Abstract
Disturbances of circadian rhythms and mammalian clock genes have been implicated in the etiologies of many chronic illnesses, including cancer. We show that transcription factor CCAAT/enhancer-binding protein alpha (C/EBPalpha)-regulated PER2 activation is a potential tumor suppressor pathway in diffuse large B-cell lymphoma (DLBCL), one of the commonest types of mature B-cell lymphoma. Expression analysis of human B-cell lymphoma samples including DLBCL (n = 50), mantle cell (n = 21), follicular (n = 25) and Burkitt (n = 18) lymphoma revealed markedly down-regulated CEBPA and PER2 mRNA levels exclusively in DLBCL samples compared to control lymphatic tissue. We demonstrated direct regulation of the circadian core clock gene PER2 by C/EBPalpha in the pro-B cell line Ba/F3, and forced expression of PER2 resulted in decreased proliferation, GO/G1 cell cycle arrest and increased rates of apoptosis. Interestingly, treatment of human DLBCL cell lines with the histone deacetylase-inhibitor suberoylanilide hydroxamic acid (SAHA) significantly increased the expression of C/EBPalpha and Per2, accompanied by cell growth inhibition; in contrast, siRNA knockdown of CEBPA reduced the anti-proliferative effect of SAHA treatment. Our results show for the first time that C/EBPalpha with its associated direct core clock gene target, PER2, are highly deregulated in DLBCL, suggesting an important tumor suppressive pathway in the pathogenesis of this lymphoma entity.
Keywords: Transcription factor, circadian rhythm, mature B-cell lymphoma
Introduction
Mature B-cell neoplasm represents a heterogeneous group of lymphomas according to the 2008 World Health Organization (WHO) classification, with a male predominance and worldwide steadily increasing incidence [1–3]. Certain endemic geographical factors appear to influence the development of B-cell neoplasms in specific areas; for example, in Africa, endemic Burkitt lymphoma (BL) accounts for a substantial proportion, whereas follicular lymphoma (FL) is more common in North America and Europe [3,4]. Diffuse large B-cell lymphoma (DLBCL) is one of the most common among adult lymphomas, and, despite increasing understanding of lymphoma pathogenesis in recent years, with an unknown etiology in most cases [4]. Nonetheless, progress has been made in improving survival for patients with DLBCL, with novel combinations of chemotherapy and immunotherapy, including addition of the monoclonal antibody, rituximab [4].
Circadian rhythms are recurring fluctuations with a period of about 24 h that can be observed in the mammalian physiology and behavior [5–7]. Circadian oscillation of clock controlled gene expression is mainly regulated at the transcriptional level. Heterodimers of Clock and Bmal1 act as activators of target gene transcription; however, interactions of Per and Cry proteins with the heterodimer abolish its transcriptional activation capacity. Per and Cry are, therefore, referred to as negative regulators of the circadian clock [8,9]. Overall, the universal circadian rhythm influences many fundamental and cancer-related biological processes, and alterations of circadian rhythm may cause abnormal immune cell trafficking and cell proliferation cycles [10–12]. We have previously shown that PER2 is a downstream CCAAT-enhancer-binding protein (C/EBP)-target gene, and its disruption might be involved in the initiation and progression of acute myelogenous leukemia (AML) [13]. In addition, recent findings provide molecular and epidemiologic evidence supporting a role of an aberrant circadian rhythm in lymphomagenesis [14– 16]. Therefore, exploration of core clock genes might further add important insights into the role of the circadian rhythm in mature B-cell lymphoma and facilitate the development of novel therapeutic biomarkers.
In the present study, we examined the transcription factor C/EBPalpha and circadian core clock gene PER2 in B-cell lymphoma. We show for the first time that C/EBPalpha-regulated PER2 activation is a potential tumor suppressor pathway in DLBCL that might be beneficial for therapeutic manipulation.
Materials and methods
Human samples
Fresh biopsy-derived lymphoma samples from patients at first diagnosis with: (a) Epstein - Barr virus (EBV)-negative DLBCL (n = 50) composed of cells resembling germinal center centroblasts; (b) mantle cell lymphoma (MCL, n = 21); (c) follicular lymphoma grade 3 (n = 25); and (d) Burkitt lymphoma (n = 18); as well as (e) normal, non-inflammatory lymphoid tissue (tonsils, n = 8) were gained from the Department of Pathology and Laboratory Medicine, UCLA Medical Center, Los Angeles, CA, after approval from the Cedars-Sinai Medical Center Institutional Review Board and written patient consent according to the Declaration of Helsinki. Pathological specimens were classified according to the WHO classification [1]. Genetic analysis for MYC, BCL1, BCL2 or BCL6 rearrangements was not performed.
Cell lines, culture conditions and compounds
The following cell lines were selected for this study: human DLBCL cell lines (SUDHL-4, −6, −16, and OCI-Ly1, -Ly4, -Ly7, -Ly10); MCL cell lines (SP-49, Jeko-1, NCEB-1); FL cell line (FLK-1); BL cell line (Daudi); as well as Ba/F3 (pro-B cells) and the adherent cell line 293T (obtained from the American Type Culture Collection [ATCC, Rockville, MD] and the German Collection of Microorganisms and Cell Cultures [DSMZ, Braunschweig, Germany]). All cells were cultured in RPMI-1640 medium (Sigma, St. Louis, MO) and 10% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products, Calabasas, CA), except OCI-Ly1, -Ly4, -Ly7, -Ly10, which were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Fisher Scientific Co., LLC, Santa Clara, CA) and 20% heat-inactivated FBS. The murine interleukin-3 (IL-3) dependent Ba/F3 cells were cultured in 10% heat-inactivated FBS and 10% conditioned medium of the WEHI-3B cell line. 293T cells were cultured as a monolayer in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA).
All cell lines were maintained at 37° C in a humidified chamber of 95% air and 5% CO 2, and cells growing as a monolayer were detached from the flask surface using 2.5% trypsin-ethylenediaminetetraacetic acid (EDTA) solution. Cell counts were determined using a hemocytometer (Allegiance Healthcare, Waukegan, IL), and only cells in the logarithmic phase of growth were used for all studies.
The histone deacetylase inhibitor (HDACi) suberoylanilide hydroxamic acid (SAHA/vorinostat), generously provided by Dr. V. M. Richen (previously from Merck Pharmaceuticals, Whitehouse Stataion, NJ), was dissolved in dimethylsulfoxide (DMSO) at a stock concentration of 100 mg/mL and stored at −20° C. Fresh dilutions in cell culture medium were made for each experiment.
Vectors, transient transfection and viability assays
Lymphoid cells (4 ×106) were transfected (Amaxa Nucleofector; Lonza, Köln, Germany) either with pcDNA3.1 V5-tagged Per2 expression vector (generous gift from S. Lei, University of Massachusetts Medical School, Worcester, MA) or with pcDNA3.1 empty vector, and selected with G418 (neomycin, 0.8 µg/mL) for 5 days. Equal numbers of transfected cells (5 × 104 /mL) were plated in fresh medium.
The zinc-inducible C/EBPalpha expression vector, pMT_C/alpha, was constructed by cloning human C/EBPalpha into the pMTCB6 + vector as previously described [13]. For the induction of C/EBPalpha, cells were incubated with ZnSO 4 (100 µM) for 16 h The trypan blue exclusion assay was used (0.4% of trypan blue [Gibco, Invitrogen, Carlsbad, CA]) and viability of cells was determined in triplicate using a hemacytometer with Neubauer grids.
For siRNA experiments, cells were transiently transfected with the customized siRNA plasmid psiRNA-h7SKGFPze (InvivoGen, San Diego, CA) containing either CEBPA DNA (target sequence: 5’-GGAGCTGACCAGTGACAATGA-3’) or control (scrambled DNA; target sequence: 5’-GGACAGCGATG CATAGGATCA-3’). We used 150–200 µg/mL phleomycin (InvivoGen, Toulouse, France) for antibiotic selection.
Cell proliferation, cell cycle and apoptosis assays
Proliferation of Ba/F3 cells transduced either with pcDNA3.1 human PER2 expression vector or with empty vector was determined by methyl-thiazol tetrazolium (MTT) assay as recommended by the manufacturer (Roche Diagnostics, Mannheim, Germany), and as previously described [17]. Transfected cells were examined by trypan blue exclusion method to test for viability. For cell cycle measurements, cells were fixed in cold ethanol, stained with propidium iodide and analyzed by flow cytometry (FACS CyAn ™ ADP; Dako, Carpinteria, CA) using ModFitLT V2.0 software (Verity Inc., Topsham, ME). Apoptosis analysis was performed with an annexin V–fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (BD PharMingen, San Diego, CA) according to the manufacturer’s instructions.
Quantitative real-time RT-PCR analysis
Quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) analyses were performed in triplicate as previously described [18]. Glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as an internal control. The results of qRT-PCR are presented as the mean ± standard deviation (SD) out of three independently performed experiments. Primer sequences have been reported previously [13].
Detection of CpG methylation by bisulfite sequencing
Genomic DNA of SUDHL-16, SUDHL-6 and Ly1 was extracted using DNAzol (Invitrogen), and modifed by sodium bisulfate using an EZ DNA Methylation Kit according to the manufacturer’s instructions (Zymo Research, Orange, CA). The CpG island in the promoter region of the PER2 gene (−1379 to + 330, ATG codon considered as + 1) was amplified from the bisulfate-modified genomic DNA with specific primers (sense primer: 5’-AAGTGGATGAGATTATTAGG-3’, anti-sense primer: 5’-TCCAACAACCCAAAAAACTTC-3’). For PCR amplification, a total volume of 10 mL was used containing modified genomic DNA, 0.5 mM of each primer, 5.0 mL of FailSafe PCR 2_PreMixe E (Epicentre Biotechnologies, Madison, WI) and platinum Taq (Invitrogen). Polymerase chain reaction products were subcloned into the pCR 2.1 vector (Invitrogen) and sequenced. A total of 20 CpG sites of the CpG islands of the PER2 promoter were analyzed.
Western blotting
Lymphoid cells were harvested for Western blotting, and proteins were extracted with radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecylsulfate [SDS] and 50 mmol/L Tris-HCl pH 7.5) containing protease inhibitor cocktail (Roche Molecular Biochemicals, Mannheim, Germany). Western blotting was performed as previously described [18]. Immunoblots were incubated with either the Per2 antibody H-90 (sc-25363; Santa Cruz Biotechnology, Santa Cruz, CA) or C/EBPalpha antibody (sc-61; Santa Cruz Biotechnology), and anti-GAPDH (sc-51906; Santa Cruz Biotechnology). Each Western blot shown is representative of three independently performed experiments. Densitometry of gels was performed using Quantity One software 4.6.3 (Bio-Rad Laboratories, Hercules, CA).
Chromatin immunoprecipitation
A chromatin immunoprecipitation (ChlP) assay kit (Upstate Biotechnology, Lake Placid, NY) was used, and chromatin was prepared for IP as instructed by the manufacturer. The sonicated chromatin was immunoprecipitated with 5 µg of either anti-acetylated histone H3 antibody (Upstate Biotechnology), anti-C/EBPalpha antibody (sc-61; Santa Cruz) or normal rabbit immunoglobulin G (IgG) antibody (Upstate Biotechnology) as a negative control. Immuno-precipitated DNA was subsequently analyzed by PCR using primers specific for the CEBPA or PER2 promoter region, respectively; input chromatin was analyzed for GAPDH mRNA as a positive control. Primer sequences (available upon request) were denatured at 95°C for 1 min and annealed at 60°C for 1 min, followed by elongation at 72°C for 1 min; each product was amplified in 35 cycles. PCR products were analyzed by 2.5% agarose/ethidium bromide gel electrophoresis. Densitometry of all agarose gels was performed using Quantity One software 4.6.3 (Bio-Rad Laboratories). Experiments were performed three times in order to validate the results.
Reporter gene assay
The promoter region of murine PER2 was isolated and cloned in a pGL3 Basic Vector (Promega, Madison, WI) as described previously [13]. Ba/F3 cells were co-transfected with the respective vectors: (a) pGL3_Per2; (b) pGL3_ Per2 + pMT_C/alpha; (c) pGL3; or (d) pGL3 + pMT_C/alpha, as described previously [13]. Experiments were performed three times.
Statistical analysis
Unless stated otherwise, data were statistically analyzed using the two-tailed Student’s t-test. The method of estimation included ± SD of the sample distribution; p-values < 0.05 were considered statistically significant. Asterisks shown in the figures indicate significant differences between the experimental and control conditions (*p < 0.05, **p < 0.01; ***p < 0.001).
Results
Expression levels of CEBPA and PER2 are highly reduced in DLBCL
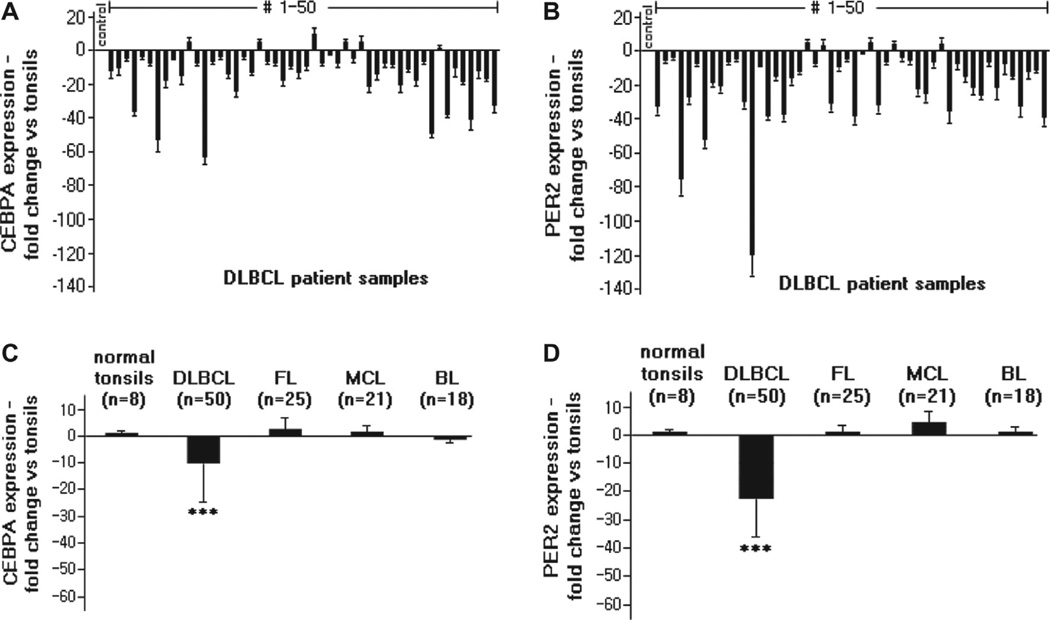
In order to analyze the expression levels of CEBPA and PER2 in human mature B-cell lymphoma, we isolated mRNA from freshly frozen human samples diagnosed with DLBCL (n = 50), MCL (n = 21), FL (n = 25) and BL (n = 18), as well as from normal tonsil samples (n = 8). Analysis by qRT-PCR revealed a markedly down-regulated expression of both CEBPA [Figures 1(A) and 1(C)] and PER2 [Figures 1(B) and 1(D)] specifically in DLBCL compared to normal tonsil samples (both p < 0.001). Also, the mRNA and protein expression of these two genes were significantly decreased in seven human DLBCL cell lines (SUDHL-4, −6, −16 and OCI-Lyl, -Ly4, -Ly7, - Ly10), as well as in the pro-B lymphoma cell line Ba/F3 [data for CEBPA not shown; and representative data for expression levels of PER2 are indicated in Supplementary Figure 1(A)]. In clear contrast, mRNA expression levels of CEBPA showed no significant change in patient samples of either MCL (p = 0.46), FL (p = 0.61) or BL (p = 0.8) compared to normal tonsils [Figure 1(C); mean expression levels of CEBPA vs. GAPDH are indicated in Supplementary Figure 2(A)]. Likewise, PER2 mRNA expression levels were not significantly different in the MCL (p = 0.42), FL (p = 0.28) or BL (p = 0.74) samples compared to normal tonsils [Figure 1(D); mean expression levels of PER2 vs. GAPDH are indicated in Supplementary Figure 2(B)]. Similar findings at the mRNA and protein levels were noted for MCL (SP-49, Jeko-1, NCEB-1), FL (FLK-1) and BL (Daudi) cell lines (data not shown). PER1, another important circadian rhythm gene of the Period family, showed no significant differences of expression in any of the lymphoma samples, including DLBCL, compared to tonsil controls [Supplementary Figure 1(B)].
Figure 1.
PER2 and CEBPA mRNA expression in B-cell lymphoma samples. (A) Expression of CEBPA was significantly down-regulated in B-cell lymphoma (DLBCL, n = 50) samples in comparison to normal tonsils (control; mean of n = 8) as analyzed by qRT-PCR. (B) Expression of PER2 was significantly down-regulated in DLBCL (n = 50) samples as analyzed by qRT-PCR. (C, D) Expression of CEBPA (C) and PER2 (D) was significantly down-regulated in DLBCL (n = 50) samples in comparison to follicular lymphoma (FL, n = 25), mantle cell lymphoma (MCL, n = 21) and Burkitt lymphoma (BL; n = 18) samples as analyzed by qRT-PCR. Normal tonsils (n = 8) were used as external controls, GAPDH as internal control. Results represent mean ± SD (indicated by bars); ***p < 0.001.
Status of methylation in PER2 promoter region in DLBCL cell lines
We have previously shown that cancer cells can silence the PER genes by methylation of their promoter region [19]. Here, we investigated the methylation status of the PER2 promoter region in human DLBCL cell lines, such as SUDHL-16, SUDHL-6 and Lyl. After analyzing a total of 20 CpG sites of a CpG island of the PER2 promoter, only a minority of the sites (≤ 6%) were methylated in up to six clones from each cell line (Supplementary Figure 3). Therefore, transcriptional gene silencing of PER2 by promoter methylation did not appear to be the underlying cause for its significantly diminished mRNA expression in DLBCL.
C/EBPalpha directly binds to PER2 promoter and induces its expression in pro-B lymphoma cell line
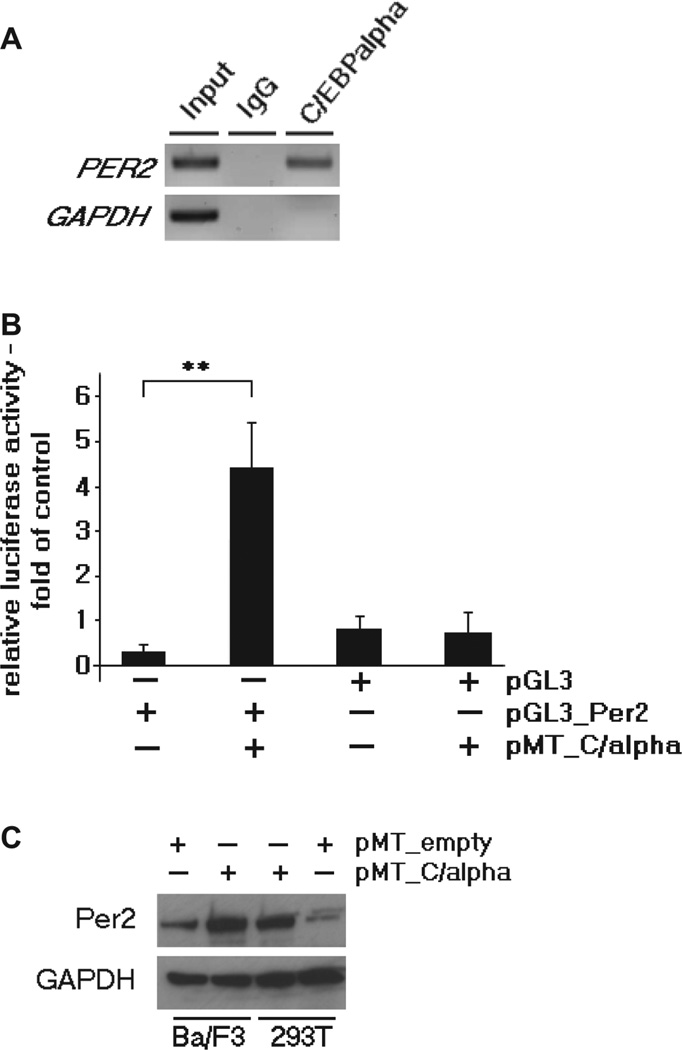
Recently, using an expression based profile, we found several C/EBP transcriptional binding motifs in the promoter region of PER2 [13]. Therefore, in the context of our present study, we analyzed the binding capacity of C/EBPalpha protein to the promoter region of PER2 in the pro-B cell line Ba/F3. These cells constitutively express low levels of C/EBPalpha (Supplementary Figure 4). As indicated by ChIP assay, C/EBPalpha protein clearly bound to the promoter region of PER2 in Ba/F3 cells [Figure 2(A)]. Moreover, reporter gene assays were performed in Ba/F3 cells with a luciferase reporter construct of the PER2 promoter sequence containing the putative C/EBP-binding site (pGL3_Per2). Transcriptional activity of pGL3_Per2 was greater than 3.5-fold in Ba/F3 cells co-transfected with pMT_C/alpha vector as compared to pGL3_Per2 alone [p = 0.01; Figure 2(B)]. Ba/F3 cells transfected with pGL3_empty vector either with or without pMT_C/alpha showed no difference regarding their luciferase activity [Figure 2(B)]. Finally, we showed that forced expression of C/EBPalpha increased the expression of Per2 in Ba/F3 cells [Figure 2(C)]. These results are in line with our previous results using a murine fibroblast cell line [13]. Taken together, C/EBPalpha appeared to enhance expression of PER2 in lymphoid cells.
Figure 2.
C/EBPalpha directly regulates the PER2 promoter and induces its expression in a murine pro-B cell line. (A) Chromatin immunoprecipitation was performed from Ba/F3 cells using rabbit C/EBPalpha antibody. IgG was used as negative control, input as positive control. Samples were analyzed by PCR using primers specific for the C/EBP site in the murine PER2 promoter, showing that C/EBPalpha protein bound to the promoter region of PER2. Chromatin was analyzed for GAPDH mRNA as an internal control. (B) Reporter gene assay: Ba/F3 cells were co-transfected with the murine Per2 reporter vector (pGL3_Per2) and pMT_C/alpha vector, and showed a mean of 3.5-fold higher transcriptional activity of pGL3_Per2 in comparison to pGL3_Per2 alone (p = 0.01), as well as in comparison to Ba/F3 cells transfected with empty vector pGL3 either with or without pMT_C/alpha. Results represent mean + SD of triplicate transfection; **p < 0.01. (C) Western blot indicates the expression of Per2 in Ba/F3 cells either with empty vector (pMT_ empty) or with pMT_C/alpha. 293T cells transfected with pMT_C/alpha were used as external positive control, GAPDH as internal control.
PER2 has significant influence on proliferation, apoptosis and cell cycle in Ba/F3 cells
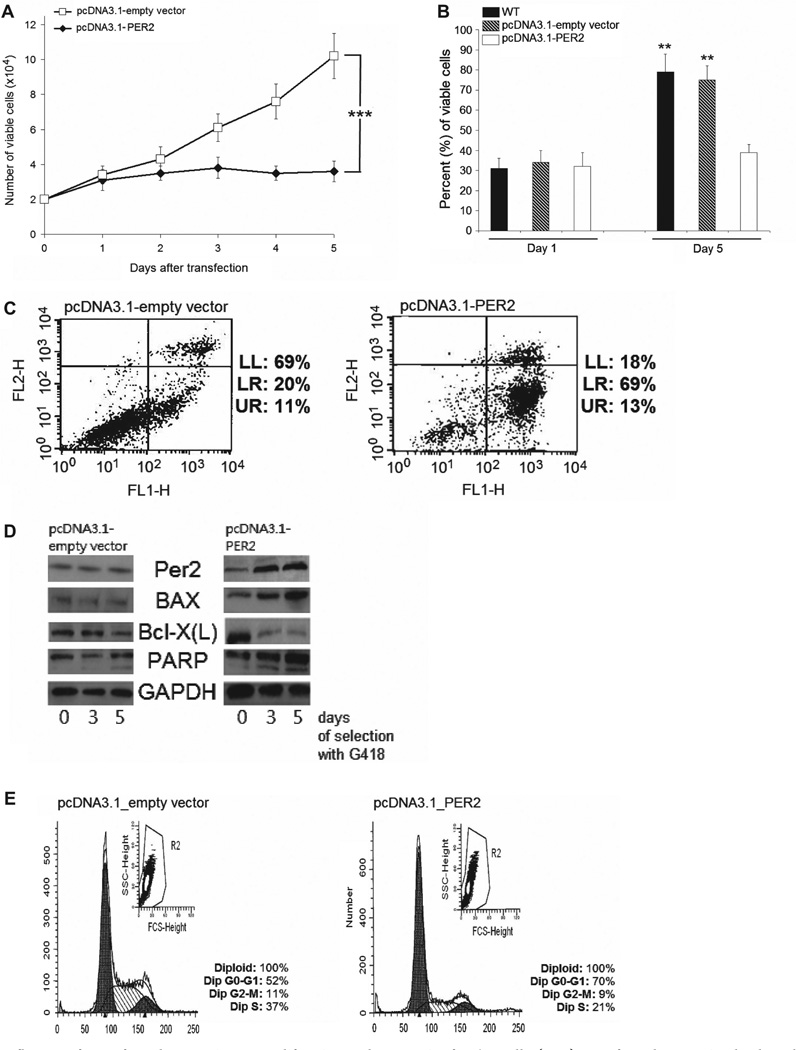
Since circadian core clock regulator PER2 is suggested to have tumor suppressor properties [11,13], we transfected Ba/F3 cells with pcDNA3.1_empty vector or pcDNA3.1 containing PER2, and selected with G418 for 5 days. As indicated by MTT assay, forced expression of the transcription factor PER2 resulted in significantly decreased growth of Ba/F3 cells in comparison to empty vector (p = 0.004) over 5 days of G418 selection [Figure 3(A)]. These findings were confirmed by counting viable cells on day 1 and day 5 using the trypan blue exclusion assay [Figure 3(B)]. In addition, using the annexin V–FITC apoptosis assay, we detected significantly increased levels of apoptotic Ba/F3 cells on day 5 of forced expression of PER2 (74 ± 18%) in comparison to vector control (35 ± 10%; p = 0.008). Representative results of one of three independent experiments are shown in Figure 3(C).
Figure 3.
Influence of PER2 forced expression on proliferation and apoptosis of Ba/F3 cells. (A, B) PER2 forced expression leads to decreased levels of cell proliferation and viability: after transfection of either pcDNA3.1_empty vector or human pcDNA3.1_PER2 into Ba/F3 cells and G418 selection for 5 days, cell proliferation was determined by (A) MTT-assay (day 1 to day 5; results represent mean ± SD; ***p < 0.001; two-way ANOVA; and (B) trypan blue cell counting on day 1 and day 5; untreated Ba/F3 cells (WT) were used as further control; results represent mean + SD; **p < 0.01. (C) PER2 forced expression leads to increased levels of apoptosis in Ba/F3 cells: Ba/F3 cell line was transfected with either pcDNA3.1_empty vector (left panel) or human pcDNA3.1_PER2 (right panel). After selection with G418 (neomycin) for 5 days, annexin V apoptosis assay with FITC/PI labeling was performed by flow cytometry. Lower left quadrant: FITC/PI negative cells; lower right quadrant: FITC positive and PI negative cells, representing early apoptotic cells; upper right quadrant: FITC/PI positive cells, representing late apoptotic and necrotic cells. (D, E) After transfection of either pcDNA3.1_empty vector or human pcDNA3.1_PER2 into Ba/F3 cells, and selection with G418, (D) Western blot was performed using antibodies against indicated proteins on days 0, 3 and 5 of selection; (E) cell cycle was determined by flow cytometry after 5 days of selection. Analysis represents one of three independent experiments, each with comparable results.
Moreover, forced expression of PER2 altered the protein levels of apoptosis-related genes. The anti-apoptotic protein Bcl-X(L) was down-regulated in Ba/F3 cells transfected with PER2, whereas levels of the pro-apoptotic Bax and poly(ADP-ribose) polymerase (PARP) cleavage activity were up-regulated in comparison to control [Figure 3(D)]. Also, overexpression of PER2 in Ba/F3 led to a decreased S-phase (22 ±3%) compared to control (39 ±5%; p = 0.033), and arrest of cells in the G0/G1 phase of the cell cycle (68 ± 9%) in comparison with empty vector (45 ± 7%; p = 0.03). Figure 3(E) indicates one representative result out of three independent cell cycle experiments.
SAHA treatment causes C/EBPalpha-dependent increase of PER2 expression and inhibition of growth in DLBCL
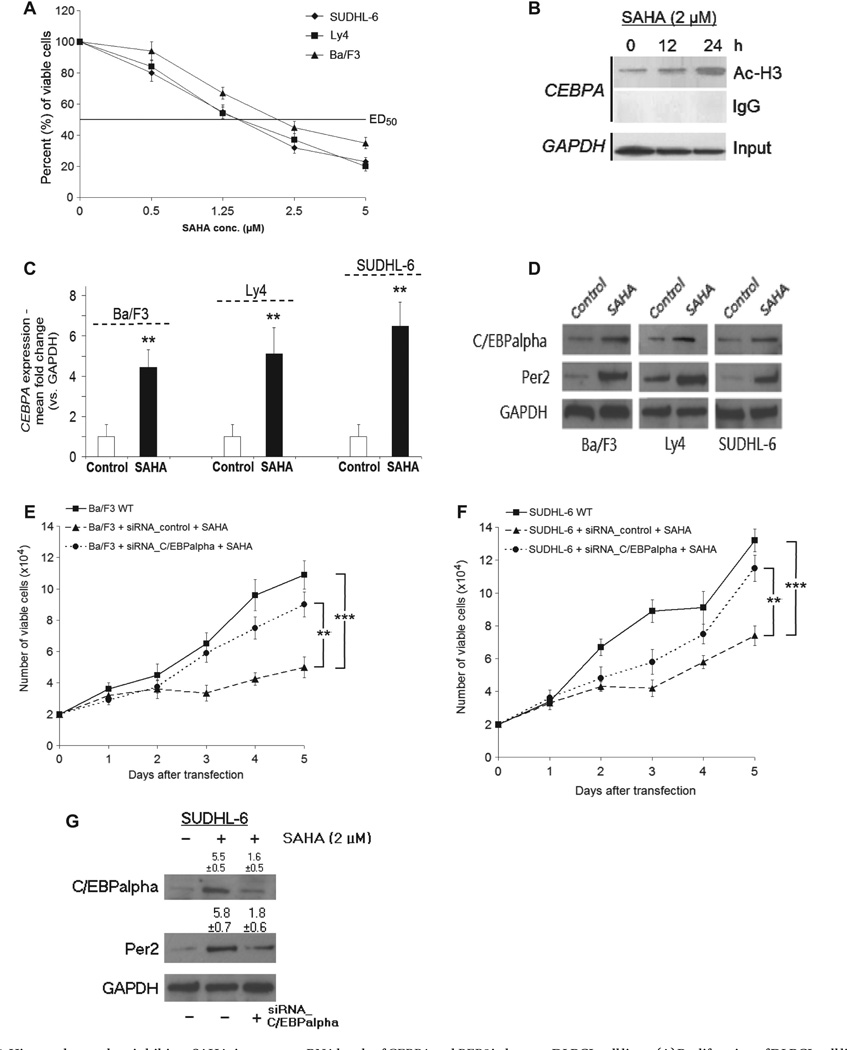
HDACi are a new class of antineoplastic agents with demonstrable preclinical anti-tumor activity in both in vitro and in vivo studies in a wide range of malignancies [20,21]. Application of increasing concentrations of SAHA for 4 days significantly inhibited growth of DLBCL cells (Ly4, SUDHL-6) and Ba/F3 as demonstrated by MTT assay [50% effective dose, ED50:1.5–2.0 µM; Figure 4(A)]. Interestingly, treatment of DLBCL cell lines (Ly4, SUDHL-4, SUDHL-6) with SAHA (2.0 µM) was associated with increased H3 acetylation in the CEBPA promoter region [Figure 4(B) shows representative results for Ly4] and, consecutively, with increased CEBPA mRNA expression [Figure 4(C)], as well as C/EBPalpha and Per2 protein expression levels [Figure 4(D)]. In contrast, plasmid-derived siRNA knock-down of CEBPA in both Ba/F3 and SUDHL-6 cells reduced the anti-tumor effect of SAHA treatment [Figures 4(E) and 4(F)] with a Per2 level almost similar to control [representative data for SUDHL-6 in Figure 4(G)].
Figure 4.
Histone deacetylase inhibitor, SAHA, increases mRNAlevels of CEBPA and PER2 inhuman DLBCLcell lines. (A) Proliferation of DLBCL cell lines and Ba/F3 exposed to suberoylanilide hydroxamic acid (SAHA; 0.5,1.25,2.5 and 5 µM) for 4 days was tested via MTT assay, ED50:1.5–2.0 µM (p < 0.01). Exposing respective cells to DMSO as control for 4 days resulted in no significant change of proliferation as measured by MTT assay (data not shown). (B) The DLBCL cell line, Ly4, was cultured in medium ± SAHA (2.0 µM) and cells were harvested after 0,12 and 24 h. Chromatin immunoprecipitation was performed using Ac-H3 antibody to measure H3 acetylation in the CEBPA promoter region by PCR. IgG was used as negative control, input as positive control; chromatin was analyzed for GAPDH mRNA as internal control. (C, D) mRNA and protein were prepared and expression of CEBPA and PER2 was analyzed by qRT-PCR. Results represent mean ± SD; **p < 0.01; for Western blot analysis, antibodies against either C/EBPalpha or Per2 and GAPDH were applied. (E–G) For siRNA experiments, Ba/F3 and SUDHL-6 cells were transfected with plasmid-derived siRNA against CEBPA, siRNA_C/EBPalpha or control (scrambled DNA), and cultured in medium ± SAHA (2.0 µM). (E, F) Trypan blue cell counting was performed on days 1–5 under phleomycin selection. Results represent mean ± SD; **p < 0.01; **p < 0.001 (two-way ANOVA). (G) SUDHL-6 cells were harvested on day 5 of phleomycin selection and Western blot analysis was performed either with anti-C/EBPalpha, anti-Per2 or anti-GAPDH antibody. Western blot analysis shown is representative for one out of three independently performed experiments. Number ± SD indicates fold change of either C/EBPalpha or Per2 as measured by densitometry out of three independent experiments; vehicle-treated SUDHL6 cells transfected with vector control were set as 1. SAHA-treated cells transfected with control showed significantly increased expression of both C/EBPalpha (p = 0.02) and Per2 (p = 0.018), whereas SAHA-treated cells transfected with siRNA_C/EBPalpha showed no significant change in protein expression (both p > 0.05).
Discussion
Mature B-cell lymphoma is a heterogeneous clinicopathologic entity characterized by distinct cells of origin and specific cytogenetic and molecular aberrations [1–4]. The etiology of mature B-cell lymphoma is largely unknown, but disruption of the circadian rhythm is one of the factors suggested to predispose to this disease [14–16]. Recent studies have revealed that a large number of genes are controlled by the circadian clock in a tissue-specific manner [5,6,22,23]. The circadian clock shares common features with the cell cycle, and disturbance of the circadian rhythms can lead to failure in control of the cell cycle causing not only sleep disorders or depressive disorders, but also cancers [11–14,19,24–27]. For instance, Per2 and Perl modulate beta-catenin and cell proliferation in colon and non-colon cancer cells [28]. Also, the knockdown of NPAS2, the largest known circadian gene, affects the expression of cancer-related genes, such as FANCG, MSH2 or MAPK12, reflecting its potential role as a tumor suppressor [29]. Interestingly, a large Finnish study revealed that night-time shift workers have a significantly increased risk of non-Hodgkin lymphoma than the population on average [15], but one limitation of this cohort study is that the lymphoma sub-entities were not defined.
Previously, we discovered with the aid of cDNA microarray analysis a link between C/EBP transcription factors and the circadian clock pathway, showing that PER2 expression is up-regulated by C/EBPalpha in leukemic cancer cell lines [13]. The purpose of the present study was to investigate the role of PER2 and its correlation to CEBPA in human mature B-cell lymphoma. In contrast to lymphoma samples of MCL, FL or BL, the expression of both CEBPA and PER2 was low in DLBCL samples, and associated with increased cellular proliferation. However, using a relatively small number (n = 8) of normal tonsils as external control limits the validity of this finding.
Fu et al. showed that mice deficient in the PER2 gene exhibited a neoplastic phenotype, with 15% dying due to lymphoma [11]. Another group found that the expression levels of both PER1 and PER2 and other circadian regulators were significantly impaired in samples of both the chronic phase and blast crisis of human chronic myeloid leukemia (CML) [30]. Additionally, the CpG islands of PER2 were also highly methylated in 40% of these CML cases. In line with previously published data indicating a tumor suppressive role for PER2, we noted that forced expression of this core clock gene in a pro-B lymphoma cell line was associated with inhibition of tumor growth, increased rates of apoptosis and cell cycle arrest. However, DNA methylation did not appear to represent a mechanism leading to de novo repression of PER2 according to the finding that CpG sites in the promoter of PER2 were not significantly methylated in DLBCL cells. Therefore, we suggest a disease-specific relationship of C/EBPalpha and Per2. Indeed, in congruence with our previously published data in myeloid leukemia [13], overexpression of this transcription factor directly stimulated the expression of Per2 in DLBCL, which was associated with decreased tumor cell proliferation.
Epigenetic active drugs such as HDACi are emerging as an exciting new class of potential anti-cancer agents in hematologic malignancies [20,21]. They act via reactivation of a large panel of genes, including tumor suppressor genes responsible for cell cycle arrest, differentiation and apoptosis. Deacetylation of histones results in a condensed chromatin structure and repression of gene transcription, whereas acetylated histones are associated with the activation of gene transcription [21]. SAHA (vorinostat) is a potent inhibitor of HDAC activity and is capable of inducing differentiation and/or apoptosis in several tumor cell lines [21]. This compound has been approved for use in cutaneous T-cell lymphoma by the US Food and Drug Administration [31]. In a phase II study of patients with relapsed mature B-cell lymphoma, eight (40%) of 20 patients with FL demonstrated a response to SAHA [32]. In the present study, we showed that C/EBPalpha is silenced in DLBCL, and forced expression of this transcription factor activates the circadian family member Per2. By means of functional experiments, we provided data that HDACi can slow the growth of DLBCL associated with stimulation of C/EBPalpha and Per2. Moreover, under the same experimental conditions, C/EBPalpa was silenced and, subsequently, Per2 expression decreased and DLBCL growth increased. Consequently, it appears that, at least in part, the anti-tumor effect of SAHA is caused by changing the expression of CEBPA with transcriptional control of PER2. The regulatory effect of SAHA on the acetylation levels of histones surrounding the promoter of PER2 has still to be investigated. In contrast to DNA hypermethylation, which was absent in the investigated lymphoma cell lines, histone deacetylation may be a further mechanism of PER2 gene silencing. Other molecular targets that could be influenced by the diminished expression of CEBPA in lymphoma are cyclin-dependent kinases (cdk). It has been suggested that the proline-histidine rich region of C/EBPalpha may play a role in the regulation of cell proliferation through the inhibition of both cdk2 and cdk4 that drive cell cycle progression [33]; but in vivo experiments did not support this observation [34]. Further potential mechanisms of C/EBPalpha-mediated growth arrest include up-regulation of p21cipl/wafl and the SWI/SNF chromatin-remodeling complex [35,36], as well as direct inhibition of the E2F complex [20,34,37].
In summary, we suggest that the circadian core clock gene PER2 is an essential downstream target of the transcription factor C/EBPalpha, with tumor suppressor characteristics. To our knowledge, this is the first study implicating an association of abnormalities of C/EBPalpha and its downstream target PER2 with the pathogenesis of DLBCL. Further elucidating the anti-tumor effect of HDACi could also benefit the development of new therapeutic strategies for patients with B-cell lymphoma.
Supplementary Material
Acknowledgements
This work was in part funded by the Deutsche Forschungs-gemeinschaft (DFG, Bonn, Germany: TH 1438/1-1; N.H.T.), Deutsche Krebshilfe e.V (N.H.T., G.B.T.), the National Institutes of Health (NIH, Bethesda, MD: grant 5R01CA026038-32; H.P.K.) and an A*STAR grant of Singapore (National University of Singapore, Singapore; H.P.K). H.P.K. is the holder of the Mark Goodson endowed Chair in Oncology Research at Cedars-Sinai Medical Center and is a member of the Jonsson Cancer Center and the Molecular Biology Institute, UCLA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at www.informahealtcare.com/lal.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon, France: IARC Press; 2008. [Google Scholar]
- 2.Clarke CA, Glaser SL. Changing incidence of non-Hodgkin lymphomas in the United States. Cancer. 2002;94:2015–2023. doi: 10.1002/cncr.10403. [DOI] [PubMed] [Google Scholar]
- 3.Müller AMS, Ihorst G, Mertelsmann R, et al. Epidemiology of non- Hodgkin’s lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol. 2005;84:1–12. doi: 10.1007/s00277-004-0939-7. [DOI] [PubMed] [Google Scholar]
- 4.Flowers CR, Sinha R, Vose JM. Improving outcomes for patients with diffuse large B-cell lymphoma. CA Cancer J Clin. 2010;6:393–408. doi: 10.3322/caac.20087. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 7.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 9.Kume K, Zylka MJ, Sriram S, et al. mCRYl and mCRY2 are essential components of the negative limb of the circadianclock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo T, Yamaguchi S, Mitsui S, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 11.Fu L, Pelicano H, Liu J, et al. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 12.Mormont MC, Levi F. Circadian-system alterations during cancer processes: a review. Int J Cancer. 1997;70:241–247. doi: 10.1002/(sici)1097-0215(19970117)70:2<241::aid-ijc16>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 13.Gery S, Gombart AF, Yi WS, et al. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Leaderer D, Guss C, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int I Cancer. 2007;120:432–435. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahti TA, Partonen T, Kyyronen P, et al. Night-time work predisposes to non-Hodgkin lymphoma. I Cancer. 2008;123:2148–2151. doi: 10.1002/ijc.23566. [DOI] [PubMed] [Google Scholar]
- 16.Micke O, Schafer U, Wormann B, et al. Circadian variations of interleukin-2 receptors, serum thymidine kinase and beta-2-microglobulin in patients with non-Hodgkin’s lymphoma and normal controls. Anticancer Res. 1997;17:3007–3010. [PubMed] [Google Scholar]
- 17.Thoennissen NH, Iwanski GB, Doan NB, et al. Cucurbitacin B induces apoptosis by inhibition of the IAK/STAT pathway and potentiates antiproliferative effects of gemcitabine on pancreatic cancer cells. Cancer Res. 2009;69:5876–5884. doi: 10.1158/0008-5472.CAN-09-0536. [DOI] [PubMed] [Google Scholar]
- 18.Iwanski GB, Lee DH, En-Gal S, et al. Cucurbitacin B, a novel in vivo potentiator of gemcitabine with low toxicity in the treatment of pancreatic cancer. Br J Pharmacol. 2010;160:998–1007. doi: 10.1111/j.1476-5381.2010.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gery S, Komatsu N, Kawamata N, et al. Epigenetic silencing of the candidate tumor suppressor gene Perl in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 20.Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 22.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 23.Kornmann B, Preitner N, Rifat D, et al. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 2001;29:E51. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez-Ferrer S, Lucas D, Battista M, et al. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 25.Chen ST, Choo KB, Hou MF, et al. Deregulated expression of the PERI, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 26.Gery S, Komatsu N, Baldjyan L, et al. The circadian gene perl plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 27.Gery S, Virk R, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 28.Wood PA, Yang X, Hrushesky WJ. Clock genes and cancer. Integr Cancer Ther. 2009;8:303–308. doi: 10.1177/1534735409355292. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman AE, Zheng T, Ba Y, et al. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6:1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann BS, Johnson JR, Cohen MH, et al. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1250. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 32.Kirschbaum M, Popplewell L, Nademanee A, et al. A phase 2 study of vorinostat (suberoylanilide hydroxamic acid, SAHA) in relapsed or refractory indolent non-Hodgkin’s lymphoma: a California Cancer Consortium study. Haematologica. 2009;94(Suppl. 2) Abstract 166. [Google Scholar]
- 33.Wang H, Iakova P, Wilde M, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 34.Porse BT, Pedersen , Hasemann MS, et al. The proline-histidinerich CDK2/CDK4 interaction region of C/EBPalpha is dispensable for C/EBPalpha-mediated growth regulation in vivo. Mol Cell Biol. 2006;26:1028–1037. doi: 10.1128/MCB.26.3.1028-1037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timchenko NA, Wilde M, Nakanishi M, et al. CCAAT/enhancer- binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-l/CIP-l/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen TA, Kowenz-Leutz E, Leutz A, et al. Cooperation between C/EBP alpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slomiany BA, DArigo KL, Kelly MM, et al. C/EBP alpha inhibits cell growth via direct repression of E2F–DP-mediated transcription. Mol Cell Biol. 2000;20:5986–5997. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.