SUMMARY
Immunological memory in vertebrates is often exclusively attributed to T and B cell function. Recently it was proposed that the enhanced and sustained innate immune responses following initial infectious exposure may also afford protection against reinfection. Testing this concept of “trained immunity,” we show that mice lacking functional T and B lymphocytes are protected against reinfection with Candida albicans in a monocyte-dependent manner. C. albicans and fungal cell wall β-glucans induced functional reprogramming of monocytes, leading to enhanced cytokine production in vivo and in vitro. The training required the β-glucan receptor dectin-1 and the noncanonical Raf-1 pathway. Monocyte training by β-glucans was associated with stable changes in histone trimethylation at H3K4, which suggests the involvement of epigenetic mechanisms in this phenomenon. The functional reprogramming of monocytes, reminiscent of similar NK cell properties, supports the concept of “trained immunity” and may be employed for the design of improved vaccination strategies.
INTRODUCTION
It is a general assumption that immunological memory is exclusively mediated by T and B cells. However, protection against reinfection is present in both plants (Durrant and Dong, 2004) and insects (Pham et al., 2007; Rodrigues et al., 2010) who lack specific immunity. Similarly, prototypic mammalian innate immune cells such as NK cells can build immunological memory, leading to protection against reinfection with viral pathogens (Cooper et al., 2009; O’Leary et al., 2006; Paust et al., 2010; Sun et al., 2009). We proposed the term “trained immunity” for the enhanced state of the innate immune responses following exposure to certain infectious agents, which may result in an increased resistance to reinfection (Netea et al., 2011).
Protection from infection in a macrophage-dependent manner was demonstrated after a previous infection with avirulent Candida species in mice deficient in T and B cells (Bistoni et al., 1986), yet the mechanism responsible for this effect remained unknown. Similarly, β-glucans from C. albicans cell wall are potent immunomodulators and are used in some countries for enhancing immunity against infections and cancer (Brown and Williams, 2009; Vetvicka, 2011).
The aim of the present study was to investigate the “training” effects exerted by C. albicans on monocytes and macrophages, the cells responsible for the T/B cell-independent protective effect during secondary Candida infection (Bistoni et al., 1986). We demonstrate that C. albicans can protect Rag1-deficient mice from reinfection, an effect that is dependent on the functional reprogramming of monocytes. The mechanism of training requires the β-glucan receptor dectin-1 and the noncanonical Raf-1 pathway and is associated with stable and genome-wide changes in histone methylation, suggesting a putative role of epigenetic programming as an underlying mechanism.
RESULTS
A Nonlethal Candida albicans Infection Induces Protection from Secondary Lethal Infection through a T/B Lymphocyte-Independent, Monocyte-Dependent Mechanism
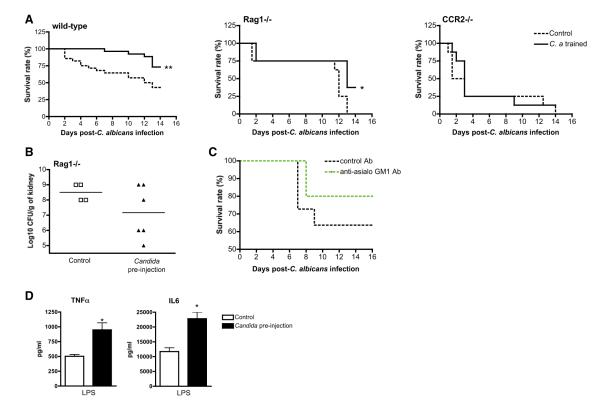
Preinjection of a low dose of live C. albicans 7 days prior to a second lethal infection increased the survival rate of wild-type (WT) mice (Figure 1A). The organ fungal loads and circulating cytokine concentrations were not measurable on day 7 after injection of the low inoculum of Candida (data not shown). The protection given by preinfection was also conferred to T/B cell-defective Rag1-deficient mice (Figures 1A and 1B), but not to monocyte-defective Ccr2-deficient mice (Figure 1A). The protective effect of Candida preinfection observed in mice treated with anti-asialo GM1 antibodies (depletion from 7.5% to 0.6%) was independent of the presence of NK cells (data not shown). Moreover, mice depleted of NK cells had a better survival than the mice treated with an unrelated antibody (Figure 1C), making it highly unlikely that NK cells play an important role in the protection. These results highlight the importance of monocytes in the C. albicans-conferred protection.
Figure 1. Nonlethal Candidiasis Protects Mice through a Macrophage-Dependent Mechanism.
(A) Survival rate of WT, Rag1−/−, and Ccr2−/− mice to systemic candidiasis, following vaccination with PBS (control) or a nonlethal dose of live C. albicans (Candida, preinjection) 7 days earlier (n ≥ 8 per group, two independent experiments).
(B) Fungal burdens in kidneys from Rag1−/− mice 14 days after C. albicans infection.
(C) Survival rate of WT mice preinjected with anti-asialo GM1 antibody or an unrelated antibody (control) prior to the inoculation of lethal C. albicans (n ≥ 10 per group).
(D) In vivo training of mice (n = 5 per group) with heat-inactivated C. albicans (black bars), 7 days prior to the LPS injection, enhanced proinflammatory cytokines release compared to PBS-treated control mice (white bars). *p < 0.05 versus control animals.
To assess the effect of monocyte training on cytokine production, we investigated the effect of Candida pretreatment in an endotoxemia model. Induction of cytokines by lipopolysaccharide (LPS) was enhanced when mice were pretreated 1 week earlier with low amounts of heat-killed C. albicans (Figure 1D).
Candida albicans Primes the Production of Proinflammatory Cytokines in Monocytes
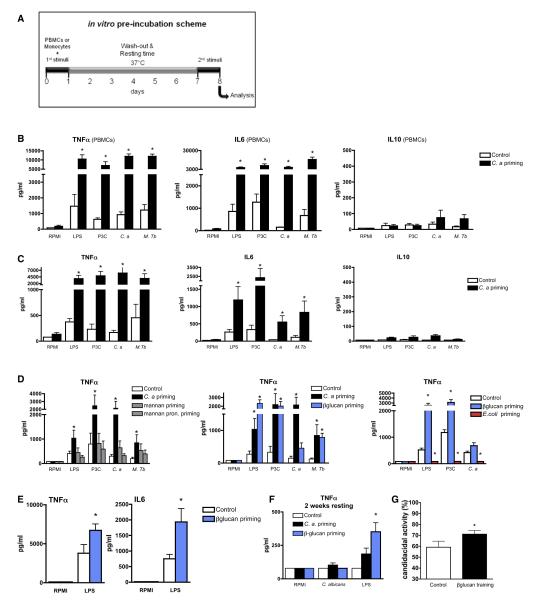
Mononuclear phagocytes are the main cell population responsible for proinflammatory cytokine production during infection. We assessed whether priming of human peripheral blood mononuclear cells (PBMCs) in vitro with a low dose of C. albicans would increase the response of cells to a secondary stimulation (Figure 2A). Preincubation of cells with 104 microorganisms/ml heat-killed Candida did not induce cytokine production after 24 hr (data not shown). In contrast, preincubation of cells with C. albicans increased the concentration of TNF-α and IL-6 in the supernatant after the secondary stimulation with C. albicans, as well as with various pattern recognition receptors (PRRs), ligands, or bacteria (Figure 2B).
Figure 2. Candida albicans and β-Glucans Prime the Production of Proinflammatory Cytokines.
(A) Diagram showing the course of the in vitro preincubation experiment.
(B and C) C. albicans training in vitro using freshly isolated human PBMCs (B) or monocytes (C) and different PRR ligands for restimulation (P3C, Pam3ys; C.a, C. albicans; M.tb, Mycobacteria tuberculosis).
(D) The training effects induced by purified β-glucans (blue), but not with mannans (gray). E. coli preincubation induced immune tolerance (red).
(E) CD14+-selected monocytes are also primed by β-glucan.
(F) The priming effect obtained after 1 week of resting period was also seen after 2 weeks.
(G) Candidacidal activity of cell culture medium (control) or β-glucans-preincubated monocytes was assessed after 5 hr of incubation with live opsonized C. albicans (n = 6). In (B)–(G), *p < 0.05 (mean ± SD, n = 5–8). See also Figure S1.
To rule out a role for lymphocytes in this effect, we repeated these experiments with purified monocytes. Exposure of monocytes to Candida for 24 hr primed the production of the proinflammatory cytokines TNF-α and IL-6, but not of the anti-inflammatory IL-10 (Figure 2C). Cell viability was identical in all conditions and above 98% (LDH release). Microscopic assessment showed no remaining intact Candida during the stimulation period.
β-Glucans Enhance the Proinflammatory State of Monocytes and Prime the Production of Proinflammatory Cytokines
β-glucans and mannans are major components of the skeletal cell wall of C. albicans (Netea et al., 2008). The priming effects induced by C. albicans could be reproduced with purified β-glucan, but not with mannans or Toll-like receptor (TLR) ligands (Figure 2D and data not shown) (Foster et al., 2007). Similarly, pretreatment of either positively selected CD14+ monocytes or CD4+ T lymphocyte-depleted PBMCs led to similarly enhanced production of proinflammatory cytokines (Figure 2E and Figure S1A). In contrast to the monocytes, pretreatment of CD56+-isolated NK cells with either C. albicans or β-glucans failed to induce an enhanced production of proinflammatory cytokines (data not shown).
While the most robust effect was seen after 24 hr of preincubation, a priming effect could be detected also when monocytes were preincubated for shorter periods of time (Figure S1B). Moreover, preincubation during 24 hr with C. albicans or β-glucans enabled monocytes to enhance their production of cytokines for 2 weeks after the training period (Figure 2F and Figure S1C). Incubation longer than 2 weeks was not achievable due to declining viability of monocytes thereafter. In addition, the enhanced production of proinflammatory cytokine by trained monocytes upon a second stimulation was dose dependent (Figure S1D). Interestingly, the initial dose of C. albicans and of β-glucan also influenced the amplitude of the training effect in a dose-dependent manner (Figure S1E).
In addition, 24 hr preincubation with β-glucans enabled monocytes to enhance their candidacidal activity (Figure 2G) and to efficiently inhibit the C. albicans outgrowth after 24 hr of culture (47% ± 9% more inhibition compared to nonprimed control monocytes).
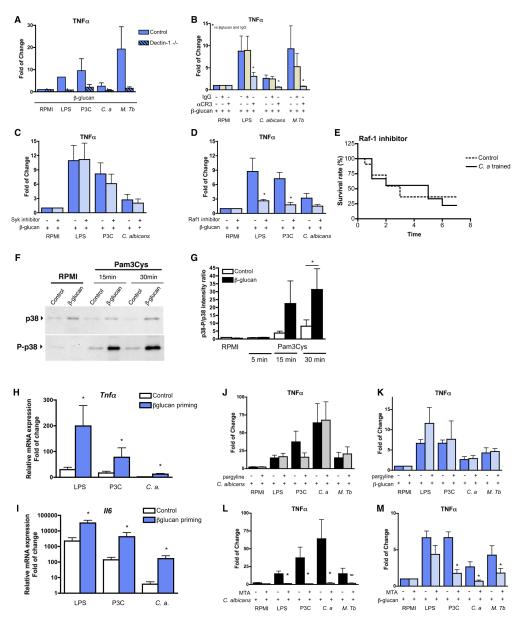
The Dectin-1/Raf-1 Pathway and the Complement Receptor 3 Are Required to Induce Training of Monocytes
The recognition of β-glucan by human cells is dependent on dectin-1 and complement receptor 3 (CR3) (Netea et al., 2008). Inhibition of dectin-1 with laminarin inhibited the priming by β-glucans (Figure S2), and monocytes isolated from a dectin-1-deficient individual (Ferwerda et al., 2009) could not be effectively trained with β-glucan (Figure 3A). Only a partial inhibition of the training of monocytes was observed when the CR3 receptor was blocked (Figure 3B). Dectin-1 activates cellular responses through the spleen tyrosine kinase and caspase-recruitment domain protein 9 (Syk/CARD9) (Drummond et al., 2011) and through the noncanonical serine-threonine kinase Raf-1 pathway (Gringhuis et al., 2009). Incubation of monocytes with a Raf-1 inhibitor, but not a Syk inhibitor, prior to priming significantly reduced the training by β-glucans (Figures 3C and 3D). Moreover, in vivo inhibition of Raf-1 in mice during the training abolished the protection conferred by the Candida preinjection (Figure 3E).
Figure 3. The Role of β-Glucan Receptors for Monocyte Training.
(A–D) Cytokine production in supernatants of adherent monocytes primed for 24 hr with either cell culture medium or β-glucans and restimulated with different PRR ligands in healthy volunteers (control) and cells isolated from a dectin-1-deficient volunteer (A), in the presence or absence of anti-CR3 antibody (B), and in the presence or absence of Syk kinase inhibitor (C) or Raf-1 inhibitor GW5074 (D).
(E) Survival rate of WT mice to C. albicans infection. All mice were initially treated with a Raf-1 inhibitor. Mice were then preinjected with either PBS (control) or a nonlethal dose of live C. albicans (Candida, preinjection) 7 days prior to inoculation of the lethal C. albicans dose (n ≥ 10 per group).
(F) Western blot of total and phosphorylated p38 in cells primed with RPMI (controls) or β-glucan and subjected to 5, 15, or 30 min stimulation with RPMI or Pam3Cys.
(G) Quantification of the effect of β-glucan priming on the phosphorylation of p38.
(H and I) Fold changes in Tnfa (H) and Il6 (I) mRNA expression obtained by quantitative real-time PCR in adherent monocytes after stimulation with LPS, Pam3Cys, or C. albicans. Cells were primed with either nutrient-rich medium (RPMI, control) or with β-glucan.
(J–M) Cytokine production in supernatants of adherent monocytes primed 24 hr with either cell culture medium or C. albicans (J and L) or β-glucans (K and M) in the absence or presence of the histone demethylase inhibitor pargyline (J and K) and the histone methyltransferase inhibitor MTA (5′-deoxy-5′(methylthio) adenosine) (L and M) and restimulated with different PRR ligands. In (A)–(D) and (J)–(M), the ratios of cytokine production by β-glucan primed versus nonprimed monocytes are presented. Data are shown as mean ± SD. n = 6 for (B), (D), and (J)–(M) and n = 5 for (C). Data presented in (A) are means of two independent experiments. n ≥ 6 for (H) and (I). *p < 0.05. See also Figure S2.
Molecular Regulation of Trained Monocytes
Cellular response to extracellular stimuli is mediated through intracellular signaling cascades such as the p38 MAPK (Zarubin and Han, 2005). Both the expression of p38 and the p38 phosphorylation (i.e., activation) upon TLR2 stimulation were stronger in the monocytes primed with β-glucan than in the controls (Figures 3F and 3G). Subsequently, increased gene transcription of Tnfα and Il6 mRNA upon restimulation was observed (Figures 3H and 3I). The long-lasting effect of the β-glucan pretreatment on gene expression prompted us to investigate whether epigenetic changes such as histone methylation (Carson et al., 2011; Foster et al., 2007; Ishii et al., 2009) are associated with the training of monocytes. Inhibition of histone demethylases by a specific inhibitor had no effect on the training of monocytes (Figures 3J and 3K). In contrast, inhibition of histone methyltransferases using MTA inhibited monocyte training by C. albicans or β-glucan (Figures 3L and 3M), supporting the hypothesis that histone methylation is likely to be involved in the training of monocytes.
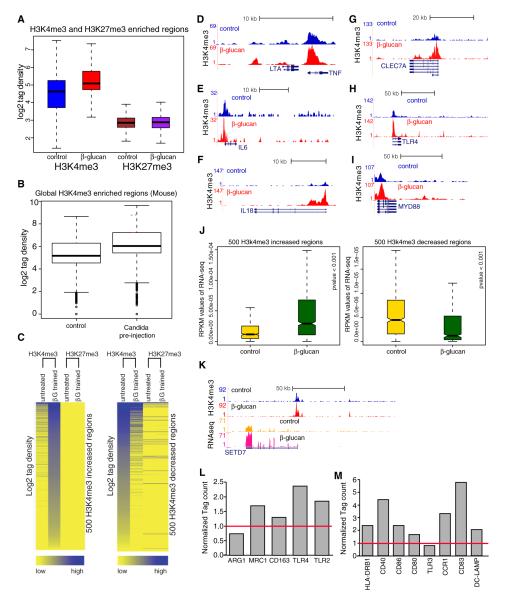
To confirm this hypothesis, we assessed the genome-wide binding pattern upon β-glucan training of H3K4me3 and H3K27me3, two histone modifications associated, among others, with the epigenetic regulation of immune-related genes (Barski et al., 2007; Buratowski and Kim, 2010; Foster et al., 2007; Ishii et al., 2009; Mikkelsen et al., 2007; Min et al., 2011; Sims et al., 2007; Wen et al., 2008). The two independent genome-wide ChIP-seq data in monocytes showed well-defined H3K4me3 peaks, while H3K27me3 was present in more diffuse blocs (Figure S3A) (Martens et al., 2010). A global increase was seen in H3K4me3 signal in monocytes after β-glucan treatment, while hardly any changes were observed in H3K27me3 regions (Figure 4A).
Figure 4. β-Glucan Alters the Epigenetic Landscape in Trained Monocytes, and This Correlates with β-Glucan-Induced Transcriptomal Changes.
(A) Box plot of log2 tag density of H3K4me3 and H3K27me3 signal in control versus trained monocytes.
(B) Box plot of log2 tag density for genome-wide H3K4me3 enriched regions in peritoneal macrophages retrieved from untreated (control, saline) and Candida preinjected mice.
(C) Intensity plots showing the normalized tag density for H3K4me3 and H3K27me3 on the 500 H3K4me3 enhanced and decreased regions.
(D–I) Genome browser screen shot showing H3K4me3 binding over TNF-α (D), IL-6 (E), IL-18 (F), DECTIN-1 (G), TLR4 (H), and MYD88 promoter regions (I) before and after the treatment with β-glucan.
(J) Box plot showing RNA expression (RPKM) of the genes associated with top and bottom 500 regions with H3K4me3 changes.
(K) H3K4me3 tag density as well as RNA-seq expression data over SETD7 histone methyltransferase before and after the treatment with β-glucan.
(L and M) RPKM values of genes after β-glucan treatment of monocytes. The genes are markers of classically and alternatively activated macrophages (L) and of inflammatory Tip-DCs (M). See also Figure S3.
To validate the physiological relevance of the β-glucan-induced epigenetic modifications observed in vitro, we performed genome-wide ChIP-seq assay for H3K4me3 in peritoneal macrophages recovered from WT mice 7 days after injection of either saline (control) or a low dose of C. albicans, showing a global increase in H3K4me3 signal in macrophages after C. albicans training (Figure 4B). Genome-wide ChIP-seq data showed well-defined H3K4me3 peaks (Figures S3B and S3C), and an increase in H3K4me3 was observed at the level of the promoter of genes associated with immune signaling pathway (e.g., Myd88, Figure S3B), or associated with the training mechanism (e.g., Raf-1, Figure S3C).
The H3K4me3 peak regions were ranked on the basis of signal changes between nontrained control and β-glucan-trained monocytes in vitro (Figure S3D). We separated 500 peak regions that showed the highest increase in H3K4me3 occupancy after β-glucan treatment (Figure S3E) and 500 peak regions that showed decrease in H3K4me3 levels after treatment (Figure S3F). The normalized tags’ density in the separated peak regions revealed significant changes for H3K4me3 levels, with fewer alterations for H3K27me3 between trained and nontrained monocytes (Figure 4C). More specifically, H3K4me3 was elevated at the promoters of important target genes such as the proinflammatory cytokines TNF-α, IL-6, and IL-18 after β-glucan treatment (Figures 4D and 4F). Interestingly, β-glucan treatment of monocytes also upregulated H3K4me3 at the promoter of DECTIN-1 (3.3-fold, Figure 4G), but also at the promoters of other C-type lectin receptors and TLRs (Figure 4H). Finally, training the monocytes with β-glucan increased the H3K4me3 at the promoter of intracellular signaling molecules, such as the adaptor molecule MYD88 (Figure 4I).
Additional genome-wide H3K4me3 ChIP-seq were performed before exposure (day 0) and 24 hr after exposure (day 1) to either cell culture medium or β-glucan. Hierarchical clustering of the top and bottom 500 regions with H3K4me3 variations at day 7 (Figure 4C) revealed that many changes noted in day 7 samples were already apparent after 24 hr of β-glucan incubation (Figure S3G). This indicates that a stable, β-glucan-induced epigenetic program could be maintained for at least a week (Figure S3H).
Transcriptome Changes in Trained Monocytes
Two independent genome-wide RNA-sequencing experiments revealed global changes in gene expression upon β-glucan training, with more than 5,800 upregulated and 1,400 downregulated genes (2-fold threshold). Analysis confirmed a strong correlation between the increase in H3K4me3 occupancy and the increase in expression (Figure 4J). Importantly, β-glucan also induced increased gene expression of several histone methyltransferases after training, including SETD7, which has been previously related to H3K4me3 (Nishioka et al., 2002) (Figure 4K).
Activation Markers in Trained Monocytes
Monocytes differentiate into classically activated (M1) or alternatively activated (M2) macrophages (Gordon, 2003; Mantovani et al., 2004). Transcriptome analysis of β-glucan-trained monocytes showed increased RNA level for TNF-α and IL-6 (Figures 3H and 3I) and for TLR2 and TLR4, but less for ARG-1 (Figure 4L). However, the M2 markers MRC1 and CD163 mRNA were also upregulated in the trained monocytes. Beside macrophages, monocytes can also differentiate in dendritic cells (DCs) such as TNF and iNOS producing DCs (Tip-DC) (Aldridge et al., 2009; Serbina et al., 2003). Except for TLR3, transcriptome analysis of β-glucan-trained monocytes also showed an increased RNA level for most of the costimulatory molecules and chemokine receptors associated with Tip-DC phenotypes (Figure 4M). Thus, β-glucan-trained monocytes display increased activation markers for M1 and M2 macrophages, as well as for Tip-DC, arguing against simple skewing of macrophage differentiation and more likely for a global pan-activation of the cells.
DISCUSSION
The nonspecific adaptive features of innate immunity (Bowdish et al., 2007) that have been demonstrated in plants, invertebrates, and mice and termed “trained immunity” (Netea et al., 2011) have not been investigated mechanistically in mammalian cells. In the present study, we demonstrate that monocytes can be trained to exhibit an enhanced and lasting response to microbial components after pre-exposure to C. albicans or β-glucans. This effect was mediated by a dectin-1 receptor/Raf-1 pathway, leading to more rapid and effective activation of signaling molecules such as p38. Monocyte training leads to enhanced gene transcription, which correlates with epigenetic changes at the level of H3K4me3, but not of H3K27me3, in innate immunity genes. This combination of signaling and molecular events leads to enhanced production of proinflammatory cytokines and candidacidal properties.
An important question concerns the molecular mechanism responsible for monocyte reprogramming. First, signaling pathways important for inflammation such as p38 activation are increased (Zarubin and Han, 2005). Second, the trimethylation profile at the level of H3K4 is enhanced. The combination of in vivo and in vitro data suggests that these changes may indeed represent an important component underlying training of monocytes. This observation is reminiscent of previously reported epigenetic mechanisms mediating LPS- and TNF-induced tolerance (Foster et al., 2007). Additionally, chromatin remodeling has been shown to be a key mechanism in the regulation of the polarization toward classically and alternatively activated macrophage subtypes (Ishii et al., 2009; Takeuch and Akira, 2011).
The identification of adaptive features for the innate immune responses induced in mononuclear phagocytes has important conceptual and practical consequences. Recently, NK cells, another prototypical type of innate immune cells, have been demonstrated to possess memory characteristics (O’Leary et al., 2006; Paust et al., 2010; Sun et al., 2009). The observation that NK cells and mononuclear phagocytes share properties of nonspecific immunological memory (or can be “trained”) strongly suggests that these features are a fundamental component of host defense (Netea et al., 2011). At this moment we have little information on the duration of the protection that can be provided by trained immunity, even though epigenetic modifications have been reported to correlate with long-term maintained mechanisms (Margueron et al., 2005; Wen et al., 2008). The conferred protection might be quite long, as epidemiologic studies in children have shown that BCG vaccination exerts nonspecific protection of children from nonmycobacterial infections for at least the duration of early childhood (Garly et al., 2003; Rosenthal et al., 1961). If demonstrated to be long-lived, it is to be expected that trained immunity will have important consequences for the design of vaccination strategies that may exploit both trained immunity and classical T/B lymphocyte-immunological memory.
One may also speculate about the evolutionary aspects of Candida-induced trained immunity. The observation that C. albicans can prime and enhance the proinflammatory immune response of monocytes may represent a possible reason why C. albicans became a commensal in humans (Pfaller and Diekema, 2007). It is tempting to hypothesize that C. albicans not only colonizes, but also trains innate immune cells at mucosal barriers, thereby providing enhanced immune control at the sites where the host is most often in contact with potentially dangerous pathogens. However, this enhanced innate immune response might be harmful in individuals with genetic predispositions for autoimmune or autoinflammatory conditions such as Crohn’s disease. In this respect, it is interesting to observe that Crohn’s disease is characterized by the presence of specific anti-Candida antibodies (McKenzie et al., 1990; Sutton et al., 2000).
In conclusion, in the present study we demonstrate that C. albicans, and β-glucans in particular, induce epigenetic reprogramming of monocytes through a dectin-1/Raf-1 pathway, resulting in enhanced cytokine production. The epigenetic modulation of innate immune responses is likely to represent an important characteristic of the protection against reinfection conferred by what it has been recently described as “trained immunity.”
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J, B6.129S7-Rag1tm1Mom/J, and B6.129S4-Ccr2tm1lfc/J female mice (8–12 weeks) were used (Jackson Laboratories). Experiments were approved by the Ethics Committee on Animal Experiments of the University of Athens. Mice were injected with live C. albicans blastoconidia (2 × 104 cfu/mouse) or pyrogen-free phosphate-buffered saline (PBS) alone. Seven days later, mice were infected intravenously with a lethal dose of live C. albicans (2 × 106 cfu/mouse). Survival was monitored and kidney fungal burden assessed on day 14. Raf-1 activation was inhibited by 25 mg/kg Sorafenib orally administered 7 days before and on the day of the low dose of C. albicans blastoconidia injection. In the endotoxemia model, mice were injected intravenously with heat-inactivated C. albicans blastoconidia (1 × 105 cfu/mouse) or PBS alone and 7 days later with an intraperitoneal dose of 10 μg purified LPS (E. coli serotype 055:B5, Sigma-Aldrich). Blood was collected 90 min later to assess the circulating TNF-α and IL-6 concentrations.
Healthy Volunteers
PBMCs were isolated from buffy coats obtained from healthy volunteers (Sanquin Bloodbank, Nijmegen, the Netherlands).
Reagents
Ficoll-Paque (GE Healthcare) was used to isolate PBMCs by differential centrifugation. CD14+ monocytes, CD56+ NK cells, and CD4− subsets were purified using MACS isolation.
Candida albicans mannans and β-1,3-(D)-glucan (β-glucan) were kindly provided by Professor David Williams. Reagents used were as follows: Pam3Cys (EMC microcollections, L2000), LPS (Sigma-Aldrich, E. coli serotype 055:B5), Laminarin (Sigma-Aldrich, L-9634), α-CR3 (R&D, AF 1730), isotype control goat IgG (R&D, AB-108-c), Syk inhibitor (EMD, 574711), Raf-1 inhibitor (GW5074, Sigma-Aldrich, 6416), histone demethylase inhibitor Pargyline (Sigma-Aldrich, P8013), and histone methyltransferase inhibitor (MTA, Sigma-Aldrich, D5011).
Microorganisms
C. albicans ATCC MYA-3573 (UC 820) and E. coli strain ATCC35218 were heat-inactivated for 30 min at 95°C. M. tuberculosis H37Rv suspensions were sonicated for 10 min on ice.
Stimulation Experiments
PBMCs (5 × 105 cells) were incubated for 1 hr at 37°C in 5% CO2 and adherent monocytes were selected by washing out nonadherent cells with warm PBS. CD14+-purified monocytes were diluted to a concentration of 1 × 105/100 μl. For training, cells were preincubated with heat-killed C. albicans (1 × 104/ml), β-glucan (10 μg/ml), mannans (10 μg/ml), or E. coli (1 × 106/ml) for 24 hr. After 7 days cells were stimulated with various stimuli: LPS, 10 ng/ml; Pam3Cys, 10 μg/ml; heat-killed C. albicans, 1 × 105/ml; M. tuberculosis, 1 μg/ml. After 24 hr, supernatants were collected and stored at −20°C.
For inhibition, before priming with C. albicans or with β-glucan, monocytes were preincubated for 1 hr with laminarin (dectin-1 inhibitor, 100 μg/ml), anti-CD3 antibody and control anti-IgG (10 μg/ml), Syk inhibitor (50 nM), Raf-1 inhibitor (1 μM), Pargyline (3 μM), and MTA (1 mM). TNF-α, IL-6, and IL-10 were measured using ELISA (IL-6, IL-10: Sanquin; TNF-α: R&D).
Quantitative RT-PCR
Isolated RNA was reverse-transcribed into complementary DNA using iScript cDNA Synthesis Kit (Bio-Rad), and quantitative PCR was performed as described in the Supplemental Information.
Chromatin Immunoprecipitation
For chromatin immunoprecipitation (ChIP), adherent monocytes were cultured as described in the Supplemental Information. For the mouse samples, mice were injected intraperitoneally with either PBS or 5 × 103 C. albicans 7 days prior the collection of the peritoneal macrophages. ChIP was performed using antibodies against H3K4me3 (Diagenode) and H3K27me3 (Millipore). ChIPed DNA was further processed for high-throughput sequencing using Illumina Genome Analyzer II as described before (Martens et al., 2010). High-quality sequenced reads were uniquely mapped to human hg18 and mouse mm9 using ELAND.
For H3K4me3 data sets, peaks were called using “Genomatix” tool NGS from RegionMiner ChIP-seq workflow. Individual peaks within 500 bp region were further merged. For H3K27me3 enriched blocs, peaks were called as previously described (Pauler et al., 2009). Biological replicates were produced for all conditions. For the differential, regions tags were counted in merged peak file from both trained and nontrained (RPMI control) monocytes and to rank peaks as a function of the magnitude of change, the difference for each peak in fractional tag count between control and trained condition was divided by the difference between 1 and their sum (fT – fC)/(1 – fT – fC).
Genome Annotation
Genes were annotated for the top 500 increased and 500 decreased regions in H3K4me3 (http://pinkthing.cmbi.ru.nl/cgi-bin/index52.pl).
RNA-Seq
RNA was processed as described in Supplemental Information. Briefly, double-stranded cDNA were used for Illumina sample prepping and sequenced. RNA-seq reads were uniquely mapped to hg18 and used for bioinformatic analysis. RPKM (reads per kilobase of gene length per million reads) values for RefSeq genes were computed using tag counting scripts and used to analyze the expression level of the genes in nontreated (RPMI) and β-glucan-trained monocytes. Biological replicates were sequenced and processed in the same manner.
Western Blots
For western blotting of p38 MAPK (total and phosphorylated), training was performed as described in stimulation experiments. A detailed western blot methodology is described in Supplemental Information.
Candidacidal Activity
Priming of the cells was performed in 96-well plates as described. A total of 1 × 104 opsonized C. albicans (see Supplemental Information) were incubated together with RPMI-primed monocytes, β-glucan-primed monocytes, or no cells (control well) for 5 and 24 hr at 37°C. After the incubation time, the content of each well was recovered in sterile water, serially diluted, and plated on Sabouraud plates. After 24 hr at 29°C, the cfu were counted. The candidacidal activity was calculated as a ratio of Candida growth in RPMI- or β-glucan-primed cells versus control wells (Candida alone, no cells).
Statistical Analysis
The differences between groups were analyzed using the Wilcoxon signed-rank test (unless otherwise stated). Statistical significance of survival experiment was calculated using the product limit method of Kaplan and Meier. The level of significance was defined as a p value of < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
J.Q. and M.G.N. were supported by a Vici Grant of the Netherlands Organization for Scientific Research (to M.G.N.). S.S. was supported by the Higher Education Commission of Pakistan and J.H.A.M. by a Vidi Grant of the Netherlands Organization for Scientific Research. D.C.I. was supported by the All-Fun EU-FP7 grant. C.L. was supported by the “DNA-in-action” consortium, Foundation for Fundamental Research on Matter (FOM), Netherlands Organisation for Scientific Research (NWO). R.J.X. was supported by grants (AI 062773, DK 043351 DK 83756) from the US National Institutes of Health and the Helmsley Trust. J.Q., S.S., C.W., J.H.A.M., C.L., J.W.M.v.d.M., and M.G.N. designed and analyzed the experiments. J.Q., S.S., M.G.N., E.J.G.-B., L.A.B.J., and T.J. performed the experiments. D.C.I. contributed to some of the experiments. J.Q., S.S., R.J.X., J.W.M.v.d.M., H.G.S., and M.G.N. wrote the manuscript, and all authors contributed to the manuscript preparation. M.G.N. supervised the project.
Footnotes
ACCESSION NUMBERS Data are available through the GEO database (GSE34260).
SUPPLEMENTAL INFORMATION Supplemental Information includes three figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2012.06.006.
REFERENCES
- Aldridge JR, Jr., Moseley CE, Boltz DA, Negovetich NJ, Reynolds C, Franks J, Brown SA, Doherty PC, Webster RG, Thomas PG. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. USA. 2009;106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bistoni F, Vecchiarelli A, Cenci E, Puccetti P, Marconi P, Cassone A. Evidence for macrophage-mediated protection against lethal Candida albicans infection. Infect. Immun. 1986;51:668–674. doi: 10.1128/iai.51.2.668-674.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish DM, Loffredo MS, Mukhopadhyay S, Mantovani A, Gordon S. Macrophage receptors implicated in the “adaptive” form of innate immunity. Microbes Infect. 2007;9:1680–1687. doi: 10.1016/j.micinf.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Brown GD, Williams DL. (1,3)-β-Glucans in Innate Immunity: Mammalian Systems. In: Bacic A, Fincher GB, Stone BA, editors. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides. Academic Press; Amsterdam: 2009. pp. 579–619. [Google Scholar]
- Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harb. Symp. Quant. Biol. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson WF, Cavassani KA, Dou Y, Kunkel SL. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics. 2011;6:273–283. doi: 10.4161/epi.6.3.14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Saijo S, Iwakura Y, Brown GD. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur. J. Immunol. 2011;41:276–281. doi: 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl. J. Med. 2009;361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Garly ML, Martins CL, Balé C, Baldé MA, Hedegaard KL, Gustafson P, Lisse IM, Whittle HC, Aaby P. BCG scar and positive tuberculin reaction associated with reduced child mortality in West Africa. A nonspecific beneficial effect of BCG? Vaccine. 2003;21:2782–2790. doi: 10.1016/s0264-410x(03)00181-6. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Wevers B, Bruijns SC, Geijtenbeek TB. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat. Immunol. 2009;10:203–213. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WF, 4th, Cavassani KA, Li X, Lukacs NW, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, Altucci L, Stunnenberg HG. PML-RARalpha/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia. Cancer Cell. 2010;17:173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- McKenzie H, Main J, Pennington CR, Parratt D. Antibody to selected strains of Saccharomyces cerevisiae (baker’s and brewer’s yeast) and Candida albicans in Crohn’s disease. Gut. 1990;31:536–538. doi: 10.1136/gut.31.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal SR, Loewinsohn E, Graham ML, Liveright D, Thorne MG, Johnson V. BCG vaccination in tuberculous households. Am. Rev. Respir. Dis. 1961;84:690–704. doi: 10.1164/arrd.1961.84.5P1.690. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton CL, Yang H, Li Z, Rotter JI, Targan SR, Braun J. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies in affected and unaffected relatives of patients with Crohn’s disease. Gut. 2000;46:58–63. doi: 10.1136/gut.46.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuch O, Akira S. Epigenetic control of macrophage polarization. Eur. J. Immunol. 2011;41:2490–2493. doi: 10.1002/eji.201141792. [DOI] [PubMed] [Google Scholar]
- Vetvicka V. Glucan-immunostimulant, adjuvant, potential drug. World J Clin Oncol. 2011;2:115–119. doi: 10.5306/wjco.v2.i2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111:1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.