Abstract
Background
African Americans suffer from higher prevalence and severity of atherosclerosis compared to Whites, highlighting racial and ethnic disparities in cardiovascular disease. Previous studies have pointed to the role of vascular inflammation and platelet activation in the formation of atherosclerotic lesions.
Methods and Results
We explored the role of genetic variation in four chemokine/chemokine receptor genes (CX3CR1, CX3CL1, CXCR3 and PF4) on systemic inflammation and platelet activation serum biomarkers (fractalkine, platelet P-selectin, PF4 and TNFα). In total, 110 SNPs were tested among 1,042 African Americans and 763 Whites. The strongest association with serum PF4 levels was observed for rs168449, which was significant in both racial groups (P-value: African Americans=0.0017, Whites=0.014, Combined=1.2×10−4), and remained significant after permutation-based multiple corrections (Pc-value: Combined=0.0013). After accounting for the effect of rs168449, we identified another significant SNP (rs1435520) suggesting a second independent signal regulating serum PF4 levels (conditional P-value: African Americans=0.02, Whites=0.02). Together these SNPs explained 0.98% and 1.23% of serum PF4 variance in African Americans and Whites, respectively. Additionally, in African Americans, we found an additional PF4 variant (rs8180167), uncorrelated with rs168449 and rs1435520, associated with serum TNFα levels (P-value=0.008, Pc-value=0.048).
Conclusions
Our study highlight the importance of PF4 variants in the regulation of platelet activation (PF4) and systemic inflammation (TNFα) serum biomarkers.
Keywords: association study, atherosclerosis, inflammation, platelets, Chemokines, PF4, TNF-alpha
Introduction
Over the past decade, epidemiological studies comparing African Americans and other minority groups to Whites have demonstrated persistent disparities in morbidity and mortality.1, 2 In particular, a disproportionately high prevalence and severity of atherosclerotic cardiovascular disease in African Americans is observed. The reasons for this health disparity are multiple and largely unresolved.3 Atherosclerosis is a lifelong progressive disease, which leads to myocardial infarction (MI), stroke, and higher mortality rates.4 This disease was formerly characterized by the accumulation of lipids and fibrous elements in large arteries; but in recent years, substantial advances in basic and experimental science have illuminated the importance of vascular inflammation5, 6 and platelet activation in disease etiology.7 However, the underlying cellular and molecular mechanisms that contribute to atherogenesis are not fully known. It has been suggested that continuous immigration and infiltration of activated macrophages and T cells into and within atherosclerotic lesions are critical events during the initial phase of plaque formation, 8 progression, and even in acute complications such as plaque rupture and thrombosis.9 The recruitment of these cells to lesions is largely guided by chemokines, and contributes to the accumulation of inflammatory cells in the atherosclerotic plaque.10
Chemokines constitute a family of structurally related chemotactic cytokines and classified into subgroups (CC, CXC, C, CX3C) based on the position of the conserved cysteine residues in the amino-terminal region of the molecule. In addition to the regulation of trafficking of macrophages and T cells, chemokines are also involved in platelet activation.11, 12 Fractalkine (CX3CL1) is not produced by endothelial cells under physiologic conditions in vivo, however its production is induced via inflammatory signals (i.e. TNFα), vascular injury, or atherosclerosis. Monocytes and platelets are known to interact directly with membrane-bound CX3CL1, and fractalkine receptor (CX3CR1) and CXCR3 are well documented in the recruitment of inflammatory cells. Previous studies have demonstrated the presence of a functional CX3CR1 on human platelets.5 Blood platelets are mainly involved in hemostasis and acute thrombus formation, but also have pro-inflammatory and growth-regulatory properties that contribute to the progression of atherosclerosis.13, 14 The activation of platelets leads to the release of a wide range of growth factors, inflammatory mediators (including chemokines, e.g., platelet factor 4), and adhesion molecules (e.g., P-selectin). Previous studies have suggested that platelet activation also induces the expression of pro-inflammatory TNFα and chemokines in monocytes/macrophages.15 Compelling evidence from functional16–18 and genetic studies19, 20 also affirms a link between chemokines and the progression of atherosclerosis. Prior studies have shown that activated platelets can stimulate endothelial cells, leading to an increase in leukocyte rolling along the vessel wall.21 However, the mechanisms by which platelets activate endothelial cells and vascular inflammation is not well understood. Platelet factor 4 (PF4, or CXCL4) is a chemokine stored in platelet alpha-granules and released during platelet activation.10, 22–24. Our prior work demonstrated PF4 role in T cell trafficking and development of experimental cerebral malaria.25
Given the importance of systemic inflammation and platelet activation to the development of atherosclerosis, we explored the role of genetic variants in regulating these processes. Indeed, these two processes yield predictive and prognostic information of considerable clinical utility. Based on our current understanding, we assume a model in which inflammatory mediators can lead to platelet activation, which in turn intensify the inflammatory response, thus forming an atherogenic amplification loop. The key inflammatory mediators on which we focused are chemokines and chemokine receptors; fractalkine receptor (CX3CR1), fractalkine (CX3CL1), chemokine (C-X-C motif) receptor 3 (CXCR3) which serves as a receptor for PF4, and platelet factor (PF4). The systemic inflammation and platelet activation serum biomarkers; fractalkine, platelet-selectin (P-selectin), platelet factor 4 (PF4) and tumor necrosis factor-alpha (TNFα) were assessed as vascular risk factors for atherosclerosis. The overall goal of this study is to explore the influence of genetic variations in CX3CR1 and CXCR3 and determine if they are asymmetrically distributed in African Americans compared to Whites, or if they modulate in the chemokine/chemokine receptors on the systemic inflammation and platelet activation serum biomarkers in African Americans and Whites.
Materials and Methods
Study Cohort
Study samples were drawn from participants of Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS) study of the National Institute on Aging Intramural Research Program.26 HANDLS is a prospective longitudinal study of approximately 4,000 African American and White adults from Baltimore City, Maryland. The purpose of the HANDLS study is to unravel the effects of race and socioeconomic status (SES) on the development of age-associated health disparities over a 20-year period. Subjects were enrolled from 2004 to 2008 by household screenings from an area probability sample of 13 neighborhoods defined by contiguous US Census tracts in the city of Baltimore. These tracts were selected because they were likely to yield representative distributions of individuals who were 30 – 64 yrs old, African Americans or Whites, males and females, and had a household income either < 125% or >= 125% of the federal poverty level. Multi-ethnic individuals were included provided they identified themselves as African Americans or Whites, but not both. To be included in the HANDLS study, participants must (1) be within age range of 30 – 64 yrs at baseline, (2) be able to give informed consent, (3) be able to perform at least 5 measures of the following evaluations: laboratory evaluation, medical history, physical examination, physical performance, cognitive testing, dietary recall, audio questionnaire, body composition, carotid Doppler, or pulse wave velocity assessment, (4) have valid picture identification, and (5) have a verifiable address at the time of entry. Exclusions included participants who were pregnant at the time of entry, had a diagnosis of AIDS, and were within 6 months of active treatment of cancer (chemotherapy, biologic, or radiation). Details of the study design are given elsewhere.26 Briefly, the data used herein consists of 1,920 community-dwelling African American and White Baltimore City residents, who were enrolled after providing written informed consent and had available serum biomarker data. The MedStar Institutional Review Board approved this protocol.
Assessment of serum biomarkers
Measurements of the serum biomarkers were evaluated on cryopreserved (−80 C°) serum aliquots. The serum samples were divided into four aliquots and each aliquot was used for the assessment of one of the four biomarkers (fractalkine, P-selectin, PF4 and TNFα). Both serum biomarker data and DNA genotypes were available for 1,920 HANDLS participants (Whites, n=828; African Americans, n=1092). Commercial MSD kits (Meso Scale Discovery, Gaithersburg, Maryland 20877) were used for the measurement of serum fractalkine (Customized Fractalkine single-Plex assay kit, Cat # N45ZA-1), PF4 (Customized PF4 Single-Plex assay kit, Cat # N45ZA-1), P-selectin (Cataloged P-Selectin Single-Plex assay kit, Cat # N451ENB-1) and TNFα (Cataloged Human ProInflammatory 9-Plex Assay Ultra-Sensitive Kit, Cat # K15007C-4) levels. Dilution factors were determined using naive serum samples before the sample testing and measurements were made according to manufacturer’s instructions. All the serum assessments were performed in duplicate.
SNP Selection, Genotyping and Quality Control
For each gene, we selected SNPs using two criteria. First, all the known missense and previously associated SNPs were selected for genotyping, and then additional tag SNPs27 were selected from HapMap phase I, II & III, using YRI and CEU datasets. All the tag SNPs were captured from the 20 kb flanking 5′ and 3′ regions of the CX3CL1, CX3CR1, CXCR3 and PF4 genes with minor allele frequency (MAF) >1% and r2 ≥0.8. Given the proximity of nearby genes to our genes of interest, some of the flanking SNPs lie in nearby genes. A total of 137 tag SNPs from the 4 genes (CX3CL1, n=30; CX3CR1, n=68; CXCR3, n=9; and PF4, n=30) were selected and grouped in 4 separate pools for genotyping using iPLEX Gold single base-pair extension with MALDI-TOF mass spectrometry (Sequenom MassArray). Quality control (QC) for both samples and SNPs was performed separately for each Sequenom genotyping pool, and the following inclusion/exclusion criteria were applied: SNPs with >5% missingness (23 SNPs) and monomorphic SNPs (4 SNPs) were excluded, and after removal of these SNPs, samples with >50% missingness (reflecting poor quality DNA and/or poor genotyping) were removed. A sensitivity analysis using a stringent sample missingness exclusion threshold of >5% demonstrated no meaningful difference in results (data not shown). After QC, 110/137 SNPs were analyzed in 1,805 (94%) individuals (Whites, n=763; African Americans, n=1042). The allele frequency comparison of the directly genotyped 110 SNPs with the HapMap (phase I, II & III) CEU and ASW population, confirmed the quality of the genotyped data and all the SNPs are in Hardy-Weinberg equilibrium.
Statistics
Before assessing genetic associations, serum biomarker levels in the studied samples were evaluated for normality and appropriate transformation was applied on each biomarker (fractalkine - log transformed, P-selectin - square root transformed, PF4 - log transformed, TNFα-inverse log transformed). Association analysis and quality control measures were performed using PLINK software package28 version 1.06 (http://pngu.mgh.harvard.edu/purcell/plink/). To assess the influence of genetic variation in the chemokine/chemokine receptors on the systemic inflammation and platelet activation serum biomarkers, all analyses were stratified by race and adjusted for age and sex in a multivariate linear regression, assuming an additive genetic model of inheritance. To create a more comprehensive fine map of the locus, imputation was performed using Hidden Markov model as implemented in the MaCH software29(version 1.0.16) (http://www.sph.umich.edu/csg/abecasis/MACH/). The CEU and CEU-YRI combined panel, from the 1000 Genomes Project,30 were used as reference populations for Whites and African American, respectively. Imputation was performed in two steps and QC was performed both before and after imputation. In step 1, model parameters were estimated using 100 iterations and, thereafter, allele dosage and maximum likelihood genotypes were imputed in step 2. To account for the uncertainty of the imputed data, the estimated allele dosage for each SNP was analyzed using ProABEL31 under a linear regression framework. Standard quality metrics were applied and only SNPs with high quality score (RSq > 0.8) were analyzed. Linkage disequilibrium (LD) patterns within the surrounding region of the significant SNPs were constructed using the solid spine method, as implemented in Haploview32 (version 4.1) (http://www.broad.mit.edu/mpg/haploview/index.php). For each defined gene region, permutation-based multiple test correction was performed using 5000 permutations. To determine independent genetic effects, multivariate conditional regression analyses were performed using allele dosage data. To identify known functional regulatory variants within or in proximity to the region of interest, the GTEx (Genotype-Tissue Expression) expression quantitative trait loci (eQTL) database was queried (http://www.ncbi.nlm.nih.gov/gtex/test/GTEX2/gtex.cgi).33 Further, a long-range haplotype test was utilized to detect recent selection and the inferences were made using integrated haplotype score (iHS), as described elsewhere.34
Results
Detailed demographic and clinical characteristics of the studied 1,805 samples are shown in Table 1 and Supplementary Table 1. The average age of the studied cohort was 48 yrs, with ~45% males.
Table 1.
Demographic and clinical characteristics
| Characteristics | African Americans | Whites | P-value* |
|---|---|---|---|
| Samples, n (%) | 1042 (57.7) | 763 (42.3) | |
| Sex, n (%) | |||
| Males | 472 (45.3) | 341 (44.7) | 0.79 |
| Age (in years), mean ± SD | 48.18 ± 8.9 | 48.39 ± 9.3 | 0.51 |
| Poverty Status¶, n (%) | |||
| Below 125% | 518 (49.7) | 261 (34.2) | <0.001 |
| Serum Biomarkers, mean ± SD | |||
| Fractalkine (pg/mL) | 895.7 ± 438.8 | 835.9 ± 334.7 | < 0.001 |
| P-selectin (ng/mL) | 104.1 ± 34.4 | 108.7 ± 37.1 | 0.018 |
| Platelet factor 4 (ng/mL) | 18098 ± 10796 | 20285 ± 19480 | < 0.001 |
| TNFα† (pg/mL) | 14.1 ± 22.4 | 13.98 ± 23.1 | 0.002 |
| Diabetes, n (%) | 138 (13.2) | 98 (12.8) | 0.58 |
| Hypertension, n (%) | 406 (38.9) | 227 (29.7) | <0.001 |
| Cigarette smoking, n (%) | 0.54 | ||
| Never smoked | 195 (18.7) | 165 (21.6) | |
| Smoked, never regularly | 91 (8.7) | 65 (8.5) | |
| Former smoker | 186 (17.8) | 143 (18.7) | |
| Current smoker | 452 (43.4) | 319 (41.8) | |
| Not known | 118 (11.3) | 71 (9.3) | |
| Total Cholesterol | 185.6 ± 44.4 | 191.2 ± 42.5 | 0.005 |
| LDL‡ (mg/dL) | 108.1 ± 38.6 | 112.9 ± 36.7 | 0.004 |
| HDL§ (mg/dL) | 55.94 ± 18.6 | 48.35 ± 14.1 | <0.001 |
P-values for continuous and categorical variable comparisons were generated using wilcoxon rank sum and pearson chi-square test, respectively.
Poverty status was determined whether a participant reported an annual household income <125% or >125% of the 2004 Department of Health and Human Services poverty guideline (http://aspe.hhs.gov/poverty/04poverty.shtml).
Tumor necrosis factor-alpha
Low-density lipoprotein
High-density lipoprotein
Association of PF4 genetic variation with serum PF4 levels
Initially, 24 SNPs chosen to tag the PF4 gene (see Methods) were tested for association with PF4 serum levels using linear regression models, adjusting for age and sex. In African Americans 3 SNPs were nominally associated with PF4 levels (P-value <0.05), and in Whites, 8 SNPs were nominally associated, with 3 SNPs in common, all showing the same direction of effect in both populations, suggesting the presence of a shared genetic association in African Americans and Whites (Table 2). In a joint analysis, adjusting for age, sex, and race, 6 of the SNPs were associated with PF4 levels, with the most significant effect observed for rs183028 (P-value: 2.7×10−4) (Table 2). The ancestral allele (A) of rs183028 is the minor allele and is significantly associated with higher serum PF4 levels in both racial groups (African American: β=0.06, SE=0.02; Whites: β=0.09, SE=0.04). The frequency of this allele (A) in Whites was 9%, compared to 41% in African Americans. Given the large allele frequency difference, we explored the possibility of population-specific selection pressure at this locus. To detect signal for recent positive selection, we used iHS statistics,34 based on differential levels of linkage disequilibrium (LD) surrounding a positively selected allele compared to the background allele at the same position, and no evidence of selection was observed (data not shown).
Table 2.
PF4 region SNPs and genetic association with serum PF4 levels
| SNP* | Position | Coded/Noncoded Allele | African Americans
|
Whites
|
Combined
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coded Allele Freq | β† | SE | P | Coded Allele Freq | β† | SE | P | β‡ | SE | P | |||
| rs1429637 | 75,052,310 | T/C | 0.036 | −0.11 | 0.05 | 0.02 | 0.060 | −0.12 | 0.05 | 0.01 | −0.12 | 0.03 | 6.3×10−4 |
| rs351997 | 75,052,923 | T/C | 0.087 | 0.004 | 0.03 | 0.91 | 0.090 | 0.09 | 0.04 | 0.02 | 0.04 | 0.02 | 0.15 |
| rs6813952 | 75,063,296 | C/A | 0.073 | −0.04 | 0.04 | 0.23 | 0.059 | 0.12 | 0.05 | 0.01 | 0.07 | 0.03 | 0.01 |
| rs1435520 | 75,064,281 | C/A | 0.019 | −0.16 | 0.07 | 0.02 | 0.053 | 0.13 | 0.05 | 0.009 | 0.14 | 0.04 | 3.9×10−4 |
| rs442155 | 75,070,096 | G/A | 0.059 | 0.07 | 0.04 | 0.08 | 0.084 | 0.10 | 0.04 | 0.02 | 0.08 | 0.03 | 0.0035 |
| rs183028 | 75,070,782 | A/G | 0.411 | 0.06 | 0.02 | 0.003 | 0.092 | 0.09 | 0.04 | 0.03 | 0.06 | 0.02 | 2.7×10−4 |
| rs409336 | 75,076,522 | C/A | 0.537 | 0.02 | 0.02 | 0.18 | 0.104 | 0.09 | 0.04 | 0.01 | 0.04 | 0.02 | 0.02 |
| rs2437285 | 75,079,102 | T/C | 0.410 | 0.01 | 0.02 | 0.54 | 0.098 | 0.09 | 0.04 | 0.01 | 0.03 | 0.02 | 0.09 |
Positions are in reference to NCBI build 36.3
Only the SNPs which are nominally associated in atleast one racial group are shown in the table
Effect size of the coded allele based on the multivariate linear regression adjusted for age and sex
Effect size of the coded allele based on the multivariate linear regression adjusted for age, sex and race
Most significant SNPs and the P-values for their association are highlighted in bold
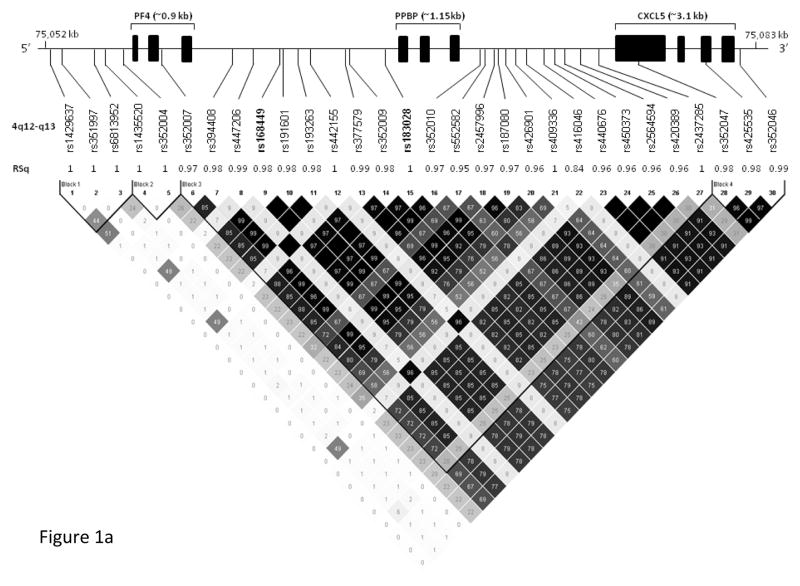
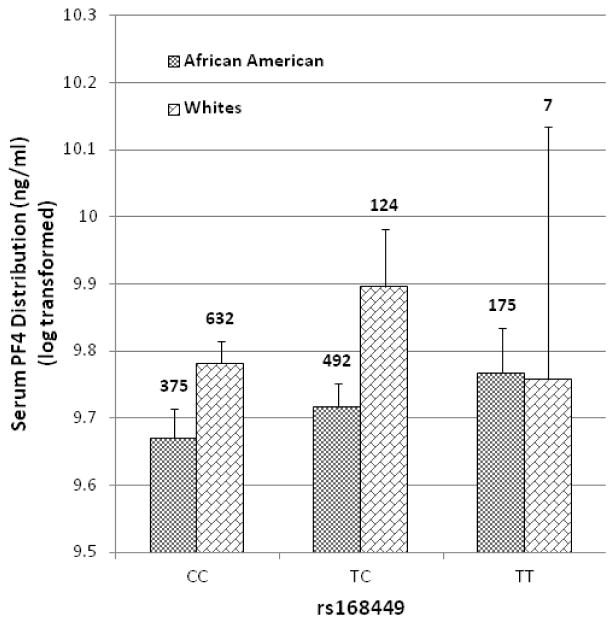
To fine map the observed genetic effect and potentially identify additional independent effects, we imputed ungenotyped SNPs over a 30-kb interval centered on rs183028 using the 1000 Genomes reference panels.30 In total, 66 SNPs were imputed, and after removing poorly imputed SNPs (RSq < 0.8), 44 SNPs were analyzed. The strongest evidence for association was observed in a region of high LD, encompassing the PF4, PPBP (pro-platelet basic protein) and CXCL5 (C-X-C motif ligand 5) genes (Figure 1a, 1b). In this region, 30 SNPs showed nominal association (P-value < 0.05) with PF4 serum levels (Table 3). In African Americans, the most significant association was observed for rs168449 (P-value = 0.0017), where the T allele (frequency 41%) was associated with higher PF4 serum levels (β= 0.05, SE=0.02). This SNP is ~1.3 kb upstream of rs183028, with which it is highly correlated in both racial groups (African Americans: r2=0.97; Whites: r2=1.0) (Figure 1a, 1b). The association of rs168449 with serum PF4 levels remained significant (Pc-value = 0.02) even after gene-wide permutation-based multiple correction (Table 3). Similarly, in Whites, this SNP showed nominal significance (P-value = 0.014) with the same direction effect (T allele, β= 0.09, SE=0.04) (Table 3). The serum PF4 variance explained by rs168449 is 0.58% and 0.64% in African Americans and Whites, respectively. In a joint analysis of both racial groups, rs168449 was the most significant SNP (P-value = 1.2×10−4, Pc-value = 0.0013) from this region, (Table 3), and the distributions of the serum PF4 levels within each genotype group of rs168449 are shown in Figure 2. Individuals who were homozygous for the T allele have higher serum levels compared to those homozygous for the major allele (C) (Figure 2). Given the small number of individuals in the T allele homozygous group, we also tested the association under a dominant genetic model. In both racial groups rs168449 maintains significance assuming a dominant model of inheritance (P-value: African Americans=0.02, Whites= 0.009, Combined=0.001). To test for genetic effects independent of rs168449, we performed conditional multivariate regression analysis. After accounting for the effect of rs168449, rs1429637 and rs1435520 still showed significant association (Table 3). Notably, these SNPs are not in LD with rs168449 (r2=0) in both racial groups, but are in strong LD with each other (r2=0.88 and 0.51 in Whites and African Americans, respectively); thus likely representing the same genetic signal (Figure 1a, 1b). These two SNPs are directly genotyped and rs1435520 showed greater significance in both African Americans (conditional P-value: 0.022) and Whites (conditional P-value: 0.02). Given the small number of homozygous rare allele individuals observed for rs1435520, we also tested genetic association of serum PF4 levels under a dominant genetic model, combining the rare homozygotes and heterozygous individuals (Supplementary Table 2). As seen with the additive model, both SNPs, rs1429637 (P-value: African Americans=0.06; Whites=0.011 and Combined=0.0016) and rs1435520 (P-value: African Americans=0.045; Whites=0.0084 and Combined=0.0009), maintain a significant association. In both racial groups, rs1435520 explains more PF4 variability than rs1429637, explaining 0.46% and 0.77% of the variance in African Americans and Whites, respectively. Since, rs168449 is an imputed SNP, we also performed conditional multivariate analysis with a directly genotyped SNP which is in complete or high correlation with rs168449, rs183028 (African Americans: r2=0.97; Whites: r2=1.0). As expected, we only observed significant associations with rs1429637 and rs1435520, confirming the presence of two independent loci (rs168449 and rs1435520) regulating serum PF4 levels (Supplementary Table 3). Combined, these two loci explain 0.98% (African Americans) and 1.23% (Whites) of the variability in serum PF4 levels.
Figure 1.
Schematic representation and LD pattern of the 4q12-q31 region SNPs. The upper panel shows the location of the PF4, PPBP and CXCL5 genes and neighboring significant SNPs (n=30). In the middle panel, imputation quality of these SNPs is shown as RSq (squared correlation between imputed and true genotypes) and directly genotyped SNPs are marked as 1. (1a) The bottom panel shows LD pattern of these SNPs in African Americans, constructed using Haploview (version 4.1) software (http://www.broad.mit.edu/mpg/haploview/index.php). Numbers within diamonds and degree of shading represents the magnitude of the pair-wise LD (measured as r2, black to white gradient reflecting higher to lower LD values). The observed most significant SNP with serum PF4 levels is highlighted in bold. For Whites, the pair-wise correlation of these SNPs is shown in Figure 1b.
Table 3.
Association and conditional analysis of the genotyped and imputed SNPs from the PF4 region
| SNPs | Position | Coded/Noncoded Allele | African Americans | Whites | Combined | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||
| Coded Allele Freq | Rsq* | β† | SE | P | Pc§ | rs168449 Conditional P¶ | Coded Allele Freq | Rsq* | β† | SE | P | Pc§ | rs168449 Conditional P¶ | β ‡ | SE | P | Pc§ | rs168449 Conditional P¶ | |||
| rs1429637 | 75,052,310 | T/C | 0.036 | 1 | −0.11 | 0.05 | 0.022 | 0.18 | 0.030 | 0.060 | 1 | −0.12 | 0.04 | 0.011 | 0.06 | 0.02 | −0.12 | 0.03 | 0.0006 | 0.005 | 0.0014 |
| rs351997 | 75,052,923 | T/C | 0.087 | 1 | 0.003 | 0.03 | 0.91 | 0.99 | 0.653 | 0.090 | 1 | 0.09 | 0.04 | 0.015 | 0.08 | 0.58 | 0.04 | 0.02 | 0.13 | 0.63 | 0.76 |
| rs6813952 | 75,063,296 | C/A | 0.073 | 1 | −0.04 | 0.03 | 0.23 | 0.86 | 0.112 | 0.059 | 1 | −0.12 | 0.04 | 0.013 | 0.07 | 0.03 | −0.07 | 0.03 | 0.01 | 0.10 | 0.06 |
| rs1435520 | 75,064,281 | C/A | 0.019 | 1 | −0.16 | 0.08 | 0.015 | 0.14 | 0.022 | 0.053 | 1 | −0.13 | 0.05 | 0.009 | 0.06 | 0.02 | −0.14 | 0.04 | 0.0004 | 0.002 | 0.0009 |
| rs352004 | 75,065,434 | T/C | 0.253 | 1 | −0.03 | 0.02 | 0.15 | 0.69 | 0.828 | 0.008 | 1 | −0.32 | 0.13 | 0.077 | 0.86 | 0.10 | −0.04 | 0.02 | 0.10 | 0.58 | 0.83 |
| rs352007 | 75,066,156 | G/C | 0.440 | 0.97 | 0.05 | 0.02 | 0.0048 | 0.04 | 0.355 | 0.092 | 0.95 | 0.09 | 0.04 | 0.016 | 0.12 | 0.97 | 0.06 | 0.02 | 0.0003 | 0.003 | 0.33 |
| rs394408 | 75,068,216 | T/C | 0.407 | 0.98 | 0.05 | 0.02 | 0.0079 | 0.05 | 0.364 | 0.091 | 0.98 | 0.09 | 0.04 | 0.015 | 0.09 | 0.64 | 0.06 | 0.02 | 0.0006 | 0.005 | 0.54 |
| rs447206 | 75,068,787 | A/G | 0.060 | 0.99 | 0.07 | 0.04 | 0.07 | 0.41 | 0.277 | 0.088 | 0.99 | 0.10 | 0.04 | 0.014 | 0.08 | 0.68 | 0.08 | 0.03 | 0.002 | 0.02 | 0.13 |
| rs168449 | 75,069,487 | T/C | 0.407 | 0.98 | 0.05 | 0.02 | 0.0017 | 0.02 | - | 0.091 | 0.99 | 0.09 | 0.04 | 0.014 | 0.09 | - | 0.06 | 0.02 | 1.2×10−4 | 0.0013 | - |
| rs191601 | 75,069,569 | C/T | 0.406 | 0.98 | 0.05 | 0.02 | 0.0077 | 0.05 | 0.493 | 0.091 | 0.99 | 0.09 | 0.04 | 0.015 | 0.09 | 0.99 | 0.06 | 0.02 | 0.0002 | 0.006 | 0.53 |
| rs193263 | 75,069,687 | T/A | 0.406 | 0.98 | 0.05 | 0.02 | 0.0077 | 0.05 | 0.458 | 0.091 | 0.99 | 0.09 | 0.04 | 0.015 | 0.09 | 0.97 | 0.06 | 0.02 | 0.0002 | 0.006 | 0.51 |
| rs442155 | 75,070,096 | G/A | 0.060 | 1 | 0.07 | 0.04 | 0.06 | 0.41 | 0.266 | 0.088 | 1 | 0.10 | 0.04 | 0.014 | 0.08 | 0.71 | 0.08 | 0.03 | 0.002 | 0.02 | 0.13 |
| rs377579 | 75,070,524 | T/C | 0.412 | 0.99 | 0.06 | 0.02 | 0.0029 | 0.03 | 0.720 | 0.091 | 0.99 | 0.09 | 0.04 | 0.015 | 0.09 | 0.82 | 0.06 | 0.02 | 0.0002 | 0.002 | 0.31 |
| rs352009 | 75,070,557 | G/A | 0.406 | 0.98 | 0.05 | 0.02 | 0.0074 | 0.05 | 0.780 | 0.091 | 0.99 | 0.09 | 0.04 | 0.015 | 0.09 | 0.84 | 0.06 | 0.02 | 0.0005 | 0.006 | 0.40 |
| rs183028 | 75,070,782 | A/G | 0.412 | 1 | 0.06 | 0.02 | 0.0029 | 0.03 | 0.720 | 0.091 | 1 | 0.09 | 0.04 | 0.015 | 0.09 | 0.83 | 0.06 | 0.02 | 1.9×10−4 | 0.0016 | 0.61 |
| rs352010 | 75,074,095 | A/G | 0.409 | 0.97 | 0.05 | 0.02 | 0.0075 | 0.05 | 0.798 | 0.094 | 0.96 | 0.09 | 0.04 | 0.015 | 0.08 | 0.99 | 0.06 | 0.02 | 0.0005 | 0.005 | 0.83 |
| rs552582 | 75,074,722 | A/G | 0.497 | 0.95 | 0.04 | 0.02 | 0.03 | 0.42 | 0.813 | 0.100 | 0.96 | 0.09 | 0.04 | 0.016 | 0.11 | 0.77 | 0.05 | 0.02 | 0.003 | 0.06 | 0.98 |
| rs2457996 | 75,075,399 | C/T | 0.409 | 0.97 | 0.05 | 0.02 | 0.0078 | 0.05 | 0.951 | 0.091 | 0.99 | 0.09 | 0.04 | 0.015 | 0.08 | 0.98 | 0.06 | 0.02 | 0.0006 | 0.005 | 0.95 |
| rs187080 | 75,075,527 | T/C | 0.416 | 0.97 | 0.05 | 0.02 | 0.01 | 0.09 | 0.780 | 0.093 | 0.98 | 0.10 | 0.04 | 0.011 | 0.08 | 0.39 | 0.06 | 0.02 | 0.0006 | 0.006 | 0.96 |
| rs426901 | 75,076,379 | G/A | 0.455 | 0.96 | 0.03 | 0.02 | 0.10 | 0.64 | 0.100 | 0.098 | 0.98 | 0.10 | 0.04 | 0.008 | 0.06 | 0.28 | 0.04 | 0.02 | 0.01 | 0.09 | 0.23 |
| rs409336 | 75,076,522 | C/A | 0.538 | 1 | 0.02 | 0.02 | 0.20 | 0.83 | 0.270 | 0.104 | 1 | 0.09 | 0.04 | 0.013 | 0.07 | 0.54 | 0.04 | 0.02 | 0.02 | 0.17 | 0.46 |
| rs416046 | 75,076,546 | A/G | 0.053 | 0.84 | 0.09 | 0.04 | 0.06 | 0.49 | 0.254 | 0.072 | 0.81 | 0.12 | 0.05 | 0.013 | 0.07 | 0.59 | 0.10 | 0.03 | 0.002 | 0.02 | 0.12 |
| rs440676 | 75,076,834 | C/A | 0.424 | 0.96 | 0.04 | 0.02 | 0.03 | 0.42 | 0.189 | 0.095 | 0.98 | 0.10 | 0.04 | 0.009 | 0.06 | 0.36 | 0.05 | 0.02 | 0.002 | 0.04 | 0.39 |
| rs450373 | 75,076,915 | G/A | 0.424 | 0.96 | 0.04 | 0.02 | 0.03 | 0.42 | 0.181 | 0.095 | 0.98 | 0.10 | 0.04 | 0.009 | 0.06 | 0.35 | 0.05 | 0.02 | 0.002 | 0.04 | 0.38 |
| rs2564594 | 75,077,164 | T/A | 0.424 | 0.96 | 0.04 | 0.02 | 0.03 | 0.42 | 0.170 | 0.095 | 0.98 | 0.10 | 0.04 | 0.009 | 0.06 | 0.35 | 0.05 | 0.02 | 0.002 | 0.04 | 0.37 |
| rs420389 | 75,077,352 | G/C | 0.424 | 0.96 | 0.04 | 0.02 | 0.03 | 0.42 | 0.160 | 0.095 | 0.98 | 0.10 | 0.04 | 0.009 | 0.06 | 0.34 | 0.05 | 0.02 | 0.002 | 0.04 | 0.35 |
| rs2437285 | 75,079,102 | T/C | 0.412 | 1 | 0.01 | 0.02 | 0.56 | 0.99 | 0.405 | 0.102 | 1 | 0.09 | 0.04 | 0.011 | 0.07 | 0.45 | 0.03 | 0.02 | 0.09 | 0.49 | 0.71 |
| rs352047 | 75,080,954 | C/G | 0.427 | 0.98 | 0.02 | 0.02 | 0.22 | 0.90 | 0.070 | 0.098 | 0.98 | 0.11 | 0.04 | 0.009 | 0.06 | 0.11 | 0.04 | 0.02 | 0.02 | 0.15 | 0.07 |
| rs425535 | 75,082,861 | T/C | 0.416 | 0.98 | 0.02 | 0.02 | 0.27 | 0.91 | 0.070 | 0.099 | 0.99 | 0.11 | 0.04 | 0.009 | 0.06 | 0.10 | 0.04 | 0.02 | 0.02 | 0.15 | 0.07 |
| rs352046 | 75,083,414 | G/C | 0.425 | 0.99 | 0.02 | 0.02 | 0.27 | 0.89 | 0.070 | 0.100 | 0.99 | 0.11 | 0.04 | 0.009 | 0.06 | 0.10 | 0.04 | 0.02 | 0.02 | 0.15 | 0.07 |
Positions are in reference to NCBI build 36.3
Squared correlation between imputed and true genotypes. Directly genotyped SNPs are marked as 1
Effect size of the coded allele based on the multivariate linear regression adjusted for age and sex (using allele dosages)
Effect size of the coded allele based on the multivariate linear regression adjusted for age, sex and race (using allele dosages)
Corrected p-values (Pc) were generated by gene-wide multiple test correction using 5000 permutations
Uncorrected conditional P-values
Significant SNPs and the P-values for their association are highlighted in bold
Figure 2.
Distribution of serum PF4 levels. The histogram displays the mean values of transformed serum PF4 levels stratified by genotype at the rs168449 locus. The 95% confidence interval (CI) around the mean is represented as vertical bars and number above the bars indicate individual counts observed for the respective genotype group.
Association of PF4 genetic variation and serum TNFα levels
We also assessed the effect of PF4 variants on three additional serum biomarkers (fractalkine, P-selectin and TNFα), and we observed significant association with TNFα serum levels. The most significant association was seen in African American for rs8180167 (β= 0.016, SE=0.006, P-value = 0.008), however, this SNP did not show an effect in Whites (β= 0.003, P-value = 0.66) (Supplementary Table 4). rs8180167 is ~12.0 kb and ~17.2 kb upstream of rs1435520 and rs168449, respectively, and is largely uncorrelated within African Americans (rs1435520 r2=0 and rs168449 r2=0.04) (Supplementary Figure 1). The association of rs8180167 remained significant after multiple test correction (Pc-value = 0.048), and no other SNPs in the region show association after accounting for the effect of rs8180167 (Supplementary Table 4). rs8180167 explains ~1.3% of serum TNFα variance.
CX3CL1, CX3CR1 and CXCR3 genetic variations and serum biomarker levels
In total, 86 SNPs tagging the remaining 3 genes were tested for all 4 serum biomarkers. We did not detect any influence of these SNPs on biomarker levels in either of the racial groups, or in the combined analysis after gene-wide permutation corrections. In addition, the CX3CR1 gene, which encodes a chemokine receptor for fractalkine, contains two non-synonymous SNPs, rs3732378 (Thr280Met) and rs3732379 (Val249Ile), that were previously associated with reduced prevalence of atherosclerosis and acute coronary events.35,36 In Whites, we observed a nominally significant association of serum fractalkine levels with Thr280Met (P-value = 0.05) (Supplementary Table 5). Given a priori evidence of this variant, we reported the uncorrected P-value and results should therefore be interpreted with caution. Surprisingly, the observed effect estimates for rs3732378 (Thr280Met) is in the opposite direction as those previously reported for serum fractalkine. As opposed to the known athero-protective effect of 280Met, the allele A (Met) was observed associated with higher serum fractalkine levels (β=0.05, SE=0.02) (Supplementary Table 5). Neither SNP showed any significance with the other serum biomarkers.
Discussion
Atherosclerosis is a systemic disease that is responsible for most cardiovascular events and stroke. 4 Given the importance of the inflammatory and platelet activation cascade in the pathogenesis of disease, 5–7 clinical interest has focused on the identification of biomarkers for atherosclerotic risk prediction. Epidemiological studies have demonstrated that African Americans have higher prevalence and disease severity compared to Whites.1, 2 The reason for this differential atherogenic susceptibility are multiple and largely unresolved. In this study, we investigated the influence of genetic variation in the chemokine/chemokine receptors on systemic inflammation and platelet activation serum biomarkers.
We performed a comprehensive genetic screen of 4 genes (CX3CL1, CX3CR1, CXCR3 and PF4), and report PF4 locus variants associated with the modulation of serum PF4 and TNFα levels. For serum PF4 levels, the most significant association was observed for rs168449, which showed consistent effect in both African American and White populations. In African Americans, allele T (frequency 41%) was associated with higher levels (Table 3), whereas, in Whites, this allele showed the same direction effect but had a ~4.5-fold lower frequency (T allele frequency 9%). In the combined dataset, rs168449 maintained a robust association, and suggested the potential involvement in the modulation of serum PF4 levels in both racial groups. While accounting for the effect of rs168449, we also report another SNP (rs1435520) as a second independent signal regulating serum PF4 levels from this region. Together these SNPs explained 0.98% and 1.23% of serum PF4 variance in African Americans and Whites, respectively. Our study also identified the association of another independent SNP (rs8180167) regulating pro-inflammatory serum TNFα levels. In race stratified analysis, this association was observed only in African Americans, and revealed significant differences in allele frequencies between the two cohorts (Supplementary Table 4). While the frequencies of the ancestral allele (A) in African American and Whites were substantially different, 75% and 18%, respectively, we did not observe any evidence for recent selection at this locus. In an attempt to identify potentially functional variants in this locus, we looked for known expression quantitative trait loci (eQTLs) using the Genotype-Tissue Expression (GTEx) database, which queries lymphoblastoid, liver, brain cerebellum, frontal cortex, temporal cortex tissues, but found no known eQTLs in this region.
As shown in Figure 1a and 1b, these three PF4 region SNPs (rs168449, rs1435520 and rs8180167) are intergenic SNPs and lie close to the PPBP and CXCL5 genes, which are known to encode inflammatory cytokines. The PPBP gene encodes platelet-derived growth factor and function as a potent chemo-attractant and activator of neutrophils. Neutrophils are the most prominent leukocytes in acute inflammatory reactions and contribute in a number of inflammatory conditions. Recently, Rotzius et al examined the role of neutrophils in a mouse model of atherosclerosis and noticed that neutrophils accumulate in atherosclerotic lesions.35 Moreover, neutrophils are the predominant immune cells in the high inflammatory shoulder regions of plaques, suggesting that these cells may play an important role in the immunological processes of atherogenesis. Similarly, CXCL5 (C-X-C motif ligand 5) is also implicated in the chemotaxis of inflammatory cells and previously shown to be a recruiter of neutrophils and involved in their activation. In accordance to these reports, our results suggest the plausible involvement of PF4, PPBP, and/or CXCL5 genes in the modulation of serum PF4 and pro-inflammatory TNFα levels.
Our study also confirms the role of a CX3CR1 missense variant (Thr280Met) that has previously been reported associated with reduced prevalence of atherosclerosis and acute coronary events.36,37 Previously, the CX3CR1-280Met mutant was demonstrated to have an athero-protective effect 38 and showed a marked reduction in the kinetics of fractalkine binding. 38 The mutant form of CX3CR1 was also associated with reduced fractalkine-induced chemotactic activity 39 and dysfunction of the receptor.39–42 In this study, we observed a significant association of serum fractalkine levels with Thr280Met, but surprisingly the observed effect of 280Met allele is in the opposite direction as those reported previously. In Whites, we observed higher fractalkine levels associated with this allele, whereas no significant association was observed in African Americans.
Previously, it has been shown that PF4 is localized to fatty streaks and atherosclerotic lesions. 43 To assess the role of PF4 in the formation of atherosclerotic lesions, Sachais et al investigated the effects of knocking out PF4 expression in mice and showed a significant decrease in lesion formation in the absence of PF4. 44 This finding provides in vivo evidence that establishes that elimination of PF4 is athero-protective. Given the current knowledge available for the role of PF4 in atherosclerosis, our study demonstrates the importance of PF4 gene variants in the modulation of serum PF4 and pro-inflammatory TNFα levels. In our study, we assumed a model in which inflammatory responses lead to platelet activation, which can in turn intensify the inflammatory responses. This model contains regulatory elements that are known risk factors for atherosclerotic cardiovascular disease but has several important limitations that merit discussion. First, given the importance of the platelet activation and inflammatory mediators in the progression of atherosclerosis, we only tested 4 genes in our study and possibly other genes have a greater biological impact on these alterations. Through this study, we observed that PF4 SNPs explain a small proportion (~1%) of genetic variability for serum PF4 and TNFα levels; hence, suggesting the involvement of other genes contributing alterations in serum levels. Second, our study is only limited to the association of the systemic inflammation and platelet activation serum biomarkers, whereas exploring sub-clinical measures of atherosclerosis (carotid intima media thickness, pulse wave velocity, heart rate variability etc.) might have high biological relevance in context to atherosclerosis. Furthermore, while we have performed multiple test correction for each gene, we have not additionally corrected for the four traits studied. Knowing that the genetic puzzle of atherosclerotic cardiovascular disease will be complex and involve genes/loci from multiple biologic pathways, the next step will be to understand the relationships between disease risk loci important in innate and adaptive immunity, inflammation, blood pressure control and vascular reactivity and other factors. However, most critical will be an approach that keeps in mind that the relative risks of disease have different weights in the process of disease development and considerable influence especially when taken in the context of gene-environment interactions.45–48 Examining those demonstrated to affect the risk of disease in African Americans might allow us to work towards a model that includes blood pressure control loci SLC24A4 (rs11160059; rs17783630) and incident coronary heart disease loci PFTK1 variant (rs1859023) identified to be important in African Americans.49,50 Our finding in PF4 highlights the possible relevance of markers of systemic inflammation in atherosclerotic heart disease among African Americans. These findings should be put in a model with other loci important in modulating levels of acute phase reactants like C-reactive protein and other inflammatory molecules. The disparity in the incidence and severity of atherosclerotic cardiovascular disease is linked to unique inter-relationships between differentially expressed gene loci from different molecular pathways and ultimately with their interaction with the environment.
In conclusion, our results overall indicate that PF4 genetic variants are likely involved in the alterations of serum PF4 levels in both African American and White populations. We also demonstrate that another PF4 variant (rs8180167) has a race specific effect in the regulation of serum TNFα levels in African Americans. Taken together, these results highlight the importance of PF4 genetic variation in the regulation of systemic inflammation (TNFα) and platelet activation (PF4) serum biomarkers.
Supplementary Material
Given the importance of systemic inflammation and platelet activation to the development of atherosclerosis, we explored the role of genetic variants in regulating these processes. We performed a comprehensive genetic screen of 4 genes (CX3CL1, CX3CR1, CXCR3 and PF4), and report PF4 locus variants associated with the modulation of serum PF4 and TNFα levels. For serum PF4 levels, the most significant association was observed for rs168449, which showed consistent effect in both African American and White populations. In African Americans, allele T was associated with higher serum levels of PF4, whereas, in Whites, this allele showed the same direction effect but had a several fold lower frequency. Our study also identified the association of another independent SNP (rs8180167) regulating pro-inflammatory serum TNFα levels. In conclusion, our results overall indicate that PF4 genetic variants are likely involved in the alterations of serum PF4 levels in both African American and White populations. We also demonstrate that another PF4 variant (rs8180167) has a race specific effect in the regulation of serum TNFα levels in African Americans. Taken together, these results highlight the importance of PF4 genetic variation in the regulation of systemic inflammation (TNFα) and platelet activation (PF4) serum biomarkers. Knowing that the genetic puzzle of atherosclerotic cardiovascular disease will be complex and involve genes/loci from multiple biologic pathways, the next step will be to understand the relationships between disease risk loci important in innate and adaptive immunity, inflammation, blood pressure control and vascular reactivity and other factors.
Acknowledgments
The authors thank the staff, clinicians and participants of the Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS) study.
Funding Sources: This publication was made possible by Grant Number 1UL1RR025005 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Re-engineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Kington RS, Smith JP. Socioeconomic status and racial and ethnic differences in functional status associated with chronic diseases. Am J Public Health. 1997;87:805–810. doi: 10.2105/ajph.87.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng-Mak DS, Dohrenwend BP, Abraido-Lanza AF, Turner JB. A further analysis of race differences in the National Longitudinal Mortality Study. Am J Public Health. 1999;89:1748–1751. doi: 10.2105/ajph.89.11.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark LT. Issues in minority health: atherosclerosis and coronary heart disease in African Americans. Med Clin North Am. 2005;89:977–1001. doi: 10.1016/j.mcna.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health: National Heart, Lung and Blood Institute. NHLBI Morbidity and Mortality Chart Book. National Heart, Lung and Blood Institute; 2009. [online], http://www.nhlbi.nih.gov/resources/docs/2009_ChartBook.pdf. [Google Scholar]
- 5.Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 6.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 7.Von Hundelshausen P, Weber C. Platelets as immune cells: bridging inflammation and cardiovascular disease. Circ Res. 2007;100:27–40. doi: 10.1161/01.RES.0000252802.25497.b7. [DOI] [PubMed] [Google Scholar]
- 8.Gerrity RG. The role of the monocyte in atherogenesis: II. Migration of foam cells from atherosclerotic lesions. Am J Pathol. 1981;103:191–200. [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno PR, Falk E, Palacios IF, Newell JB, Fuster V, Fallon JT. Macrophage infiltration in acute coronary syndromes: implication for plaque rupture. Circulation. 1994;90:775–778. doi: 10.1161/01.cir.90.2.775. [DOI] [PubMed] [Google Scholar]
- 10.Reape TJ, Groot PH. Chemokines and atherosclerosis. Atherosclerosis. 1999;147:213–225. doi: 10.1016/s0021-9150(99)00346-9. [DOI] [PubMed] [Google Scholar]
- 11.Kowalska MA, Ratajczak MZ, Majka M, Jin J, Kunapuli S, Brass L, et al. Stromal cell-derived factor-1 and macrophage-derived chemokine: 2 chemokines that activate platelets. Blood. 2000;96:50–57. [PubMed] [Google Scholar]
- 12.Abi-Younes S, Sauty A, Mach F, Sukhova GK, Libby P, Luster AD. The stromal cell-derived factor-1 chemokine is a potent platelet agonist highly expressed in atherosclerotic plaques. Circ Res. 2000;86:131–138. doi: 10.1161/01.res.86.2.131. [DOI] [PubMed] [Google Scholar]
- 13.Nieswandt B, Aktas B, Moers A, Sachs UJ. Platelets in atherothrombosis: lessons from mouse models. J Thromb Haemost. 2005;3:1725–1736. doi: 10.1111/j.1538-7836.2005.01488.x. [DOI] [PubMed] [Google Scholar]
- 14.Gawaz M, Langer H, May AE. Platelets in inflammation and atherogenesis. J Clin Invest. 2005;115:3378–3384. doi: 10.1172/JCI27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyrich AS, McIntyre TM, McEver RP, Prescott SM, Zimmerman GA. Monocyte tethering by P-selectin regulates monocyte chemotactic protein-1 and tumor necrosis factor-alpha secretion. Signal integration and NF-kB translocation. J Clin Invest. 1995;95:2297–2303. doi: 10.1172/JCI117921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boring L, Gosling J, Cleary M, Charo IF. Decreased lesion formation in CCR2−/− mice reveals a role for chemokines in the initiation of atherosclerosis. Nature. 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 17.Gu L, Okada Y, Clinton SK, Gerard C, Sukhova GK, Libby P, et al. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficientmice. Mol Cell. 1998;2:275–281. doi: 10.1016/s1097-2765(00)80139-2. [DOI] [PubMed] [Google Scholar]
- 18.Boisvert WA, Santiago R, Curtiss LK, Terkeltaub RA. A leukocyte homologue of the IL-8 receptor CXCR-2 mediates the accumulation of macrophages in atherosclerotic lesions of LDL receptor-deficient mice. J Clin Invest. 1998;101:353–363. doi: 10.1172/JCI1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szalai C, Duba J, Prohászka Z, Kalina A, Szabó T, Nagy B, et al. Involvement of polymorphisms in the chemokine system in the susceptibility for coronary artery disease (CAD). Coincidence of elevated Lp(a) and MCP-1–2518 G/G genotype in CAD patients. Atherosclerosis. 2001;158:233–239. doi: 10.1016/s0021-9150(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 20.Valdes AM, Wolfe ML, O’Brien EJ, Spurr NK, Gefter W, Rut A, et al. Val64Ile polymorphism in the C-C chemokine receptor 2 is associated with reduced coronary artery calcification. Arterioscler ThrombVasc Biol. 2002;22:1924–1928. doi: 10.1161/01.atv.0000038486.48400.e7. [DOI] [PubMed] [Google Scholar]
- 21.Von Hundelshausen P, Weber C. Platelets as immune cells: bridging inflammation and cardiovascular disease. Circ Res. 2007;100:27–40. doi: 10.1161/01.RES.0000252802.25497.b7. [DOI] [PubMed] [Google Scholar]
- 22.Gerrity RG. The role of the monocyte in atherogenesis: II. Migration of foam cells from atherosclerotic lesions. Am J Pathol. 1981;103:191–200. [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno PR, Falk E, Palacios IF, Newell JB, Fuster V, Fallon JT. Macrophage infiltration in acute coronary syndromes: implication for plaque rupture. Circulation. 1994;90:775–778. doi: 10.1161/01.cir.90.2.775. [DOI] [PubMed] [Google Scholar]
- 24.Kowalska MA, Ratajczak MZ, Majka M, Jin J, Kunapuli S, Brass L, et al. Stromal cell-derived factor-1 and macrophage-derived chemokine: 2 chemokines that activate platelets. Blood. 2000;96:50–57. [PubMed] [Google Scholar]
- 25.Srivastava K, Cockburn IA, Swaim A, Thompson LE, Tripathi A, Fletcher CA, et al. Platelet factor 4 mediates inflammation in experimental cerebral malaria. Cell Host Microbe. 2008;4:179–187. doi: 10.1016/j.chom.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans MK, Lepkowski JM, Powe NR, LaVeist T, Kuczmarski MF, Zonderman AB. Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS): A longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethnicity & Disease. 2010;20:267–275. [PMC free article] [PubMed] [Google Scholar]
- 27.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, et al. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotzius P, Thams S, Soehnlein O, Kenne E, Tseng CN, Björkström NK, et al. Distinct infiltration of neutrophils in lesion shoulders in ApoE−/− mice. Am J Pathol. 2010;177:493–500. doi: 10.2353/ajpath.2010.090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moatti D, Faure S, Fumeron F, Amara Mel-W, Seknadji P, McDermott DH, et al. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97:1925–1928. doi: 10.1182/blood.v97.7.1925. [DOI] [PubMed] [Google Scholar]
- 37.McDermott DH, Halcox JP, Schenke WH, Waclawiw MA, Merrell MN, Epstein N, et al. Association between polymorphism in the chemokine receptor CX3CR1 and coronary vascular endothelial dysfunction and atherosclerosis. Circ Res. 2001;89:401–407. doi: 10.1161/hh1701.095642. [DOI] [PubMed] [Google Scholar]
- 38.Lesnik P, Haskell CA, Charo IF. Decreased atherosclerosis in CX3CR1−/− mice reveals a role for fractalkine in atherogenesis. J Clin Invest. 2003;111:333–340. doi: 10.1172/JCI15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott DH, Fong AM, Yang Q, Sechler JM, Cupples LA, Merrell MN, et al. Chemokine receptor mutant CX3CR1-M280 has impaired adhesive function and correlates with protection from cardiovascular disease in humans. J Clin Invest. 2003;111:1241–1250. doi: 10.1172/JCI16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faure S, Meyer L, Costagliola D, Vaneensberghe C, Genin E, Autran B, et al. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science. 2000;287:2274–2277. doi: 10.1126/science.287.5461.2274. [DOI] [PubMed] [Google Scholar]
- 41.Daoudi M, Lavergne E, Garin A, Tarantino N, Debré P, Pincet F, et al. Enhanced adhesive capacities of the naturally occurring Ile249–Met280 variant of the chemokine receptor CX3CR1. J Biol Chem. 2004;279:19649–19657. doi: 10.1074/jbc.M313457200. [DOI] [PubMed] [Google Scholar]
- 42.Moatti D, Faure S, Fumeron F, Amara Mel-W, Seknadji P, McDermott DH, et al. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97:1925–1928. doi: 10.1182/blood.v97.7.1925. [DOI] [PubMed] [Google Scholar]
- 43.Pitsilos S, Hunt JL, Mohler ER, Prabhakar AM, Poncz M, Dawicki J, et al. Platelet factor 4 localization in carotid atherosclerotic plaques: Correlation with clinical parameters. Thromb Haemost. 2003;90:1142–1150. doi: 10.1160/TH03-02-0069. [DOI] [PubMed] [Google Scholar]
- 44.Sachais BS, Turrentine T, Dawicki McKenna JM, Rux AH, Rader D, Kowalska MA. Elimination of platelet factor 4 (PF4) from platelets reduces atherosclerosis in C57Bl/6 and apoE−/− mice. Thromb Haemost. 2007;98:1108–1113. [PubMed] [Google Scholar]
- 45.Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegård A, et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet. 2005;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 47.Swanberg M, Lidman O, Padyukov L, Eriksson P, Akesson E, Jagodic M, et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet. 2005;37:486–494. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- 48.Dwyer JH, Allayee H, Dwyer KM, Fan J, Wu H, Mar R, et al. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N Engl J Med. 2004;350:29–37. doi: 10.1056/NEJMoa025079. [DOI] [PubMed] [Google Scholar]
- 49.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbalic M, Reiner AP, Wu C, Hixson JE, Franceschini N, Eaton CB, et al. Genome-wide association analysis of incident coronary heart disease (CHD) in African Americans: a short report. PLoS Genet. 2011;7:e1002199. doi: 10.1371/journal.pgen.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.