% which may be more suitable for a bariatric population, our sample had 80% power to detect an OR of 1.29 for a variant with an allele frequency of at least 0.05 at p<0.05.
Our data revealed that the PPP1R3B SNP rs4240624 was not significantly associated with liver steatosis, lobular inflammation, or hepatocyte ballooning. However, this variant was the only SNP in our study that tended to be associated with cirrhosis (p=0.07). In the GOLD Consortium meta-analysis, PPP1R3B was associated with fatty liver identified on CT scan, but it was not associated with histologic features of NAFLD. As hypothesized by Speliotes et al, this may suggest that PP1R3B is related to liver steatosis but not hepatocyte inflammation and fibrosis (11). Alternatively, the SNP may be associated with intrahepatic accumulation of a substance that resembles steatosis on abdominal radiographic imaging and causes cirrhosis (i.e., glycogen storage disease type IV, which is characterized by glycogen precipitation in the liver that results in cirrhosis).
Our data also replicate distinct patterns of serum lipids for some NAFLD-associated variants. The GOLD Consortium meta-analysis found no association between lipid levels and PNPLA3 rs738409[G]. Our data suggests a relationship between the NAFLD-associated G allele and lower total- and HDL-cholesterol levels in the full cohort (p=0.03). This may be a type I error or alternatively due to a difference in ascertainment; all subjects in our study were selected for extreme obesity. With regard to serum triglycerides, there was no association between this SNP and serum triglyceride levels in the overall cohort, but there was a trend toward an association with lower triglyceride levels in patients with NAFLD (p=0.08). In support of this finding, the relationship between this SNP and serum triglyceride levels has been reported by others (26, 27).
As in the GOLD Consortium, we found association of the NAFLD-associated NCAN rs2228603[T] allele with lower serum LDL-cholesterol, total cholesterol and triglyceride levels, particularly in those with NAFLD. Interestingly, this association was not found in equally obese patients without NAFLD suggesting that NAFLD and lower serum lipids may be mechanistically linked in rs22286603[T] carriers. Since NAFLD is clinically associated with higher serum lipids, this seemingly paradoxical finding suggests that more than one NAFLD subtype may exist. Elucidation of the causative gene/variant(s) at this locus may uncover a novel disease mechanism in which sequestration of triglycerides in the liver, either as a result of increased triglyceride uptake and/or decreased lipolysis result in lower serum lipid levels.
By contrast, the NAFLD-associated GCKR rs780094[T] allele, was associated with higher serum triglycerides. These findings are similar to the GOLD Consortium analysis as well as other studies (20–22). Finally, we did not detect any association between the LYPLAL1 or PPP1R3 SNPs and serum lipids. It is possible that the lack of association may be due to inadequate sample size and power, especially for PPP1R3 in which the GOLD Consortium reported increased LDL- and HDL-cholesterol.
The NCAN locus contains at least 20 genes in a 500 kb region on chromosome 19p13 (28–30). None of these genes are particularly compelling positional candidates for NAFLD or lipid homeostasis. Additional studies are needed to fine map this locus and perform functional studies. Neurocan, the protein product of NCAN, is a chondroitin sulfate proteoglycan primarily expressed in the nervous system that is thought to be involved in cell adhesion and migration (31–34). Rs2228603 is located in exon 3 of NCAN and encodes a non-conservative nonsynonymous mutation (Pro92Ser), which is predicted by the software tool PolyPhen-2 to alter protein structure and function (35). While neurocan itself has not yet been shown to be expressed in the liver or to directly affect lipid metabolism or hepatic steatosis, it is becoming increasingly recognized that the central nervous system (CNS) is an important regulator of peripheral glucose and triglyceride metabolism (36–38). The liver is highly innervated by both sympathetic and parasympathetic nerves (39). Bruinstroop et al have shown that post-prandial serum triglyceride levels were significantly elevated in parasympathetic or sympathetic denervated rats compared to sham-operated animals (40). Robertson et al demonstrated that vagal stimulation increases hepatic secretion of very low-density lipoprotein (VLDL) triglyceride (41). Since NCAN rs2228603 has been associated with increased liver fat and decreased serum triglyceride (11, 29), it is plausible that this variant is associated with a brain-liver axis that, when deregulated, increases the risk for NAFLD. Furthermore, this axis may be modulated by increased dietary fat intake, as suggested by a 50% increase in hepatic VLDL synthesis after fat intake (43).
Further evidence for central control of liver lipid metabolism comes from studies of neuropeptide Y (NPY), a 36-amino acid peptide neurotransmitter secreted by the hypothalamus that has glucose and lipid regulatory effects in the liver. Fasting rats treated with intracerebroventricular injection of NPY had increased VLDL secretion into the bloodstream by 2.5-fold (44). Intracerebroventricular administration of an NPY-Y5 receptor agonist reproduced this effect, while an NPY-Y1 receptor antagonist decreased VLDL secretion. These findings demonstrate that the CNS can control VLDL secretion.
In summary, in light of the increased risk for NAFLD conferred by obesity, we have extended findings of the GOLD Consortium to include a large population of bariatric surgery patients in whom liver steatosis, inflammation, and fibrosis were documented histologically. Specifically, we have shown that variants in PNPLA3 and NCAN are associated not only with liver steatosis, but also NASH, a more progressive and clinically ominous manifestation of NAFLD. NCAN is a gene expressed in brain and thus may identify an untapped potential role for the CNS in the mechanism of fatty liver. Alternatively variants in other genes at this locus may be responsible for the NAFLD phenotype. Fine mapping and functional studies will be required to identify the causative genes/variants, which may provide insight into a novel pathway for NAFLD development, leading to new strategies for prevention and treatment of this disease. In addition, follow up analysis of the effects of gastric bypass surgery on liver steatosis and inflammation and dyslipidemia in our bariatric cohort may provide additional insight into the potential role of these loci as determinants of metabolic responses to weight loss.
Supplementary Material
Acknowledgments
Financial Support
This work was supported by NIH R01 DK088231 and DK091601, the Obesity Research Institute of Geisinger Medical Center, the Mid-Atlantic Nutrition and Obesity Research Center (NIH P30 DK072488), and the Baltimore Veterans Administration Geriatric Research and Education Clinical Center (GRECC). Dr. Gorden was supported by NIH T32 DK067872 and is currently supported by NIH F32 DK093267. Dr. Yerges-Armstrong is supported by NIH F32 AR59469. Dr. Speliotes is supported by NIH K23 DK080145, the Biological Scholars Program and other funds from the University of Michigan.
Dr. Still receives grant and consulting support from Ethicon-Endosurgery.
Abbreviations
- ALT
alanine transaminase
- AST
aspartate transaminase
- BMI
body mass index
- CNS
central nervous system
- GOLD
Genetics of Obesity-related Liver Disease
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- SNP
single-nucleotide polymorphism
- VLDL
very low-density lipoprotein
Footnotes
Disclosures
Drs. Shuldiner, Gerhard, Chu, Yerges-Armstrong, Speliotes, Yang, and Gorden and Mr. Wood have no conflicts to report.
References
- 1.Eckel RH, Alberti KG, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2010 Jan 16;375(9710):181–183. doi: 10.1016/S0140-6736(09)61794-3. [DOI] [PubMed] [Google Scholar]
- 2.Argo CK, Caldwell SH. Epidemiology and natural history of non-alcoholic steatohepatitis. Clin Liver Dis. 2009 Nov;13(4):511–513. doi: 10.1016/j.cld.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Argo CK, Northup PG, Al-Osaimi AM, Caldwell SH. Systematic review of risk factors for fibrosis progression in non-alcoholic steatohepatitis. J Hepatol. 2009 Aug;51(2):371–379. doi: 10.1016/j.jhep.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Bellentani S, Saccoccio G, Masutti F, Croce LS, Brandi G, Sasso F, et al. Prevalence of and risk factors for hepatic steatosis in northern italy. Ann Intern Med. 2000 Jan 18;132(2):112–117. doi: 10.7326/0003-4819-132-2-200001180-00004. [DOI] [PubMed] [Google Scholar]
- 5.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, et al. Prevalence of hepatic steatosis in an urban population in the united states: Impact of ethnicity. Hepatology. 2004 Dec;40(6):1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 6.Rinella ME, Alonso E, Rao S, Whitington P, Fryer J, Abecassis M, et al. Body mass index as a predictor of hepatic steatosis in living liver donors. Liver Transpl. 2001 May;7(5):409–414. doi: 10.1053/jlts.2001.23787. [DOI] [PubMed] [Google Scholar]
- 7.Stranges S, Dorn JM, Muti P, Freudenheim JL, Farinaro E, Russell M, et al. Body fat distribution, relative weight, and liver enzyme levels: A population-based study. Hepatology. 2004 Mar;39(3):754–763. doi: 10.1002/hep.20149. [DOI] [PubMed] [Google Scholar]
- 8.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008 Dec;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romeo S, Sentinelli F, Dash S, Yeo GS, Savage DB, Leonetti F, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of european descent. Int J Obes (Lond) 2010 Jan;34(1):190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 10.Speliotes EK, Butler JL, Palmer CD, Voight BF, et al. GIANT Consortium, MIGen Consortium. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010 Sep;52(3):904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011 Mar;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood GC, Chu X, Manney C, Strodel W, Petrick A, Gabrielsen J, et al. An electronic health record-enabled obesity database. BMC Med Inform Decis Mak. 2012 May 28;12(1):45. doi: 10.1186/1472-6947-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhard GS, Chokshi R, Still CD, Benotti P, Wood GC, Freedman-Weiss M, et al. The influence of iron status and genetic polymorphisms in the HFE gene on the risk for postoperative complications after bariatric surgery: A prospective cohort study in 1,064 patients. Patient Saf Surg. 2011 Jan 10;5(1):1. doi: 10.1186/1754-9493-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999 Sep;94(9):2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 15.Still CD, Wood GC, Chu X, Erdman R, Manney CH, Benotti PN, et al. High allelic burden of four obesity SNPs is associated with poorer weight loss outcomes following gastric bypass surgery. Obesity (Silver Spring) 2011 Aug;19(8):1676–1683. doi: 10.1038/oby.2011.3. [DOI] [PubMed] [Google Scholar]
- 16.Christou N, Efthimiou E. Five-year outcomes of laparoscopic adjustable gastric banding and laparoscopic roux-en-Y gastric bypass in a comprehensive bariatric surgery program in canada. Canadian Journal of Surgery. 2009;52(6):E249–E258. [PMC free article] [PubMed] [Google Scholar]
- 17.Jakobsen GS, Hofso D, Roislien J, Sandbu R, Hjelmesaeth J. Morbidly obese patients--who undergoes bariatric surgery? Obes Surg. 2010 Aug;20(8):1142–1148. doi: 10.1007/s11695-009-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LABS Writing Group for the LABS Consortium. Belle SH, Chapman W, Courcoulas AP, Flum DR, Gagner M, et al. Relationship of body mass index with demographic and clinical characteristics in the longitudinal assessment of bariatric surgery (LABS) Surg Obes Relat Dis. 2008 Jul-Aug;4(4):474–480. doi: 10.1016/j.soard.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatrath H, Vuppalanchi R, Chalasani N. Dyslipidemia in patients with nonalcoholic fatty liver disease. Semin Liver Dis. 2012 Feb;32(1):22–29. doi: 10.1055/s-0032-1306423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi M, Kao WH, Boerwinkle E, Hoogeveen RC, Rasmussen-Torvik LJ, Astor BC, et al. Association of rs780094 in GCKR with metabolic traits and incident diabetes and cardiovascular disease: The ARIC study. PLoS One. 2010 Jul 22;5(7):e11690. doi: 10.1371/journal.pone.0011690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onuma H, Tabara Y, Kawamoto R, Shimizu I, Kawamura R, Takata Y, et al. The GCKR rs780094 polymorphism is associated with susceptibility of type 2 diabetes, reduced fasting plasma glucose levels, increased triglycerides levels and lower HOMA-IR in japanese population. J Hum Genet. 2010 Sep;55(9):600–604. doi: 10.1038/jhg.2010.75. [DOI] [PubMed] [Google Scholar]
- 22.Pollin TI, Jablonski KA, McAteer JB, Saxena R, Kathiresan S, Kahn SE, et al. Triglyceride response to an intensive lifestyle intervention is enhanced in carriers of the GCKR Pro446Leu polymorphism. J Clin Endocrinol Metab. 2011 Jul;96(7):E1142–E1147. doi: 10.1210/jc.2010-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009 Oct;50(10):2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011 Jun;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 25.Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010 Apr;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 26.Krarup NT, Grarup N, Banasik K, Friedrichsen M, Faerch K, Sandholt CH, et al. The PNPLA3 rs738409 G-allele associates with reduced fasting serum triglyceride and serum cholesterol in danes with impaired glucose regulation. PLoS One. 2012;7(7):e40376. doi: 10.1371/journal.pone.0040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer CN, Maglio C, Pirazzi C, Burza MA, Adiels M, Burch L, et al. Paradoxical lower serum triglyceride levels and higher type 2 diabetes mellitus susceptibility in obese individuals with the PNPLA3 148M variant. PLoS One. 2012;7(6):e39362. doi: 10.1371/journal.pone.0039362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008 Feb;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008 Feb;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L, Ding H, Zhang X, He M, Huang S, Xu Y, et al. Genetic variants at newly identified lipid loci are associated with coronary heart disease in a chinese han population. PLoS One. 2011;6(11):e27481. doi: 10.1371/journal.pone.0027481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch U, Feng K, Zhou XH. Neurocan: A brain chondroitin sulfate proteoglycan. Cell Mol Life Sci. 2001 Nov;58(12–13):1842–1856. doi: 10.1007/PL00000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Retzler C, Gohring W, Rauch U. Analysis of neurocan structures interacting with the neural cell adhesion molecule N-CAM. J Biol Chem. 1996 Nov 1;271(44):27304–27310. doi: 10.1074/jbc.271.44.27304. [DOI] [PubMed] [Google Scholar]
- 33.Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M. The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. J Cell Biol. 1994 May;125(3):669–680. doi: 10.1083/jcb.125.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grumet M, Milev P, Sakurai T, Karthikeyan L, Bourdon M, Margolis RK, et al. Interactions with tenascin and differential effects on cell adhesion of neurocan and phosphacan, two major chondroitin sulfate proteoglycans of nervous tissue. J Biol Chem. 1994 Apr 22;269(16):12142–12146. [PubMed] [Google Scholar]
- 35.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam TK, Gutierrez-Juarez R, Pocai A, Bhanot S, Tso P, Schwartz GJ, et al. Brain glucose metabolism controls the hepatic secretion of triglyceride-rich lipoproteins. Nat Med. 2007 Feb;13(2):171–180. doi: 10.1038/nm1540. [DOI] [PubMed] [Google Scholar]
- 37.Nogueiras R, Wiedmer P, Perez-Tilve D, Veyrat-Durebex C, Keogh JM, Sutton GM, et al. The central melanocortin system directly controls peripheral lipid metabolism. J Clin Invest. 2007 Nov;117(11):3475–3488. doi: 10.1172/JCI31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Hoek AM, Voshol PJ, Karnekamp BN, Buijs RM, Romijn JA, Havekes LM, et al. Intracerebroventricular neuropeptide Y infusion precludes inhibition of glucose and VLDL production by insulin. Diabetes. 2004 Oct;53(10):2529–2534. doi: 10.2337/diabetes.53.10.2529. [DOI] [PubMed] [Google Scholar]
- 39.Powley TL. Vagal circuitry mediating cephalic-phase responses to food. Appetite. 2000 Apr;34(2):184–188. doi: 10.1006/appe.1999.0279. [DOI] [PubMed] [Google Scholar]
- 40.Bruinstroop E, Pei L, Ackermans MT, Foppen E, Borgers AJ, Kwakkel J, et al. Hypothalamic neuropeptide Y (NPY) controls hepatic VLDL-triglyceride secretion in rats via the sympathetic nervous system. Diabetes. 2012 May;61(5):1043–1050. doi: 10.2337/db11-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson MD, Mason AO, Frayn KN. Timing of vagal stimulation affects postprandial lipid metabolism in humans. Am J Clin Nutr. 2002 Jul;76(1):71–77. doi: 10.1093/ajcn/76.1.71. [DOI] [PubMed] [Google Scholar]
- 42.Tilg H, Moschen AR. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol Metab. 2008 Dec;19(10):371–379. doi: 10.1016/j.tem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Cohn JS, Wagner DA, Cohn SD, Millar JS, Schaefer EJ. Measurement of very low density and low density lipoprotein apolipoprotein (apo) B-100 and high density lipoprotein apo A-I production in human subjects using deuterated leucine. effect of fasting and feeding. J Clin Invest. 1990 Mar;85(3):804–811. doi: 10.1172/JCI114507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stafford JM, Yu F, Printz R, Hasty AH, Swift LL, Niswender KD. Central nervous system neuropeptide Y signaling modulates VLDL triglyceride secretion. Diabetes. 2008 Jun;57(6):1482–1490. doi: 10.2337/db07-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.