Abstract
Viral miRNAs, ∼22nt RNA molecules which post-transcriptionally regulate gene expression, are emerging as important tools in immune evasion. Viral infection is a complex process that requires immune evasion in order to establish persistent life-long infection of the host. During this process viruses express both protein-coding and non-coding genes, which help to modulate the cellular environment making it more favorable for infection. In the last decade, it was uncovered that DNA viruses express a diverse and abundant pool of small non-coding RNA molecules, called microRNAs (miRNAs). These virally encoded miRNAs are non-immunogenic and therefore are important tools used to evade both innate and adaptive immune responses. This review aims to summarize our current knowledge of herpesvirus- and polyomavirus-encoded miRNAs, and how they contribute to immune evasion by targeting viral and/or host cellular genes.
Introduction
All currently identified miRNAs are described as noncoding RNA molecules ∼22 nucleotides in length that post-transcriptionally regulate gene expression by complementary binding to target mRNA transcripts. MiRNAs were first identified in 1993 by Ambros and colleagues in the nematode Ceanorhabidtis elegans and were originally thought to be a regulatory phenomenon specific for this organism [1]. Hallmark discoveries in 2000 by the Ruvkin laboratory showed that one C. elegans miRNA, let-7, was 100% conserved within the genomes of mice and humans [2]. This finding initiated a rapid discovery phase for novel miRNAs and now more than 15,000 different miRNAs have been identified in all metazoan and plant species investigated thus far. In 2004 this list was further expanded to include DNA viruses, when Tuschl and colleagues identified the first viral miRNAs encoded by Epstein-Barr virus (EBV), a human DNA tumor virus [3]. To date, over 200 viral miRNAs have now been identified, with most being encoded by herpesviruses.
Functionally, miRNAs regulate a vast set of targets that are involved in multiple cellular processes including development, immunity, and apoptosis. In mammalian species, it has been estimated that greater than half of all protein coding genes contain a miRNA target site, indicating their heavy impact on gene regulation [4]. Viral miRNAs have also been found to target a large number of host genes involved in regulating cell proliferation, apoptosis, and host immunity. In addition, viral miRNAs target their own genes, which also help the virus to remain hidden from the host immune response.
Viral miRNA biogenesis and functional targeting is 100% dependent on the host molecular miRNA maturation and silencing machinery. In brief, viral miRNA biogenesis initiates in the nucleus, where after transcription, the host RNase III endonuclease Drosha cleaves pri-miRNA hairpins into 60 to 80nts long pre-miRNAs. These pre-miRNAs are rapidly exported by the Exportin 5/Ran GTPase pathway into the cytoplasm where they are further processed by a second host RNase III endonuclease, Dicer, into a short dsRNA duplex. One strand of the duplex is incorporated into the host RNA-induced silencing complex (RISC), which guides the mature miRNA to 3′UTR's of mRNAs containing complementary sequences, leading to translational silencing and/or transcript degradation. An important parameter of targeting is complementary base pairing between the miRNA ‘seed’ sequence (5′ nucleotides 2-7) and the target transcript [5]. To date, no viral proteins have been described that directly contribute to either miRNA biogenesis or targeting mechanisms.
Based on the requirements of nuclear machinery and RNA cleavage for miRNA processing, it is no surprise that cytoplasmic replicating DNA viruses and RNA viruses have not been found to express miRNAs. In fact, out of the currently identified viral miRNAs (>200) the vast majority (>90%) are encoded by all three families of herpesviruses (α, β, and γ), which are DNA viruses that replicate in the nucleus of host cells. In addition to the herpesvirus miRNAs, a small number of miRNAs (7) have been found in other nuclear replicating DNA viruses, including adenovirus and several polyomaviruses, although for the former, miRNAs are processed in a Drosha-independent manner. Interestingly, all three of these DNA virus families establish persistent infections in a large percentage of the human population, indicating that immune evasion is essential for their lifecycles and that viral miRNAs may play an important role in promoting immune evasion. To date only herpesvirus and polyomavirus miRNA targets have been characterized (Table 1) and this review will focus on how miRNAs encoded by these viruses contribute to immune evasion.
Table 1. Immunomodulatory viral miRNAs.
| DNA Virus Family | Virus | Target | miRNA | Function | References |
|---|---|---|---|---|---|
|
| |||||
| Latency | |||||
| Alphaherpesvirus | HSV-1 | Viral ICP0 | miR-H2-3p | Immediate-early transactivator | [58] |
| Viral ICP4 | miR-H6 | Immediate-early transactivator | [58] | ||
| ICP34.5 | miR-H3 and -H4 | Lytic neurovirulence factor | [60]** | ||
| HSV-2 | Viral ICP0 | miR-III | Immediate-early transactivator | [57] | |
| ICP34.5 | miR-I and -II | Lytic neurovirulence factor | [56,57] | ||
|
| |||||
| Cell-mediated Immunity | |||||
| Betaherpesvirus | HCMV | Host MICB | miR-UL112-1 | NK cell ligand | [14] |
| Latency | |||||
| Viral IE72 | miR-UL112-1 | Immediate-early transactivator | [29,30] | ||
| Cell-mediated Immunity | |||||
| MCMV | Host CXCL16 | miR-M23-2 | Chemokine | [27] | |
|
| |||||
| Cell-mediated Immunity | |||||
| Gammaherpesvirus | EBV | Host MICB | miR-BART2-5p | NKG2D ligand | [15] |
| Host CXCL11 | miR-BHRF1-3 | Chemokine, T-cell attractant | [28] | ||
| Viral LMP1 | miR-BART1-5p | Transforming factor | [10] | ||
| miR-BART16 | |||||
| miR-BART17-5p | |||||
| Viral LMP2a | miR-BART22 | Transforming factor | [11] | ||
| Latency | |||||
| Viral BALF5 | miR-BART2 | DNA polymerase | [44] | ||
| Host Dicer | miR-BART6-5p | miRNA processing enzyme | [45] | ||
| Cell-mediated Immunity | |||||
| KSHV | Host MICB | miR-K12-7 | NKG2D ligand | [15] | |
| Host C/EBPβ (LIP) | miR-K12-3 | Inhibits IL6 and IL10 expression | [19] | ||
| miR-K12-7 | |||||
| Host TWEAKR | miR-K10a | Tumor necrosis factor receptor | [24] | ||
| Cell-mediated Immunity/Latency | |||||
| Host IKKε | miR-K12-11 | Interferon signaling molecule | [23] | ||
| Latency | |||||
| Viral RTA | miR-K12-5 | Master lytic switch | [47] | ||
| miR-K9* | [51] | ||||
| miR-K12-7-5p | [52] | ||||
| Host Rbl2 | miR-K12-4-5p | Transcriptional repressor | [47] | ||
| Host IκBα | miR-K1 | Inhibits NF-κβ | [48] | ||
| Host NFIB | miR-K3 | Transcriptional activator | [49] | ||
|
| |||||
| Cell-mediated Immunity | |||||
| Polyomavirus | SV40 | Viral Large TAg | miR-S1 | Transforming factor | [7] |
| JCV | Viral Large Tag | miR-J1 | Transforming factor | [8] | |
| Host ULBP3 | miR-J1-3p | NKG2D ligand | [18] | ||
| BKV | Viral Large Tag | miR-B1 | Transforming factor | [8] | |
| Host ULBP3 | miR-B1-3p* | NKG2D ligand | [18] | ||
Anti-viral host defenses
The host defense against viral infection is mediated by two main components: innate and adaptive immunity. Considered the first line of defense, innate immunity primarily involves early viral detection which elicits an interferon response and activation of natural killer (NK) cells. At later time points adaptive immunity is induced which elicits antigen-specific responses, including antibody production by B cells and the activation of cytotoxic T lymphocytes (CTL). To counteract these hosts defense mechanisms both herpesvirus and polyomavirus have coevolved miRNAs to regulate a variety of host-immune modulatory pathways (Fig. 1).
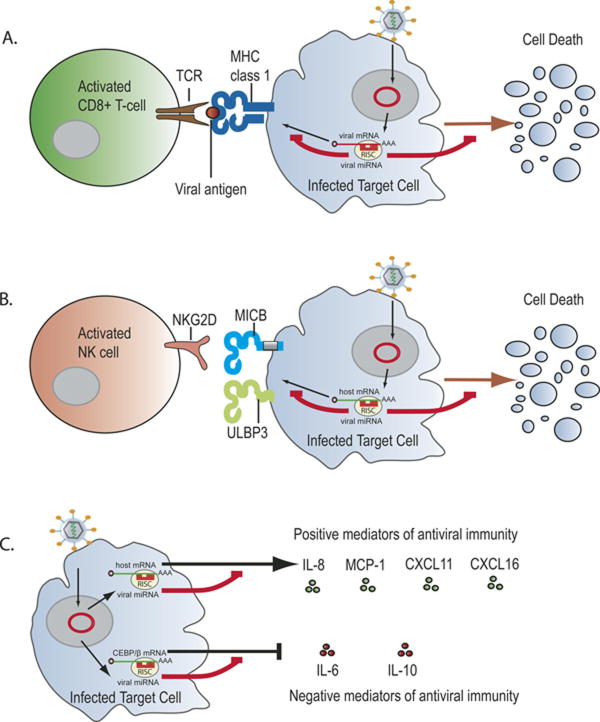
Fig. 1.
Viral miRNAs can inhibit cell mediated immunity by modulating the recognition and activity of effector cells. (a) A CTL response to virally infected cells is initiated when the T-cell receptor (TCR) of CD8+T cells recognizes viral antigenic peptides presented by class I MHC molecules on the surface of infected cells. Viral miRNAs can block this initiating step by inhibiting the expression of viral peptides before they are presented. (b) NK cell killing of virally infected cells is initiated when an activation receptor (i.e. NKG2D) engages ligands (i.e. MICB or ULBP3) expressed by virally infected cells. Viral miRNAs can block this step by inhibiting the expression of these ligands. (c) Viral miRNAs can regulate the activities of leukocytes involved in the immune response by inhibiting the production of cytokines/chemokines which promote anti-viral immunity or by inducing production of cytokines that may inhibit anti-viral immunity.
Viral miRNA inhibition of cell mediated immunity
Cell mediated immunity is essential for viral clearance of infected cells. The effectors of this mechanism involve both antigen specific CTLs and non-specific NK cells, both are cytotoxic mediators which eliminate infected cells through processes that induce apoptosis and ultimately result in cell lysis. Several viral miRNAs have been found that work to inhibit these cell mediated responses by targeting viral antigens or proteins involved in antigen presentation and activation pathways.
The clearance of virally infected cells by CTLs requires viral antigen presentation by infected target cells which are recognized by antigen specific CTL receptors (Fig. 1a). In murine models it has been shown that expression of the polyomavirus viral protein, T-antigen, can elicit such a response [6]. Studies done with the primate polyomavirus, simian virus 40 (SV40), identified miRNAs encoded antisense to the T-antigen that mediate its transcripts cleavage during infection [7]. Subsequent studies found that the human polyomaviruses JCV and BKV, as well as a murine Polyomavirus muPyV also encode miRNAs that cleave T-antigens [8]. Using mutant SV40 viruses, which do not express the miRNAs and as a result express increased levels of large T-antigens, it was shown in cell culture that these viruses had increased susceptibility to CTL lysis compared to wild type virus [7]. This study provided the first evidence that viral miRNAs can inhibit CTL recognition by directly targeting viral antigens. However, when a miRNA mutant of muPyV virus was used to infect mice there was no detectable change in the CTL response or viral clearance, indicating that more studies are needed to fully understand the function of polyomavirus miRNA mediated T-antigen regulation during natural infection [9].
MiRNAs expressed by the gammaherpesvirus EBV have also been shown to suppress the expression of viral antigens [10,11]. During latency EBV expresses the latent membrane proteins 1 and 2a (LMP1 and LMP2A); both proteins are crucial for EBV transformation of B cells and have been shown to elicit a CTL response [12,13]. LMP1 expression was found to be targeted by three EBV miRNAs (miR-BART16, miR-BART17-5p, and miR-BART1-5p) in nasopharyngeal carcinoma (NPC), an epithelial cancer associated with EBV infection [10]. However, in this study a link between miRNA targeting of LMP1 and the CTL response was not made, rather, it was shown that LMP1 downregulation was important in attenuating a pro-apoptotic affect caused by abundant LMP1 expression. Members of this same group later found that EBV miR-BART22, is responsible for targeting LMP2a [11]. Bioinformatic analysis of possible miR-BART22 targets identified high sequence complementarity (16 nucleotides) between mir-BART-22 and LMP-2a. Like LMP1 expression in NPC, low protein levels of LMP2A correlated well with high levels of miR-BART22 in patient samples, indicative of miRNA targeting within these tumors. Based on the immunogenicity of these viral proteins the authors believe that EBV miRNAs regulate their expression to inhibit immune surveillance; this hypothesis will need to be further validated by demonstrating miRNA targeting in appropriate in vitro and in vivo models.
Elimination of virally infected cells by NK cells involves NK cell receptor recognition of ligands expressed by target cells (Fig. 1b). Elegant genetic studies on the betaherpesvirus human cytomegalovirus (HCMV) demonstrated that HCMV miRNA miR-UL112-1 targets the major histocompatibility complex class l-related chain B (MICB), a stress induced ligand recognized by the NKG2D receptor expressed by NK cells and CD8+ T-cells [14]. Using a HCMV mutant virus containing a miR-UL112-1 deletion, the authors showed that mutant virus infected cells were more efficiently recognized and killed by NK cells in vitro, providing direct evidence that MICB repression mediated by this miRNA is important for NK cell evasion. Later studies discovered that gammaherpesvirus miRNAs, EBV miR-BART2-5p and Kaposi's sarcoma-associated herpesvirus (KSHV) miR-K12-7, also regulate MICB expression, and inhibition of MICB miRNA targeting with miRNA sponges in virally infected cells increased NK cell killing [15]. Interestingly, it was recently shown that miR-UL112-1 acts in combination with a host miRNA, miR-376a, to more efficiently regulate MICB expression, and that this synergistic action is important for inhibiting NK cell killing in vitro. These data strongly suggest that miRNA-dependent regulation of MICB is important for herpesviral persistence, which is further underscored by the fact that both HCMV and KSHV encode proteins that inhibit MICB surface expression [16, 17]. In addition, the observation that HCMV-, EBV-, and KSHV-encoded miRNAs target the MICB gene by completely different sequences raises a very interesting question about the co-evolution of viral miRNAs and their corresponding cellular targets.
A separate stress induced ligand (ULBP3), also recognized by the NKG2D receptor, is targeted by miR-J1-3p, a miRNA that is conserved between two different polyomaviruses, JCV and BKV [18]. Suppressing miR-J1-3p using a miRNA sponge leads to increased ULBP3 expression in JCV infected cells and subsequently enhanced NK cell killing. The identification of these multiple targets confirms that miRNAs expressed by both herpesvirus and polyomavirus represent important tools for suppressing immune surveillance by dampening NK cell target cell recognition.
In addition to inhibiting recognition by cell mediated effectors, experimental evidence suggests that herpesvirus miRNAs can inhibit cell mediated immunity by directly modulating cytokine expression (Fig. 1C). KSHV miRNAs miR-K12-3 and miR-K12-7, when ectopically expressed in human myelomonocytic and murine macrophage cell lines can increase secretion of host cytokines IL-6 and IL-10, which are highly expressed in KS lesions [19]. Bioinformatic analysis in combination with antagomir-based derepression assays demonstrated that miR-K12-3 and miR-K12-7 downregulates LIP, an isoform of C/EBPβ that functions as a negative transcriptional regulator of IL-6. Although these cytokines have broad functions in suppressing the activity of multiple immune cell types including T-cells, NK cells, and dendritic cells their impact during natural KSHV infection needs to be further tested [20-22].
Recently, it was shown that KSHV miR-K12-11 targets l-kappa-B kinase epsilon (IKKε), an important signaling molecule in the antiviral interferon response pathway [23]. To test the impact of IKKε targeting by miR-K12-11 without any confounding effects of other KSHV immune regulatory proteins, miR-K12-11 transduced lung cancer cells were infected with two RNA viruses, Sendai virus (SeV) and vesicular stomatitis virus (VSV), which strongly induce the interferon response. Results showed that upon infection, miR-K12-11 expressing cells had markedly attenuated interferon signaling and enhanced VSV titers.
An additional study demonstrated that ectopic expression of KSHV miR-K10a, in primary endothelial cells, markedly reduced production of pro-inflammatory cytokines, IL-8 and monocyte chemoattractant protein 1 (MCP-1), by targeting tumor necrosis factor (TNF)-like weak inducer of apoptosis receptor (TWEAKR), a target identified by a elegant tandem-array screen using ectopic expression in combination with miRNA inhibition in KSHV-infected lymphoma cells [24]. Curiously, these pro-inflammatory cytokines are induced by KSHV proteins (vFLIP and vGPCR) and may promote tumorigenesis [25, 26]. To integrate these paradoxical observations, the authors hypothesize that miR-K10a dependent regulation of IL-8 and MCP-1 may provide a mechanism that fine tunes cytokine expression to levels beneficial for the virus without eliciting a strong immune response. Hence, this regulatory loop is analogous to the fine tuning of LMP-1 expression by EBV miRNAs to balance proliferation and apoptotic activities.
The first in vivo phenotype of a viral miRNA knock-out was recently reported using a murine cytomegalovirus (MCMV) mutant [27]. In this study, mice infected with mutant viruses, lacking miR-M23-1 and miR-M23-2, showed a significant reduction in viral load in salivary glands. Furthermore, when both NK cells and CD4+ T-cells were depleted the ability of the mutant virus to replicate in salivary glands was partly restored, indicating that these viral miRNAs are important immune modulators. A number of potential immune targets were predicted by bioinformatics, and subsequent in vitro analysis confirmed that miR-M23-2 targets CXCL16, a host chemokine involved in T-cell activation and NK cell migration. Hence, the loss of miR-M23-2-dependent down-regulation of CXCL16 is believed to lead to rapid viral immune clearance.
Currently, the only other viral miRNA reported to modulate cytokine expression is EBV miR-BHRF1-3, which was found to target the T-cell attractant chemokine CXCL11 [28]. However, the functional relevance of this regulation during EBV infection in vivo has yet to be reported.
Herpesvirus latent infection
To avoid recognition from host immune surveillance herpesviruses have co-evolved a mode of infection known as latency, during which the virus remains hidden by expressing only a small number of genes including viral miRNAs. One mechanism for controlling latent gene expression is to utilize viral miRNAs to suppress both viral and/or cellular genes that can trigger reactivation. Unlike viral proteins, which can elicit strong immune responses, viral miRNAs are indistinguishable from their cellular counterparts, nonimmunogenic, and therefore ideal tools for latent gene regulation. Mounting evidence, summarized in this section, shows that herpesvirus miRNAs play an important role in maintaining latent infection, thereby contributing to immune evasion resulting in life-long persistent infections, a hallmark of all herpesviruses.
The first direct evidence that herpesvirus miRNAs play a role in the maintenance of latency was provided by two independent studies in HCMV, which demonstrated that HCMV miR-UL112-1 can target IE72, a major immediate early viral transactivator that promotes lytic replication [29, 30]. Furthermore, Grey et al. showed that introduction of a miR-UL112-1 mimic into cells prior to HCMV infection significantly attenuates viral replication. Additionally, miR-UL112-1 was found to target UL114, a DNA glycosylase involved in viral replication and encoded antisense to miR-UL112-1 [31]. While Grey et al. showed that miR-UL112-1 did not mediate UL114 transcript cleavage; a separate study reported that ectopic expression of miR-UL112-1 can indeed target UL114 and reduce its expression in various cell model systems [32]. However, although mutant viruses lacking UL114 lead to a moderate decrease in viral replication, probably due to decreased DNA replication, the authors suggest that miR-UL112-1 targeting of IE72 is the main mechanism for reducing viral replication.
Unlike HCMV, where miRNA expression is mainly characterized during productive infection, miRNA expression for EBV and KSHV during latent and lytic replication has been well characterized using a combination of approaches (Northern blot, tiled arrays, and sequence analysis) [3, 33-41]. While subsets of EBV miRNAs have been found to be differentially expressed during latent and lytic lifecycles, KSHV miRNAs are predominantly expressed during latency [42, 43]. Early studies of EBV miRNAs found that latently expressed miR-BART2 is encoded antisense to the viral DNA polymerase BALF5, which is only expressed during lytic replication [3]. Later studies by a separate group provided evidence that miR-BART2 mediates regulation of BALF5 by antisense directed cleavage of the BALF5 transcript [44]. While ectopic miR-BART2 expression in EBV infected 293 cells modestly reduced virus production, inhibition of miR-BART2 with antagomirs did not induce lytic reactivation. Based on these observations the authors propose that instead of playing a central role in maintaining latency the targeting of BALF5 by miR-BART2 may help to safeguard against inadvertent lytic reactivation.
Most recently, it was shown that latently expressed EBV miR-BART6-5p can target the cellular miRNA processing enzyme Dicer, leading to increased expression of the two major IE transactivators RTA and Zta [45]. However, the direct functional consequence of Dicer targeting needs to be further validated by measuring viral replication after miR-BART6-5p inhibition.
In KSHV, the viral replication and transcription activator (RTA), a master regulator of lytic reactivation, has been shown to be regulated either directly or indirectly by multiple viral miRNAs. Two independent studies, using similar KSHV bacmid 36 derived recombinant viruses that lack 10 of 12 miRNA genes, reported elevated expression of lytic genes, including RTA, during de novo infection in separate cell lines [46, 47]. To determine the mechanism leading to increased lytic gene expression Lu et al. screened the individual KSHV miRNAs, using miRNA expression plasmids, for their ability to target a RTA luciferase construct and found that miR-K5 can repress RTA expression, albeit the 3′UTR of RTA lacks a canonical miR-K5 seed sequence. Additionally, Lu et al. carried out genome wide epigenetic analysis of the miRNA knock-out virus and found drastically reduced repressive marks on histones along with a global reduction of DNA methylation, suggesting that epigenetic modifications induced by viral miRNAs may contribute to the maintenance of latency. Searching for a mechanism to explain these modifications Lu et al. found that miR-K12-5p targets retinoblastoma (Rb)-like protein 2 (Rbl2), a negative regulator of DNA methyltransferases, thereby leading to an increase in DNA methylation. This is the first reported evidence that viral miRNAs can directly impact the epigenetic status of herpesvirus genomes during latency.
In the second study, Lei et al. also found an increase in RTA mRNA expression in cells infected with a very similar KSHV miRNA knock-out virus, but they did not identify direct targeting of the RTA 3′UTR by miR-K12-5p or any other KSHV miRNA [46]. Instead, Lei et al showed that miR-K1 targets the host gene lkBα, an inhibitor of NFκB, leading to activation of NFκB, which is known to inhibit lytic reactivation and, in the case of PELs, contributes to cell survival [48].
In addition to targeting lkBα, two independent studies reported that lytic reactivation can be regulated by KSHV miR-K12-11 targeting of IKKε and nuclear factor I/B (NFIB) [23, 49]. As mentioned before miR-K12-11 targeting of IKKε leads to attenuation of the interferon response [23]. This same study found that inhibiting miR-K12-11, with an anti-miR-K12-11 sponge, leads to an increase in lytic gene expression (RTA and ORF65) in bacmid infected A549 cells. The authors also showed that IKKε overexpression enhanced lytic replication when TPA, a chemical agent that triggers lytic reactivation, was used.
Using lentiviruses to express individual KSHV miRNAs in BC3 cells, Lu et al found that miR-K1, K3, K7, and K11 were all capable of moderately decreasing RTA mRNA levels [49]. MiR-K3 showed the greatest effect on RTA, and further investigation found that it directly targets NFIB, a cellular transcription factor that had previously been shown to reactivate KSHV when overexpressed [50]. Further analysis identified that the promoter of RTA contains a putative NFIB binding site and that ectopic NFIB expression could activate an RTA promoter construct. Additionally, shRNA knockdown of NFIB resulted in decreased RTA expression. This study provides indirect evidence that miR-K3 maintains latency by targeting NFIB, but further experiments using anti-miR-K3 antagomirs or a miR-K3 knockout virus are needed to prove this mechanism.
In addition to indirectly regulating RTA expression two separate studies have demonstrated that miR-K12-9* and miR-K12-7-5p can directly target and regulate RTA expression through seed match binding [51, 52]. Using luciferase constructs, containing the 3′UTR of RTA, and KSHV miRNA mimics, Bellare et al. identified that miR-K9* directly targets RTA through a canonical 6mer seed match site. Furthermore, when miR-K9* function in latently infected cells was inhibited with specific antagomirs a moderate increase in lytic reactivation, was observed. In a separate study by Lin et al., which used KSHV miRNA expression plasmids instead of miRNA mimics, miR-K9* and miR-K12-7-5p were also found to target RTA [52]. Lin et al. further show that miR-K12-7-5p targeting of RTA is mediated through a 7mer seed match site and that ectopic expression of miR-K12-7-5p in latently infected cell lines reduces the amount of progeny virus produced. In summary, these studies lend further credence that KSHV miRNAs directly regulate RTA expression during latency. However, while some studies hypothesize that KSHV miRNAs function as major regulators of latency, Bellare et al. suggests that these miRNAs may provide a mechanism for fine tuning and/or sensitizing latently infected cells to stimuli that trigger lytic replication.
We note that most of the KSHV miRNAs found to target RTA appear to differ between studies, with the exception of miR-K9* and miR-K7 [51, 52]. One possible reason for these differing results is that the miRNA targeting screens were not done using the same parameters, miRNA expression systems, and/or identical cellular backgrounds making it difficult to directly compare these studies. Nonetheless, in summary the evidence indicates that KSHV miRNA regulation of lytic reactivation is a complex process and that KSHV miRNAs do indeed play a synergistic role in the maintenance of latency.
The human alphaherpesvirus HSV-1 and HSV-2, are closely related neurotropic viruses which establish latency in sensory neurons and have recently been found to express miRNAs that contribute to the regulation of latency. Interestingly, the majority of genes in both viruses share similar genomic positions, including the viral miRNAs, many of which are encoded antisense to protein coding genes. Early studies of HSV-1 and HSV-2 found that only one viral product, the non-coding latency-associated transcript (LAT), is abundantly expressed during latency and that its expression is important for establishing and maintaining latency in HSV-1, as well as affecting lytic reactivation in HSV-1 and HSV-2 [53-55]. However, the mechanisms governing LAT function in these processes were not fully understood. Insight into LATs function was recently uncovered by the discovery that LAT encodes a number of viral miRNAs, some of which are located antisense to genes important for lytic reactivation [56-58]. First, Umbach et al. reported that HSV-1 ICP0, an important lytic transcriptional regulator, is targeted by the antisense encoded miR-H2, but curiously it does not mediate antisense cleavage of the ICP0 transcript [58]. In addition to LAT encoded miRNAs, Umbach et al. identified a separately expressed latent miRNA, miR-H6, which targets ICP4, a major viral transactivator. Although HSV-1 and HSV-2 miRNAs are not homologous in sequence, their relative genomic positions are very similar. The HSV-2 miR-lll was found to be positionally conserved to HSV-1 miR-H2, and like miR-H2 it was also found to regulate ICP0 expression [57]. Interestingly, although HSV-2 miR-H6 is positionally conserved to the HSV-1 miR-H6, it does not target or regulate the expression of ICP4 [59, 60]. Lastly, the HSV-2 LAT miRNAs, miR-l and miR-ll, were found in similar locations to the HSV-1 miRNAs, miR-H3 and miR-H4, all being encoded antisense to ICP34.5, a lytic neurovirulence factor [56, 57]. Published reports have shown that HSV-2 miR-1 and miR-ll can target and regulate ICP34.5 while unpublished reports by Tang et al have confirmed targeting by HSV-1 miR-H3 and miR-H4 [56, 57, 60]. The antisense locations and positional conservation of HSV miRNAs indicate that these viruses have co-evolved miRNAs to maintain latency in neuronal cells; however, more studies using miRNA knockout viruses and in vivo models will be required to further delineate the role of specific alphaherpesvirus miRNAs in latency control, reactivation and viral persistence of the infected host. With respect to immune evasion alphaherpesviruses have evolved the perfect “stealth mechanism” in that they do not express any latency-associated proteins that could trigger an immune response but instead seem to establish and maintain latency exclusively by expressing non-immunogenic miRNAs.
The future of viral miRNA research and new emerging questions
Since the discovery of EBV miRNAs in 2004, our advancement in understanding viral miRNA function has been considerably rapid. With hundreds of targets now reported, and many more unreported, the impact that these small RNAs have on virus/host interactions is unquestionable. Recently developed ribonomics-based assays, that combine in vivo UV crosslinking with RISC-specific immunoprecipitation, provide powerful experimental tools to probe for miRNA/mRNA interactions under physiological conditions. Analyzing RISC complexes from virus infected cells will allow to catalogue miRNA/target gene interaction within specific cell types. HITS-CLIP (High throughput sequencing UV cross linking Immunoprecipitation) uses 254 nm UV to directly cross-link RNA protein complexes prior to immunoprecipitation [61]. In a second method, PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation), cells are first labeled with photoreactive nucleoside analogs that are incorporated into nascent mRNAs in living cells [62]. An advantage of PAR-CLIP over conventional HITS-CLIP is that upon cDNA cloning of the recovered RNA, the cross-linking induces base transition, which creates a RISC footprint within the recovered mRNA tag [62].
While these new techniques will allow to determine detailed miRNA targetomes in virally infected cells, determining the importance of each target on for virus biology remains a large task [63, 64]. Furthermore, while the identification of viral miRNA targets is an important step, the use of appropriate model systems (i.e recombinant viruses, in vivo infection models, NK killing assays) are still needed to decipher their functional relevance, especially with respect to immune evasion and other aspects of host/viral interactions. In turn, many new questions (Listed in Table 2) have been raised from published viral miRNA studies. So as new discoveries in viral miRNA function are made new questions emerge, making the complexities governing viral-host interactions, at the least, a little more transparent.
Highlights.
>We review currently known viral miRNA targets. >We examine viral miRNA function in regulating immune evasion. >The future of viral miRNA research is described.
Emerging Questions.
Is polyoma virus miRNA regulation of T-antigen important for immune evasion in vivo?
Does regulation of viral antigens and cytokines by EBV and KSHV miRNAs inhibit immune evasion in vivo?
Are HCMV miRNAs essential for maintaining latent infection?
Most studies show modest inhibition of RTA by KSHV miRNAs, so are miRNAs essential for regulation of latency or do the act more as a secondary mechanism?
Because multiple viral miRNAs can regulate the immune response how do they work synergistically?
How do viral miRNAs work synergistically with viral proteins to regulate the host immune response?
Acknowledgments
Research was supported by R01 CA88763, R01 CA 119917, and RC2CA148407 to R.R. I.B. was supported by the BMID and BEID NIH T32 training programs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mylin LM, Schell TD, Roberts D, Epler M, Boesteanu A, Collins EJ, Frelinger JA, Joyce S, Tevethia SS. Quantitation of CD8(+) T-lymphocyte responses to multiple epitopes from simian virus 40 (SV40) large T antigen in C57BL/6 mice immunized with SV40, SV40 T-antigen-transformed cells, or vaccinia virus recombinants expressing full-length T antigen or epitope minigenes. Journal of virology. 2000;74:6922–6934. doi: 10.1128/jvi.74.15.6922-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 8.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. Journal of virology. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387:157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, Chan AW, Ng EK, Lo KW, To KF. Neoplasia. Vol. 11. New York, N.Y: 2009. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22; pp. 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna R, Burrows SR, Nicholls J, Poulsen LM. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. European journal of immunology. 1998;28:451–458. doi: 10.1002/(SICI)1521-4141(199802)28:02<451::AID-IMMU451>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Meij P, Leen A, Rickinson AB, Verkoeijen S, Vervoort MB, Bloemena E, Middeldorp JM. Identification and prevalence of CD8(+) T-cell responses directed against Epstein-Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. International journal of cancer. 2002;99:93–99. doi: 10.1002/ijc.10309. [DOI] [PubMed] [Google Scholar]
- 14.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nature immunology. 2010;11:806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 16.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, Stern-Ginossar N, Lankry D, Gruda R, Mandelboim O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell host & microbe. 2011;9:93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Qin Z, Kearney P, Plaisance K, Parsons CH. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. Journal of leukocyte biology. 2010;87:25–34. doi: 10.1189/jlb.0409251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirone M, Lucania G, Aleandri S, Borgia G, Trivedi P, Cuomo L, Frati L, Faggioni A. Suppression of dendritic cell differentiation through cytokines released by Primary Effusion Lymphoma cells. Immunology letters. 2008;120:37–41. doi: 10.1016/j.imlet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. lnterleukin-10 and the interleukin-10 receptor. Annual review of immunology. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 22.Mosmann TR. Properties and functions of interleukin-10. Advances in immunology. 1994;56:1–26. [PubMed] [Google Scholar]
- 23.Liang D, Gao Y, Lin X, He Z, Zhao Q, Deng Q, Lan K. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKvarepsilon. Cell research. 2011;21:793–806. doi: 10.1038/cr.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. Journal of virology. 2010;84:12139–12151. doi: 10.1128/JVI.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz M, Murphy PM. Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor constitutively activates NF-kappa B and induces proinflammatory cytokine and chemokine production via a C-terminal signaling determinant. J Immunol. 2001;167:505–513. doi: 10.4049/jimmunol.167.1.505. [DOI] [PubMed] [Google Scholar]
- 26.Sun Q, Matta H, Lu G, Chaudhary PM. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-kappaB activation. Oncogene. 2006;25:2717–2726. doi: 10.1038/sj.onc.1209298. [DOI] [PubMed] [Google Scholar]
- 27.Dolken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 2010;6:e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grey F, Hook L, Nelson J. The functions of herpesvirus-encoded microRNAs. Medical microbiology and immunology. 2008;197:261–267. doi: 10.1007/s00430-007-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. Journal of virology. 2009;83:10684–10693. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. Journal of virology. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. Journal of virology. 2008;82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 39.Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–397. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. Journal of virology. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. Journal of virology. 2007;81:9967–9975. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X, Cullen BR. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. Journal of virology. 2006;80:2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. Journal of virology. 2005;79:14457–14464. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.lizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, Lieberman P, Nishikura K. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12:193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. Journal of virology. 2010;84:2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Oliveira DE, Ballon G, Cesarman E. NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol. 2010;18:248–257. doi: 10.1016/j.tim.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. Journal of virology. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu F, Harada JN, Brown HJ, Deng H, Song MJ, Wu TT, Kato-Stankiewicz J, Nelson CG, Vieira J, Tamanoi F, Chanda SK, Sun R. Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog. 2007;3:e44. doi: 10.1371/journal.ppat.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell host & microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PloS one. 2011;6:e16224. doi: 10.1371/journal.pone.0016224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krause PR, Stanberry LR, Bourne N, Connelly B, Kurawadwala JF, Patel A, Straus SE. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med. 1995;181:297–306. doi: 10.1084/jem.181.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawtell NM, Thompson RL. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. Journal of virology. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson RL, Sawtell NM. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. Journal of virology. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. Journal of virology. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. Journal of virology. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. Herpes Simplex Virus 2 MicroRNA miR-H6 Is a Novel Latency-Associated Transcript-Associated MicroRNA, but Reduction of Its Expression Does Not Influence the Establishment of Viral Latency or the Recurrence Phenotype. Journal of virology. 2011;85:4501–4509. doi: 10.1128/JVI.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M, Lieber D, Bailer SM, Hoffmann R, Ruzsics Z, Kremmer E, Pfeffer S, Zimmer R, Koszinowski UH, Grasser F, Meister G, Haas J. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell host & microbe. 2010;7:324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Malterer G, Dolken L, Haas J. The miRNA-targetome of KSHV and EBV in human B-cells. RNA biology. 2011;8 doi: 10.4161/rna.8.1.13745. [DOI] [PubMed] [Google Scholar]