Abstract
Heparin-immobilized microspheres were included in microdialysis sampling perfusion fluids under both in vitro and in vivo conditions to improve the recovery of different cytokines, acidic fibroblast growth factor, vascular endothelial growth factor, monocyte chemoattractant protein-1 (or CCL2), and regulation upon activation normal T cell express sequence (or CCL5). Different strategies to dissociate captured CCL2 and CCL5 from the immobilized heparin were attempted, and both cytokines could be quantitatively eluted from the beads using a phosphate buffer (pH 7.4) containing 25% (v/v) acetonitrile which did not interfere with the subsequent detection of cytokine using an ELISA assay. Using these heparin-immobilized microspheres, a two to fivefold increase of microdialysis relative recovery (RR) was achieved for the four cytokines from a quiescent solution. Enhanced microdialysis RR of CCL2 using the heparin-immobilized microspheres from microdialysis probes implanted into the peritoneal cavity of a rat was performed to test the in vivo application. This work suggests that the heparin-immobilized microspheres provide an alternative affinity agent to the previously used antibody-immobilized microspheres for enhanced microdialysis sampling of cytokines.
Keywords: Cytokines, Microdialysis sampling, Heparin-immobilized microspheres
Introduction
Cytokines are important signaling proteins that are secreted by immune cells and are present in tissue extracellular fluid [1-3]. Due to their potent activity, cytokines normally are measured in biological tissues at picomolar to femtomolar concentrations. Elevated expression of cytokines indicates the activation of cytokine pathways associated with inflammation or disease progression [4]. Thus, analytical methods that can be applied to the real-time collection and quantification of cytokines in situ are prerequisites for the study of cytokine biology and their involvement in pathology.
Microdialysis sampling is a well-established in vivo sampling technique [5, 6]. Sampling is based on passive diffusion of analytes across a semi-permeable hollow-fiber dialysis membrane. Collected dialysate samples can be directly analyzed by a variety of analytical methods, thus providing concentration and temporal information of the targeted analyte within its local tissue environment [7, 8]. Microdialysis sampling has been extensively used for in vivo collection of low-molecular-weight hydrophilic analytes in the study of drug metabolism, neuroscience, and pharmaceutics [9-11]. The efficiency of the dialysis process with respect to analyte mass transport is characterized by the analyte relative recovery (RR), shown in Eq. 1,
| (1) |
where Coutlet is the outlet analyte concentration of microdialysis probe and Csample,∞ is the analyte concentration far away from the microdialysis probe in the sample medium [12].
Microdialysis RR is highly dependent on the analyte diffusion properties which can be characterized by the analyte mass transport resistances among the three different regions it passes: sample medium, dialysis membrane, and perfusion fluid. Protein molecules, such as cytokines, usually have small RR values due to their large molecular weights and small aqueous diffusion coefficients. Typical microdialysis RR values for 10-kDa or larger proteins across 100-kDa molecular weight cutoff (MWCO) membranes range between 1% and 5% at flow rates of 0.5 and 1.0 μL/min [13-15]. The combination of low recovery of cytokines and their suspected low concentrations in vivo makes collecting these important signaling proteins challenging especially when using shorter microdialysis probes with higher flow rates (10-mm probes with 1.0-μL/min flow rate) intended for basic science studies as compared to the longer microdialysis probes with lower flow rates for clinical studies (10- or 30-mm probes with 0.3-μL/min flow rate) [16, 17].
Previously, we have reported the use of commercially available antibody-immobilized microspheres (7 μm o.d.) as affinity agents to increase cytokine RR during microdialysis sampling both in vitro and in vivo [18, 19]. The antibody–antigen interaction between cytokines and the microspheres results in an increased cytokine diffusive mass transport driving force and thus increased amounts of cytokines recovered. An average of 3 to 20 times RR enhancement was achieved for a series of cytokines using these antibody-immobilized microspheres. One problem associated with the analysis step using the antibody-immobilized microsphere approach is that we have previously observed the saturation of the antibodies when cytokine concentrations are >5,000 pg/mL, resulting in analyses that are not quantitative [19]. This difficulty is a severe concern when there is a need to quantify multiple cytokines expressed at concentrations that span a wide dynamic range. Flow cytometry analysis does not allow for repeated sample measurements; thus, if certain cytokine concentrations are out of range, there is no possibility to reanalyze the sample. For this reason, alternative approaches that will separate the cytokine recovery or capture event from the actual analysis are desired.
Heparin is a heterogeneous linear polysaccharide belonging to the family of glycosaminoglycans (GAGs) that are commonly found on the membrane of cells or within the extracellular matrix [20]. Heparin is negatively charged at physiological pH due to its highly sulfated disaccharide repeating units, sulfated uronic acid, and glucosamine. Heparin and other GAGs are of critical importance in intercellular communication in organisms due to their interactions with a wide variety of proteins [21]. Heparin affinity chromatography is commonly used in life science research for the removal or separation of different heparin-binding proteins. Several cytokines have been reported to bind heparin with dissociation constants in the nanomolar range [22-24]. Specific interactions between cytokines and heparin in vivo have been reviewed [25].
Antibody–antigen interactions typically have dissociation constants, KD, in the low nanomolar range, and their dissociation rate constants (koff) are roughly 10−5 s−1 [26, 27]. These slow dissociation kinetics generally require harsh conditions (low pH) to decouple the formed antibody–antigen complex, which subsequently interfere with immunoassay quantitation. Compared to antibody–antigen interactions, GAG (heparin)–cytokine interactions generally have faster dissociation rates (koff in the range 10−2–10−4 s−1) [28-30]. This is due to several factors, including shallow heparin binding pockets on the surface of cytokines and rapid conformational rearrangement of heparin on binding to cytokines [21]. Faster heparin/cytokine kinetics may provide an advantage for the dissociation of cytokine from heparin. Therefore, for the capture of cytokines, it is reasonable to consider decoupling the affinity capture step from the detection step using heparin-immobilized microspheres instead of the antibody-immobilized microspheres especially when during microdialysis sampling cytokine concentrations are in many cases unknown. Additionally, heparin provides the advantage of low cost, wide availability, and chemical stability compared to antibodies.
In this paper, we report an approach to use in-house prepared heparin-immobilized microspheres as affinity agents to increase the in vitro microdialysis RR of four proteins that will collectively be called cytokines, human acidic fibroblast growth factor (aFGF, pI 5.4, MW 16.0 kDa), rat vascular endothelial growth factor (VEGF, pI 8.5, MW 45.0 kDa), rat monocyte chemoattractant protein-1 (MCP-1/CCL2, pI 10.5, MW 26.2 kDa), and rat regulation upon activation normal T cell express sequence (RANTES/CCL5, pI 9.0, MW 8.0 kDa). Cytokine quantification was achieved either by bead-based flow cytometry or by ELISA after dissociation of cytokines from the microspheres using an appropriate dissociation reagent. Enhanced microdialysis RR of CCL2 from the rat peritoneal cavity using the heparin-immobilized microspheres demonstrates the in vivo application of this approach. A flowchart for the experimental approaches described in this manuscript is shown in Fig. 1.
Fig. 1.
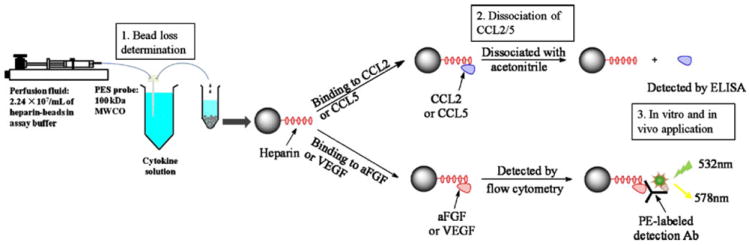
Flowchart of different approaches performed in this study
Experimental section
Chemicals
Glycerol, heparin (sodium salt from porcine intestine), neomycin, protamine, poly-l-lysine, albumin (from bovine serum, BSA), and lipopolysaccharide (LPS, from Escherichia coli K-235) were purchased from Sigma (St. Louis, MO, USA). Cell culture grade water and phosphate-buffered saline (PBS, 1X, Dulbecco’s formula, pH 7.4, consisting of CaCl2 0.90 mM, MgSO4 0.49 mM, KCl 2.68 mM, KH2PO4 1.47 mM, NaCl 136.89 mM, and Na2HPO4 8.10 mM) purchased from MP Biomedicals (Irvine, CA, USA) were sterilized and used for in vivo experiments. LPS was dissolved in sterilized PBS at a concentration of 100 μg/mL and stored at 4 °C before use. Isoflurane (liquid for inhalation anesthesia) was purchased from Hospira, Inc. (Lake Forest, IL, USA). HPLC grade water, acetonitrile (Fisher Scientific, Pittsburgh, PA, USA), and in-house prepared PBS (pH 7.4, consisting of NaCl 136.89 mM, Na2HPO4 8.10 mM, KH2PO4 1.47 mM, and KCl 2.68 mM) were used for in vitro experiments. Recombinant rat MCP-1 was from Cell Sciences (Canton, MA, USA). Rat MCP-1 ELISA kit was from BD Biosciences Pharmingen (San Jose, CA, USA). Recombinant human aFGF, biotin-conjugated anti-human aFGF polyclonal antibody, and human aFGF ELISA kit were obtained from Antigenix America, Inc. (Huntington Station, NY, USA). Recombinant rat RANTES and rat RANTES ELISA kit were purchased from Peprotech, Inc. (Rocky Hill, NJ, USA). Recombinant rat VEGF and rat VEGF ELISA kit were from R&D Systems, Inc. (Minneapolis, MN, USA). R-phycoerythrin-conjugated streptavidin was purchased from Molecular Probes, Inc. (Eugene, OR, USA).
CCL2 and CCL5 dissociation
The preparation of heparin-immobilized microspheres has been described [31]. A total of 1.0×107 heparin-immobilized microspheres were incubated with 500 μL of CCL2 or CCL5 (2.5 ng/mL) in PBS (pH 7.4) containing 0.05% (w/v) BSA at room temperature for 2 h. After centrifugation, the supernatant was removed and the cytokine content in the supernatant was determined using the corresponding rat CCL2 or rat CCL5 ELISA kit. Absorbance at 450 nm (corrected by subtraction of absorbance at 570 nm) was monitored using a Tecan SPECTRAFluor plate reader (Tecan Group Ltd., Männedorf, Switzerland). The amount of cytokine captured on the microspheres was determined by mass balance, which was the difference between the amount of cytokine added and that remaining in the reaction mixture.
Screening of appropriate reagents for cytokine dissociation from the heparin-immobilized microspheres was carried out using a parallel batch dissociation assay. Solutions (110 μL) of different dissociation reagents (Table 1) were added to 8×105 microspheres with bound CCL2 or CCL5. The suspension was equilibrated at 4 °C for 12 h. After centrifugation, the supernatant was removed and cytokine content in the supernatant was determined using the ELISA kit. The percent cytokine dissociated was calculated for each batch based on mass balance, which was the mass ratio of cytokine eluted to the total amount of cytokine loaded on the microspheres.
Table 1.
Performance of various reagents for the dissociation of CCL2 and CCL5 from the heparin-immobilized microspheres
| Dissociation reagentsa | % CCL2 eluted from the microspheresb | % CCL5 eluted from the microspheresb |
|---|---|---|
| 1 M NaCl | 4.0±0.59 | 9.2±1.30 |
| 2.22 μM neomycin | 4.9±0.31 | 5.2±0.59 |
| 30 μM neomycin | 7.2±0.25 | 12.5±0.80 |
| 2.22 μM protamine | 7.9±0.45 | 4.9±0.55 |
| 50 μg/mL poly-l-lysine | 9.8±1.06 | 7.5±0.95 |
| 2 M NaCl in 10 mM sodium carbonate buffer, pH 9.5 | 12.1±1.24 | 33.7±2.05 |
| 2 M NaCl | 18.4±2.30 | 35.6±1.77 |
| 1 M NaCl and 25% (v/v) glycerol | 20.5±1.17 | 19.6±1.83 |
| 10% (v/v) acetonitrile | 27.2±1.12 | 33.4±2.55 |
| 0.5 M CaCl2 | 29.7±1.50 | 18.4±1.51 |
| 2 M NaCl in 10 mM sodium acetate buffer, pH 4.0 | 35.0±3.44 | 39.9±2.05 |
| 20% (v/v) acetonitrile | 90.8±1.57 | 88.8±2.67 |
| 30% (v/v) acetonitrile | 92.2±2.98 | 91.5±2.56 |
Data represent mean ± SD, n=3 batch processes
Unless indicated, all dissociation reagents were dissolved or mixed in PBS, pH 7.4, containing 0.05% (w/v) BSA
Percent CCL2 or CCL5 eluted was calculated by the mass ratio of cytokine eluted to the total amount of cytokine loaded on the microspheres
Bead loss determination
CMA/20 microdialysis probes with a 10-mm polyethersulfone (PES) membrane (100-kDa MWCO, i.d. 420 μm, o.d. 500 μm, CMA Microdialysis, North Chelmsford, MA, USA) were used. The external diameter of the probe internal cannula is 350 μm. A 1000 series gastight syringe (Hamilton, Reno, NV, USA) mounted on a BAS microdialysis syringe pump with a Bee syringe pump controller (BASi, West Lafayette, IN, USA) was used to pump the perfusion fluid (PBS with 0.05% (w/v) BSA, pH 7.4) through the microdialysis probes. To determine the mass balance of microspheres passing through the microdialysis sampling system, the heparin-immobilized microspheres at an amount of 2.24±0.11×107 beads/mL in the perfusion fluid were included in the syringe and perfused through the microdialysis probe at three different flow rates (2.0, 1.0, and 0.5 μL/min). The concentration of microspheres used here was based on total heparin content determined so as to match previous work using a heparin–albumin conjugate (which was 7.7×105 beads/mL) [32]. The syringe was agitated on an in-house built rotator that can rotate 180° and make a complete cycle in 90 s [32]. The rotator served to keep the microspheres in suspension within the syringe during microdialysis sampling. Three dialysate samples (75 μL each) were collected at each flow rate. After each collection, the flow rate was increased to 5.0 μL/min and the system was perfused for 10 min. The bead amount collected in each sample was determined by manual counting on a hemacytometer (Hausser Scientific, Horsham, PA, USA).
In order to find the bead loss site through the microdialysis sampling system, the heparin-immobilized microspheres at an amount of 1.85±0.10×107 beads/mL were included in a syringe that was perfused at 0.5 μL/min for 120 min. The microspheres were collected at three different positions denoted in Fig. 2. The bead amounts collected at each position were determined by manual counting with a hemacytometer.
Fig. 2.
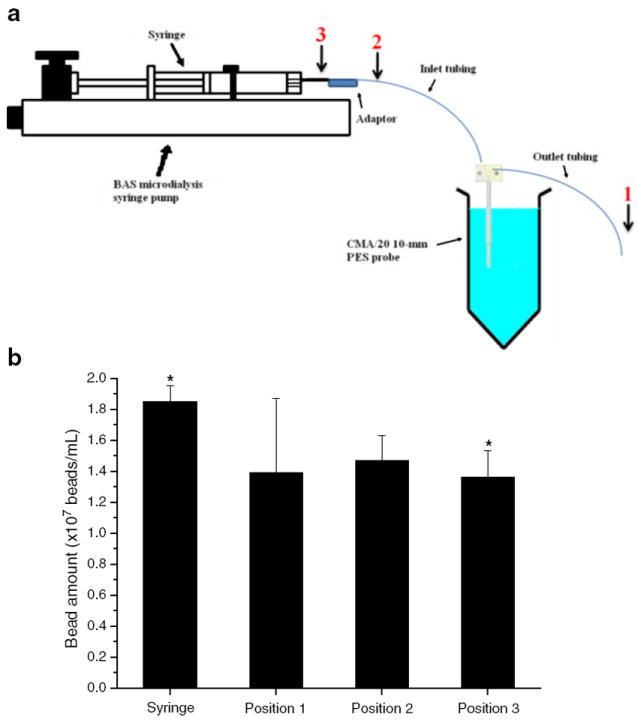
Determination of bead loss site during microdialysis sampling at a 0.5-μL/min flow rate. Approximately 2×107 beads/mL of heparin-immobilized microspheres was included in the syringe. Microspheres were collected at position 1 (probe outlet tubing), position 2 (probe inlet tubing), and position 3 (syringe outlet). The image of BAS pump and syringe was redrawn from http://www.basinc.com/products/iv/bee.html. Asterisk represents significant difference between bead amounts collected at position 3 (syringe outlet) and included in the syringe indicated by a paired t test (p<0.05, n=3)
In vitro microdialysis sampling
In vitro microdialysis sampling of cytokines was performed at ambient room temperature (21–25 °C). The perfusion fluid consisted of either 0.05% (w/v) BSA in PBS (pH 7.4) together with heparin-immobilized microspheres (2.24×107 beads/mL) for enhanced microdialysis sampling or 0.05% (w/v) BSA in PBS (pH 7.4) for the control experiment. For the enhanced microdialysis experiment that included the heparin-immobilized microspheres in the perfusion fluid, the syringe was agitated on the rotator to keep the microspheres in suspension within the syringe. Solutions containing each cytokine (200 ng/mL of aFGF or VEGF; 10 ng/mL of CCL2 or CCL5) were prepared in a buffered solution having the same composition as the perfusion fluid (PBS with 0.05% (w/v) BSA, pH 7.4). A 1-mL portion of the cytokine solution was placed into a 1.5-mL microcentrifuge tube. Two CMA/20 microdialysis probes with a 10-mm PES membrane for both control and enhanced microdialysis experiments were immersed into this quiescent solution.
During microdialysis sampling, the probes were perfused at flow rates of 2.0, 1.0, and 0.5 μL/min, and 75 μL of sample was collected at each flow rate. All the microdialysis experiments (both control and those containing heparin-immobilized microspheres in the perfusate) were performed in triplicate at each flow rate using one probe. All samples were stored at 4 °C prior to sample analysis.
Sample preparation and analysis
aFGF and VEGF The concentration of aFGF or VEGF in the dialysate collected from the control probe was measured by mixing 15 μL of dialysate with 15 μL of heparin-immobilized microspheres (4.48×107 beads/mL) followed by flow cytometric analysis as per the procedure described in the previous paper [31]. Dialysates (30 μL) containing microspheres were analyzed directly by flow cytometry. Dialysate sample of aFGF was analyzed on the BD FACSArray flow cytometer (BD Biosciences) and the VEGF sample was analyzed on the Luminex 100 system (Luminex Corp., Austin, TX, USA). Each sample was measured in duplicate and the average was taken for RR calculation. Cytokine concentration in the sample medium was determined in duplicate via splitting of a 30-μL aliquot taken before and after microdialysis sampling. The average concentration between these duplicate samples was taken as the cytokine concentration in the sample medium for RR calculation.
CCL2 and CCL5 Dialysate (60 μL) collected from the control probe or probe with microspheres was diluted to 120 μL with the perfusion fluid, which was supplemented to an acetonitrile concentration of 25% (v/v). The suspension was then incubated at 4 °C for 12 h to allow dissociation of CCL2 or CCL5 from the microspheres. After dissociation, samples containing acetonitrile were centrifuged at 700×g for 1 min. Solution (control dialysate, 110 μL) or supernatant (dialysate with microspheres, 110 μL) was further diluted to 220 μL with the assay diluent from the ELISA kit. Cytokine concentrations in the samples were determined in duplicate using the corresponding ELISA kit. Cytokine concentration in the sample medium was determined in duplicate via splitting of a 60-μL aliquot taken before and after microdialysis sampling. The average concentration between these duplicate samples was taken as the cytokine concentration in the sample medium for RR calculation.
In vivo microdialysis sampling
Male Sprague–Dawley rats (200–225 g, Taconic, NY, USA) were used for in vivo microdialysis sampling experiments. All animal experimental protocols were approved by the Albany Medical College IACUC committee and met the guidelines set forth by the NIH for the care and use of experimental animals. The rats were anesthetized with isoflurane during the whole experiment. Body temperature was maintained at 37.0±0.5 °C using a rectal temperature-controlled heating pad (CMA Microdialysis, Inc.) during the aseptic surgery and sampling process. All surgical tools were autoclaved before use.
To insert the microdialysis probes, two 0.5-cm incisions were made in the abdominal wall, one on each side. Two identical CMA/20 microdialysis probes with 10-mm PES membranes were inserted through each incision into the peritoneal cavity, and the incisions were closed using surgical staples. The probe tubing lines were taped to the animal to minimize probe movement. A tissue wetted with sterile PBS was placed on the incisions to prevent tissue dehydration. One probe served as the control probe which was perfused with sterile PBS containing 0.1% (w/v) BSA. The other probe was perfused with the same perfusion fluid as control together with heparin-immobilized microspheres (2.0×107 beads/mL) for enhanced microdialysis sampling. The syringe with the heparin-immobilized microspheres in the perfusion fluid was agitated on the rotator as used for in vitro experiment to keep the microspheres in suspension within the syringe. Dialysates were collected every 30 min after probe implantation for a total of 3 h at a flow rate of 1.0 μL/min. To elicit an immune response that would produce CCL2, the rats were intravenously administered with LPS (0.1 μg/g of body weight) 30 min post-probe implantation. At the end of the experiment, 200 μL of sterile PBS was injected into the peritoneal cavity after microdialysis probe removal and equilibrated for 1 min. The fluid was then removed from the cavity and used to determine the concentration of CCL2 in the peritoneal cavity. All samples were stored at 4 °C <2 days prior to analysis.
In a separate set of experiments, the perfusion of free heparin was compared to the heparin-immobilized microspheres for the ability to increase the in vivo microdialysis RR of CCL2. Three probes were implanted into the peritoneal cavity and perfused with sterile PBS containing either 0.1 μM of heparin in 0.1% (w/v) BSA, 4.0× 106 beads/mL (~0.1 μM heparin) in 0.1% (w/v) BSA, or 0.1% (w/v) BSA (control). Implantation and LPS dosing schedules were the same as described above.
Samples were analyzed using the same procedure as the in vitro experiment. Dialysate (30 μL) collected from control probe or probe with microspheres was diluted to 60 μL with the perfusion fluid, which was supplemented to an acetonitrile concentration of 25% (v/v). The suspension was then incubated at 4 °C for 12 h to allow dissociation of CCL2 from the microspheres. After centrifugation at 700×g for 1 min, solution (control dialysate, 55 μL) or supernatant (dialysate with microspheres, 55 μL) was further diluted to 220 μL with the assay diluent from the ELISA kit. CCL2 concentration was determined in duplicate using the rat CCL2 ELISA kit. The peritoneal washed sample was also measured in duplicate using the rat CCL2 ELISA kit.
In vitro calibration of explanted microdialysis probes
After completing the in vivo microdialysis sampling experiment, each pair of microdialysis probes implanted in each rat was calibrated. Two CMA 1-mL glass microsyringes were mounted on a CMA/102 dual-channel microdialysis pump (CMA Microdialysis Inc.) and used to pump the perfusion fluid through the microdialysis probes. The perfusion fluid had the same composition as the one used for the in vivo control experiment, which was the PBS containing 0.1% (w/v) BSA, pH 7.4. Probes explanted from the peritoneal cavity were first perfused (flow rate=1.0 μL/min) with HPLC grade water at room temperature for 1 h and stored before calibration. Solutions containing the same concentration of CCL2 as quantified in the peritoneal wash solution for each rat (49, 36, 35, 38, 77, and 32 ng/mL for rats 1 to 6, respectively; Table 4) were prepared as the sample solution. A 500-μL portion of this solution was placed into a 1.5-mL microcentrifuge tube and used as the sample medium which was quiescent and kept at 37±0.5 °C in a sand bath. Two explanted microdialysis probes from each rat were immersed into each corresponding solution. The probes were perfused at a flow rate of 1.0 μL/min using the perfusion fluid, and 30 μL of dialysate was collected. All experiments were performed in triplicate using each probe. Samples were stored at 4 °C prior to analysis.
Table 4.
Comparison of CCL2 concentrations in the control dialysates, enhanced dialysates, peritoneal washed samples, and estimated RRs
| Control (pg/mL) | Enhanced (pg/mL) | Peritoneal washed sample (pg/mL) | Estimated control RR (%) | Estimated enhanced RR (%) | |
|---|---|---|---|---|---|
| Rat 1 | 4,730 | 10,240 | 49,200 | 9.6 | 20.8 |
| Rat 2 | 4,630 | 8,310 | 36,250 | 12.8 | 22.9 |
| Rat 3 | 6,120 | 7,860 | 34,900 | 17.6 | 22.5 |
| Rat 4 | 4,510 | 8,670 | 37,550 | 12.0 | 23.1 |
| Rat 5 | 9,730 | 13,670 | 76,690 | 12.7 | 17.8 |
| Rat 6 | 2,340 | 5,460 | 31,650 | 7.4 | 17.3 |
Two microdialysis probes were implanted in the peritoneal cavity of each rat, one as control and the other as the enhanced probe. The flow rate was 1.0 μL/min. Both control and enhanced dialysate samples shown here were samples collected in the last 30 min before peritoneal wash. The peritoneal washed sample was obtained by injecting 200 μL of PBS into the peritoneal cavity right after all the dialysate samples collection. The estimated RR values were calculated as the ratio of CCL2 concentrations in the dialysate sample and the peritoneal washed sample
Statistical analysis
Statistical analysis of data including one-way analysis of variance (ANOVA), two-way ANOVA with replicate samples, and the Student’s t test was performed using Microsoft Office Excel (Edition 2007).
Results and discussion
Dissociation of cytokines from the heparin-immobilized microspheres
An important aspect of this work was to be able to capture cytokines with heparin-immobilized microspheres and release the cytokines so that standard protein detection methods could be used. An ideal dissociation reagent should be able to release a maximum amount of cytokine from the microspheres and still be compatible with ELISA or other detection methods. For this purpose, a series of judiciously chosen dissociation reagents were screened for their ability to release CCL2 and CCL5 from the heparin-immobilized microspheres. Table 1 shows the percentage of CCL2 and CCL5 released by various reagents, including different salt types at different concentrations, buffers at different pH values, buffers with organic modifiers, and highly positively charged displacers. The reagents are arranged in increasing order of percent CCL2 released from the heparin-immobilized microspheres. All the conditions tested were found to not interfere with the CCL2 and CCL5 ELISA assays. While heparin/protein interactions are usually considered to be mostly ionic [22, 33, 34], 2 M NaCl was not effective enough to completely dissociate CCL2 or CCL5 from the heparin-immobilized microspheres in this experiment. This may be explained by the batch condition used in this work as compared to the continuous flow condition commonly used in heparin-sepharose affinity chromatography for cytokine binding studies [25]. The cation-exchange displacer, neomycin, or the heparin-binding protein and polypeptide, protamine and poly-l-lysine, did not exhibit acceptable displacement efficacy either using concentrations that did not sacrifice immunoassay performance. However, the percent CCL2 or CCL5 that was eluted significantly increased using a PBS buffer supplemented with 20–30% (v/v) acetonitrile. More than 90% of bound cytokine could be dissociated from the heparin-immobilized microspheres using this method, and acetonitrile below 30% (v/v) in the dissociation buffer did not interfere with the subsequent ELISA assay. This suggests that binding of the cytokine to heparin may involve more than electrostatic interactions between the negatively charged heparin and the cytokine protein.
Bead loss determination during microdialysis sampling
It is possible for the microspheres to settle during the microdialysis sampling process. Table 2 shows the amount of microspheres collected under varying flow rate conditions. Approximate amounts of 1.7–2.0×107 beads/mL (1.31–1.52×106 beads in 75 μL of dialysate) could be collected at the probe outlet with three different flow rates (0.5, 1.0, and 2.0 μL/min) when the amount of 2.24×107 beads/mL (1.68×106 beads in 75 μL of perfusion fluid) was included in the syringe, indicating that 78–90% microspheres passed through the microdialysis sampling system. Residual microspheres were flushed out of the probe at 5.0 μL/min for 10 min. Approximately 2.65–2.82×107 beads/mL (2.0–2.9×105 more beads than expected) were collected in this 50 μL of perfusate. The calculated total bead amounts collected showed no significant difference (p<0.05) from the initial beads included inside the syringe indicating mass balance.
Table 2.
Microsphere passage through the CMA/20 microdialysis probe with a 10-mm PES membrane
| No. of microspheres in 75 μL of perfusion fluida | 1.68±0.08×106 | ||
| No. of microspheres in 75 μL of dialysates collected at different flow ratesb | 0.5 μL/min (1.31±0.12×106) | 1.0 μL/min (1.44±0.10×106) | 2.0 μL/min (1.52±0.08×106) |
| Percentage of microspheres collected at different flow rates | 78.0±8.0 | 85.7±7.2 | 90.5±6.4 |
| No. of microspheres recovered after 10-min flush at 5 μL/minb | 2.9±0.5×105 | 2.6±0.3×105 | 2.0±0.3×105 |
| Total no. of microspheres collected c | 1.60±0.13×106 | 1.70±0.10×106 | 1.72±0.09×106 |
Data represent mean ± SD, n=4 readings of bead amount using hemacytometer
Data represent mean ± SD, n=3 collections at each flow rate
The calculated total number of microspheres collected showed no significant difference (p<0.05) from the initial microspheres included inside the syringe, 1.68±0.08×106 microspheres in 75 μL of perfusion fluid
To determine where the microspheres were lost through the microdialysis sampling system, microspheres at the amount of 1.85×107 beads/mL were included in the syringe and collected at different positions shown in Fig. 2. Figure 2 shows the bead amount collected at each of the three positions at 0.5 μL/min for 120 min. A one-way ANOVA showed no significant difference (p<0.05) among the bead amounts collected at the three different positions. However, the microspheres collected at position 3 (syringe outlet) were significantly fewer than the microspheres in the syringe indicated by a paired t test (p<0.05). This suggests that most microspheres were lost at position 3 at the syringe tip rather than through the microdialysis sampling probe. Different syringes (CMA Microdialysis vs. BAS) gave the same results (data not shown).
In order to solve the problem of bead settling during the sampling process, the flow rate was increased to 5.0 μL/min at certain time intervals for 1 min. At a flow rate of 0.5 μL/min, the probes were perfused for 120 min to obtain 60 μL of sample. In this period, the flow rate was increased to 5.0 μL/min every 40 min and maintained for 1 min. Then, the flow rate was decreased back to 0.5 μL/min. Similarly, at flow rates of 1.0 and 2.0 μL/min, the probes were perfused for 60 and 30 min, and the flow rates were increased to 5.0 μL/min every 20 and 10 min, respectively. The same procedure was performed for both the control probe and the probe with microspheres. In this way, microspheres at the amount of 2.08±0.15×107 beads/mL could be collected finally at the microdialysis probe outlet, which were ~93% of the total microspheres included in the syringe.
In vitro enhanced microdialysis RR of cytokines
Table 3 shows the in vitro enhanced microdialysis RRs of four cytokines—aFGF, VEGF, CCL2, and CCL5—using heparin-immobilized microspheres as affinity agents in the perfusion fluid at three different flow rates. Paired t tests (p<0.05) between the control and the heparin-immobilized microsphere-containing probes show that the RR values are statistically different for aFGF at the flow rate of 0.5 μL/min, VEGF at all three flow rates, CCL2 at the flow rates of 1.0 and 2.0 μL/min, and CCL5 at the flow rates of 0.5 and 1.0 μL/min. An average of two to fivefold increase of RR was achieved for the four cytokines using these heparin-immobilized microspheres in the perfusion fluid, which is similar to that for antibodies [35].
Table 3.
Microdialysis relative recovery enhancement of cytokines using heparin-immobilized microspheres as affinity agents as compared to control at three different flow rates
| Cytokines (MW) | Flow rate (μL/min) | Relative recovery (%)
|
|
|---|---|---|---|
| Control | Enhanced | ||
| aFGF (16 kDa) | 0.5 | 8.4±2.3 | 29.2±4.8a |
| 1.0 | 4.9±1.9 | 9.8±0.7 | |
| 2.0 | 3.2±2.3 | 4.5±1.8 | |
| VEGF (45 kDa) | 0.5 | 2.2±0.1 | 11.3±0.7a |
| 1.0 | 1.8±0.1 | 6.5±0.7a | |
| 2.0 | 1.4±0.2 | 3.9±0.5a | |
| CCL2 (26.2 kDa) | 0.5 | 6.9±3.0 | 13.8±2.2 |
| 1.0 | 2.8±1.1 | 11.2±1.4a | |
| 2.0 | 2.0±0.4 | 8.9±1.2a | |
| CCL5 (8 kDa) | 0.5 | 9.7±1.6 | 26.3±3.3a |
| 1.0 | 7.9±1.2 | 13.0±3.2a | |
| 2.0 | 5.3±0.4 | 8.5±1.6 | |
Data represent mean ± SD, n=3
Statistically significant difference between control and enhanced relative recovery values at 95% confidence level
Control RR values differ between the cytokines shown in Table 3, and the difference is mainly due to the variations of their molecular weights, tertiary structures, and diffusion coefficients. The control RR of CCL5, a cytokine with 8-kDa MW, was significantly higher (p<0.05) than that of VEGF, a cytokine with 45-kDa MW at each corresponding flow rate. The principal mechanism for the RR enhancement is that binding of cytokines onto the heparin-immobilized microspheres decreases the concentration of free cytokines inside the microdialysis probe, thus increasing the cytokine concentration gradient and the driving force for cytokine recovery across the membrane. In general, cytokines exhibiting higher control recovery also gave higher enhanced recovery. The trend observed for the RR enhancements was not simply related to the binding affinities and kinetics of cytokines to heparin revealed in previous papers [22, 28, 30, 36]. This suggests that binding interactions between cytokines and the affinity agent only account for a part of the RR enhancement. Multiple factors may affect the enhancement including the diffusive properties of these cytokines both in the aqueous buffered solution and across the polymeric microdialysis membrane.
In vivo enhanced microdialysis RR of CCL2
Figure 3 shows the dialysate CCL2 concentrations detected every 30 min from both the control and the enhanced probes implanted in the peritoneal cavities of six rats given LPS. While there were rat-to-rat differences in the detected CCL2 concentrations, the general trend with each probe and each animal was the increased cytokine collection after LPS administration. Dialysate samples from the control probes in six rats not given LPS had CCL2 concentrations ranging between 90 and 1,020 pg/mL. Two and half hours after the administration of LPS, the control dialysate CCL2 concentrations ranged between 2,340 and 9,730 pg/mL. CCL2 production could be detected before LPS injection (0–30 min after probe implantation), which indicated that the immunological response (CCL2 production) took place right after the probe implantation because the surgery and the probe implantation initiated the foreign body response of the animal [37]. LPS administration afterwards accelerated the CCL2 production.
Fig. 3.
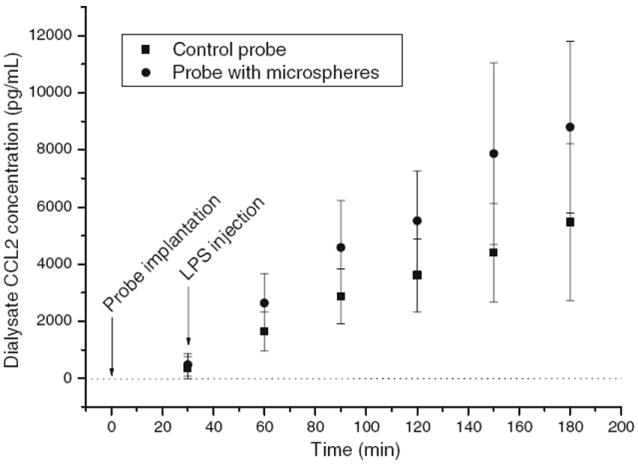
Enhanced in vivo microdialysis sampling of rat CCL2. Two identical CMA/20 10-mm PES microdialysis probes were implanted in the peritoneal cavity of a male Sprague–Dawley rat. One probe was used as a control (square) and the other was perfused with microspheres (circle). Dialysate samples were collected every 30 min continuously from six rats right after probe implantation. LPS was injected intravenously 30 min after probe implantation. Data represent mean ± SD, n=6 rats for t=30, 60, 90, 120, and 150 min; n= 5 rats for t=180 min due to the loss of one rat under anesthesia
One of the difficulties of working with endogenous cytokines and microdialysis sampling is the issue of the validation of the approach. Standard microdialysis sampling calibration procedures, such as zero net flux or approach to zero flow, require steady-state concentrations for the analyte [38]. In our experience with sampling cytokines, it is rare to achieve a steady-state concentration. Cytokine concentrations can be quite variable, and for implanted objects, the concentrations can be in the low 10- to 100-pg/mL range [39-42], which, combined with microdialysis sampling extraction efficiencies, will lead to non-quantifiable concentrations. To obtain more consistent cytokine concentrations, we chose to use LPS to induce cytokine production, as previously demonstrated in mice [43]. While this approach may produce variable cytokine concentrations, the alternative of not having measureable concentrations is even more difficult since it is hard to validate a concentration that cannot be quantified. A separate validation issue we have observed is that cytokine concentrations seem to be at or very close to the detection limits for the ELISA assays. We have unpublished experimental observations where using free heparin or antibodies in the perfusion fluid has consistently given detectable (low picograms per milliliter range) cytokine concentrations as compared to controls [44].
A two-way ANOVA with replicate measures shows that both the concentrations between the time points and the heparin-infused probes are significantly different for CCL2 collection, p<0.001 (the time points and heparin treatment). The CCL2 dialysate mean concentrations collected from heparin-infused probes were approximately twofold higher than the control dialysate concentrations. The recovery enhancement was not as high as that obtained during in vitro experiment, which was an approximately fourfold enhancement for CCL2 at a 1.0-μL/min flow rate. A possible explanation to this result is that other proteins present in vivo may also bind to heparin and thus prevent CCL2 binding to the immobilized heparin on the bead surface, thus decreasing the binding capacity of the beads to CCL2. Considering the long collection time (30 min in this study) which is required to obtain sufficient sample volume for the ELISA assay, the twofold enhancement can reduce the sampling time to 15 min. This will help monitor rapid cytokine change with better temporal resolution. It should also be noted that incomplete CCL2 dissociation from the beads (~90% recovery) and bead loss (~15% loss at 1.0-μL/min perfusion rates, as shown in Table 2, or ~7% loss at 1.0-μL/min rates with periodic 1-min ramp to 5.0 μL/min) during the sampling process may underestimate the enhanced recovery results.
Table 4 shows the CCL2 concentrations in the dialysates collected during the last 30 min before probe removal and those in the peritoneal cavity right after probe removal. There was not enough fluid within the peritoneal cavity to be collected directly for the determination of CCL2 concentration present outside the probe. A peritoneal wash with the external fluid (200 μL of PBS) was performed to get an estimate of the resident cytokine concentration within the cavity. The ratio of CCL2 concentrations in the dialysate sample and the peritoneal washed sample was used to estimate the RR during in vivo microdialysis sampling. The calculated control and enhanced RRs were approximately 12.0±3.4% and 20.7±2.6% (n=6 rats), respectively. However, it should be noted that the peritoneal wash may dilute the overall CCL2 concentration external to the probes. This indicates that the actual RR values should be lower than the estimated RRs calculated above.
In vitro calibration of microdialysis probes
In vitro calibrations were performed on the explanted probes with the results shown in Table 5. The RRs are quite similar, and there is no significant difference (p<0.05) between the two probes of each pair used for the in vivo experiment. Compared to the calculated percentage recoveries for each control probe from Table 4 (9.6%, 12.8%, 17.6%, 12.0%, 12.7%, and 7.4% for rats 1 to 6, respectively) during the in vivo experiment, the in vitro RRs are lower, indicating the dilution of CCL2 concentration external to the probes due to peritoneal wash. Additionally, the in vitro RR values in Table 5 are higher than the control RRs reported in Table 3. We have previously observed batch-to-batch differences with probe recovery from CMA/Microdialysis PES probe lots.
Table 5.
In vitro calibration of explanted microdialysis probes
| Rat 1 | Rat 2 | Rat 3 | Rat 4 | Rat 5 | Rat 6 | ||
|---|---|---|---|---|---|---|---|
| RR (%) | Probe 1 | 8.6±0.6 | 10.3±1.5 | 11.2±1.2 | 9.5±0.9 | 9.2±1.5 | 7.5±0.8 |
| Probe 2 | 9.0±0.8 | 9.9±1.1 | 10.8±1.5 | 10.2±1.0 | 9.8±0.9 | 7.2±0.8 |
Explanted probes were immersed in a solution containing the same CCL2 concentration as measured in the peritoneal washed solution for each rat. The flow rate was 1.0 μL/min. Probes 1 and 2 corresponded to the control and the enhanced probes, respectively, used for in vivo experiment. RR% was calculated by the recovered CCL2 concentration divided by the CCL2 concentration in the sample medium. Data represent mean ± SD, n=3 measurements for three dialysate samples collected using each probe
Comparison of free heparin and heparin-immobilized microspheres for enhanced microdialysis RR of CCL2 in vivo
The effect of free heparin and the heparin-immobilized microspheres on CCL2 RR enhancement was compared. Table 6 shows the CCL2 concentrations in the dialysates collected every 30 min after probe implantation and those in the peritoneal cavity right after probe removal. Generally, dialysate CCL2 concentrations collected from the probes perfused with free heparin or the heparin-immobilized microspheres were higher than those from the control probe during each 30-min time period for each rat. The calculated percentage recoveries during the last 30 min for the probes perfused with heparin and the microspheres, 15.3±2.9% and 16.5±1.5%, respectively, were significantly higher than the recovery for the control probe, 10.2±2.5% (p<0.05, n=3 rats). No significant recovery difference was observed between the probe perfused with free heparin and the probe perfused with the heparin-immobilized microspheres, indicating that the same concentration of heparin exhibited the same effect on CCL2 recovery enhancement whether the heparin is dissolved in the perfusion fluid or is immobilized on the bead surface.
Table 6.
Comparison of CCL2 concentrations collected from the control probe, probes perfused with free heparin and the heparin-immobilized microspheres, and the peritoneal washed samples
| Time (min)
|
Peritoneal wash | |||||||
|---|---|---|---|---|---|---|---|---|
| 30a | 60 | 90 | 120 | 150 | 180 | |||
| Rat 1 (pg/mL) | Control | N.D. | 380 | 620 | 850 | 1,400 | 2,340 | 29,060 |
| Heparin | N.D. | 840 | 1,430 | 2,320 | 3,700 | 4,350 | ||
| Beads | 90 | 910 | 1,870 | 2,860 | 4,290 | 4,540 | ||
| Rat 2 (pg/mL) | Control | N.D. | 320 | 610 | 1,390 | 2,350 | 3,160 | 32,860 |
| Heparin | 140 | 1,050 | 1,920 | 2,900 | 4,200 | 4,130 | ||
| Beads | 170 | 710 | 1,680 | 3,060 | 4,270 | 5,150 | ||
| Rat 3 (pg/mL) | Control | N.D. | 320 | 1,560 | 3,870 | 7,400 | 9,380 | 72,160 |
| Heparin | 120 | 760 | 2,850 | 7,270 | 12,160 | 13,260 | ||
| Beads | 280 | 2,030 | 4,960 | 8,900 | 13,010 | 13,150 | ||
N.D. not detectable
LPS was injected intravenously after the first 30-min sample collection
Even though no significant difference in dialysate cytokine concentrations was observed between free heparin and heparin-immobilized microspheres infused probes, there are still advantages to using the heparin-immobilized microsphere perfusion over free heparin. Heparin can diffuse through the microdialysis pores and has a loss of about 1.4% from the CMA/20 10-mm PES probes when 1 μM of heparin is perfused at 0.5 μL/min [45]. Such a loss could potentially lead to the displacement of cytokines or other proteins from extracellular matrix glycosaminoglycans [46], which would further complicate the analysis and data interpretation.
Conclusions
Affinity-based microdialysis sampling of cytokines using in-house prepared heparin-immobilized microspheres in the perfusion fluid has been demonstrated. Microdialysis RRs of four cytokines—aFGF, VEGF, CCL2, and CCL5—were significantly enhanced in vitro using the heparin-immobilized microspheres as affinity agents. The dialysates could be quantified either by the bead-based flow cytometric assay or by standard ELISA after the dissociation of cytokines from the microspheres using an appropriate dissociation reagent. This approach was also applied in vivo for enhanced RR of CCL2 from the rat peritoneal cavity. This work suggests that the heparin-immobilized microspheres provide an alternative affinity agent to the previously used antibody-immobilized microspheres for enhanced microdialysis sampling of cytokines.
Acknowledgments
We thank NIH EB 001441 for the funding support of this work. Part of this work was also supported by the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. Professor Robert J. Linhardt, Rensselaer Polytechnic Institute, is gratefully acknowledged for the discussion of heparin/cytokine interactions. We also thank Dr. Fuming Zhang (Rensselaer Polytechnic Institute) for the assistance with heparin immobilization chemistry and Steven Lotz and Yili Lin (Albany Medical College) for the assistance with flow cytometric analysis.
Contributor Information
Jia Duo, Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Julie A. Stenken, Department of Chemistry and Biochemistry, University of Arkansas, Fayetteville, AR 72701, USA, jstenken@uark.edu
References
- 1.Fitzgerald KA, O’Neill LAJ, Gearing AJH, Callard RE. The cytokine factsbook. 2. Academic; New York: 2001. [Google Scholar]
- 2.Mizgerd JP, Spieker MR, Doerschuk CM. J Immunol. 2001;166:4042–4048. doi: 10.4049/jimmunol.166.6.4042. [DOI] [PubMed] [Google Scholar]
- 3.Gouwy M, Struyf S, Proost P, van Damme J. Cytokine Growth Factor Rev. 2005;16:561–580. doi: 10.1016/j.cytogfr.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello CA. Chest. 2000;113:503–508. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 5.Westerink BH, Cremers TIFH. Handbook of microdialysis: methods, applications and perspectives. 1. Elsevier Academic; Amsterdam: 2007. [Google Scholar]
- 6.Watson CJ, Venton BJ, Kennedy RT. Anal Chem. 2006;78:1391–1399. doi: 10.1021/ac0693722. [DOI] [PubMed] [Google Scholar]
- 7.Jin G, Cheng Q, Feng J, Li F. J Chromatogr Sci. 2008;46:276–287. doi: 10.1093/chromsci/46.3.276. [DOI] [PubMed] [Google Scholar]
- 8.van der Zeyden M, Oldenziel WH, Rea K, Cremers TI, Westerink BH. Pharmacol Biochem Behav. 2008;90:135–147. doi: 10.1016/j.pbb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Heal DJ, Smith SL, Kulkarni RS, Rowley HL. Pharmacol Biochem Behav. 2008;90:184–197. doi: 10.1016/j.pbb.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Wotjak CT, Landgraf R, Engelmann M. Pharmacol Biochem Behav. 2008;90:125–134. doi: 10.1016/j.pbb.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Garrison KE, Pasas SA, Cooper JD, Davies ML. Eur J Pharm Sci. 2002;17:1–12. doi: 10.1016/s0928-0987(02)00149-5. [DOI] [PubMed] [Google Scholar]
- 12.Bungay PM, Morrison PF, Dedrick RL. Life Sci. 1990;46:105–119. doi: 10.1016/0024-3205(90)90043-q. [DOI] [PubMed] [Google Scholar]
- 13.Trickler WJ, Miller DW. J Pharm Sci. 2003;92:1419–1427. doi: 10.1002/jps.10410. [DOI] [PubMed] [Google Scholar]
- 14.Kjellström S, Appels N, Ohlrogge M, Laurell T, Marko-Varga G. Chromatographia. 1999;50:539–546. [Google Scholar]
- 15.Dostálová I, Pacák K, Nedvidková J. Int J Biol Macromol. 2003;32:205–208. doi: 10.1016/s0141-8130(03)00055-2. [DOI] [PubMed] [Google Scholar]
- 16.Helmy A, Carpenter KLH, Skepper JN, Kirkpatrick PJ, Pickard JD, Hutchinson PJ. J Neurotrauma. 2009;26:549–561. doi: 10.1089/neu.2008.0719. [DOI] [PubMed] [Google Scholar]
- 17.Murdolo G, Herder C, Wang Z, Rose B, Schmelz M, Jansson PA. Am J Physiol. 2008;295:1095–1105. doi: 10.1152/ajpendo.90483.2008. [DOI] [PubMed] [Google Scholar]
- 18.Ao X, Sellati TJ, Stenken JA. Anal Chem. 2004;76:3777–3784. doi: 10.1021/ac035536s. [DOI] [PubMed] [Google Scholar]
- 19.Ao X, Stenken JA. Methods. 2006;38:331–341. doi: 10.1016/j.ymeth.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Gesslbauer B, Rek A, Falsone F, Rajkovic E, Kungl AJ. Proteomics. 2007;7:2870–2880. doi: 10.1002/pmic.200700176. [DOI] [PubMed] [Google Scholar]
- 21.Capila I, Linhardt RJ. Angew Chem Int Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 22.Lau EK, Paavola CD, Johnson Z, Gaudry J, Geretti E, Borlat F, Kungl AJ, Proudfoot AE, Handel TM. J Biol Chem. 2004;279:22294–22305. doi: 10.1074/jbc.M311224200. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs RV. Adv Exp Med Biol. 2003;535:125–143. doi: 10.1007/978-1-4615-0065-0_9. [DOI] [PubMed] [Google Scholar]
- 24.Mulloy B, Rider CC. Biochem Soc Trans. 2006;34:409–413. doi: 10.1042/BST0340409. [DOI] [PubMed] [Google Scholar]
- 25.Handel TM, Johnson Z, Crown SE, Lau EK, Sweeney M, Proudfoot AE. Annu Rev Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 26.DiGiacomo RA, Xie L, Cullen C, Indelicato SR. Anal Biochem. 2004;327:165–175. doi: 10.1016/j.ab.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Quinn J, Martin A, Havard J. ICx Nomadics Inc; Oklahoma City, OK: 2010. [15 Feb 2010]. http://www.discoversensiq.com/uploads/file/applications/Direct_Immobilization_of_Antigen_for_Kinetic_Analysis.pdf. [Google Scholar]
- 28.Mach H, Volkin DB, Burke CJ, Middaugh CR, Linhardt RJ, Fromm JR, Loganathan D, Mattsson L. Biochemistry. 1993;32:5480–5489. doi: 10.1021/bi00071a026. [DOI] [PubMed] [Google Scholar]
- 29.Amara A, Lorthioir O, Valenzuela A, Magerus A, Thelen M, Montes M, Virelizier J, Delepierre M, Baleux F, Lortat-Jacob H, Arenzana-Seisdedos F. J Biol Chem. 1999;274:23916–23925. doi: 10.1074/jbc.274.34.23916. [DOI] [PubMed] [Google Scholar]
- 30.Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, Vita C. Biochemistry. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
- 31.Duo J, Stenken JA. Anal Bioanal Chem. 2010 doi: 10.1007/s00216-010-4170-1. [DOI] [Google Scholar]
- 32.Duo J, Fletcher H, Stenken JA. Biosens Bioelectron. 2006;22:449–457. doi: 10.1016/j.bios.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Kuschert GSV, Coulin F, Power CA, Proudfoot AEI, Hubbard RE, Hoogewerf AJ, Wells TNC. Biochemistry. 1999;38:12959–12968. doi: 10.1021/bi990711d. [DOI] [PubMed] [Google Scholar]
- 34.Kuschert GSV, Hoogewerf AJ, Proudfoot AEI, Chung C, Cooke RM, Hubbard RE, Wells TNC, Sanderson PN. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 35.Duo J, Espinal RF, Stenken JA. 2009 IEEE/NIH Life Science Systems and Applications Workshop (LiSSA 2009) 2009. pp. 112–115. [Google Scholar]
- 36.Gupta K, Gupta P, Wild R, Ramakrishnan S, Hebbel RP. Angiogenesis. 1999;3:147–158. doi: 10.1023/a:1009018702832. [DOI] [PubMed] [Google Scholar]
- 37.Gupta K, Gupta P, Wild R, Ramakrishnan S, Hebbel RP. Anal Chem. 2007;79:1816–1824. [Google Scholar]
- 38.Stenken JA. Anal Chim Acta. 1999;379:337–358. [Google Scholar]
- 39.Wang X, Lennartz MR, Loegering DJ, Stenken JA. Cytokine. 2008;43:15–19. doi: 10.1016/j.cyto.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldwin L, Hunt JA. Cytokine. 2008;41:217–222. doi: 10.1016/j.cyto.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Kalltorp M, Oblogina S, Jacobsson S, Karlsson A, Tengvall P, Thomsen P. Biomaterials. 1999;20:2123–2137. doi: 10.1016/s0142-9612(99)00115-5. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez A, Meyerson H, Anderson JM. J Biomed Mater Res A. 2009;89A:152–159. doi: 10.1002/jbm.a.31939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ao X, Rotundo RF, Loegering DJ, Stenken JA. J Microbiol Meth. 2005;62:327–336. doi: 10.1016/j.mimet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y. PhD Dissertation, Rensselaer Polytechnic Institute. 2009. G-quadruplex DNA and their binding proteins. [Google Scholar]
- 45.Wang Y, Stenken JA. Anal Chim Acta. 2009;651:105–111. doi: 10.1016/j.aca.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorne RG, Lakkaraju A, Rodriguez-Boulan E, Nicholson C. Proc Natl Acad Sci USA. 2008;105:8416–8421. doi: 10.1073/pnas.0711345105. [DOI] [PMC free article] [PubMed] [Google Scholar]


