Abstract
Primary malignant melanoma of sinonasal tract (PMMST) is a rare but severe form of melanoma. We retrospectively analyzed 17 cases and focused on the histological presentation, the expression of c-Kit, EGFR, cyclin-D1/Bcl-1, PS100 and HMB45 and searched for BRAF, NRAS and KIT mutations that are known to be associated with melanoma subtypes, together with amplifications of KIT, CCND1, CDK4, MDM2 and MITF using quantitative PCR. In the majority of cases (78%), an in situ component was evidenced. Invasive components were composed of diffuse areas of rhabdoid, epithelioid or spindle cells, and in most cases lacked inflammatory reaction, suggesting that an immune escape phenomenon probably develops when the disease progresses. EGFR was rarely and weakly expressed in the in situ component of 2 cases. None of the investigated case showed BRAFV600E, but one had a D594G mutation. NRAS mutations in exon 2 (G12D or G12A) were found in 3 cases (18%) and a KIT mutation in exon 11 (L576P) in one, while C-Kit was expressed at the protein level in half cases. Amplifications of CCND1 were evidenced in 5 cases, confirmed in 3 by FISH studies, but this was not always correlated with protein expression, found in 8 patients (62.5%), 3 having no significant amplification. In conclusion, PMMST are not associated with BRAFV600E mutations. Instead, NRAS or KIT mutations and CCND1 amplification can be found in a proportion of cases, suggesting that PMMST are heterogeneous at the molecular level and should not be sensitive to therapeutic approaches aiming at BRAF.
Keywords: sinonasal melanoma, Primary malignant melanoma of sinonasal tract, BRAF mutation, NRAS, KIT, CCND1 amplification, cyclin D1
Introduction
Primary melanoma of sinonasal tract (PMST) is a rare melanoma variant that primarily develops in the nasal cavity, nasopharynx or paranasal tissue and can thereafter spread in several compartments. Their histological and phenotypical characteristics have been well described, especially in the large series from Thompson and colleagues[1], but only few series of cases were published to date. The patients most often present with epistaxis and the tumor can show a broad spectrum of cytologic aspects and architectural patterns, so that the diagnosis most often relies on the immunohistochemical markers, with a similar sensitivity and specificity to cutaneous and other non cutaneous melanomas[1]. The prognosis is poor, with many patients having local recurrence and a high mortality rate, despite surgery and radiation and/or chemotherapy, with a mean 5 years overall survival of 42–43%[1, 2]. The prognosis depends on the level of local invasion, and, due to the particular anatomic localization, a specific adapted staging system was proposed[1, 2], including one that fits into the international TNM staging system requirements[1]. The oncogenic events responsible for melanoma development, in both cutaneous and non cutaneous melanomas, are better characterized. Besides genes associated with familial melanomas, such as CDKN2A, various genetic aberrations occur at the somatic level, especially involving receptors or downstream partners involved in the MAPK signalling pathway. Among them KIT, RAS and BRAF are found to be mutated at various frequencies and have been the most studied, while other genes were found to be mutated in more sporadic studies. These include amplifications of CCND1, CDK4 and MITF, or suppressor genes abnormalities (PTEN, APAF1 and TP53). BRAF mutations are found in about 50% of cutaneous melanomas, with the V600E substitution being the most frequent aberration, and are very rare in mucosal melanomas.[3] BRAF and NRAS are two upstream members of the MAPK pathway, which are activated by tyrosine kinase receptors, such as c-Kit. The KIT gene, mapped to 4q12, encodes an oncogenic transmembrane receptor tyrosine kinase, c-Kit, whose ligand is stem cell factor. C-Kit plays an important role in melanocyte migration, development, differentiation and tumorigenesis. KIT mutation are found in up to 20% of mucosal melanomas, a lower proportion of acral melanomas, and appears to be very rare in cutaneous nonacral tumors.[3]. Interestingly, it was recently shown that the percentage of c-Kit positive neoplastic cells in acral melanoma was a predictive factor of the mutational status.[4] The finding of these genetic aberrations recently allowed the setting of targeted therapies directed against these oncogenes or their signaling pathways in patients with advanced stages[5, 6] Although none has dramatically improved the management of advanced stage melanoma, encouraging results were actually obtained. While PMMST are often aggressive and locally advanced melanomas, only one recent study has focused on the oncogenic mutations associated with this melanoma subtype, showing that a proportion of cases are associated with NRAS and KIT mutations but not with CCND1 amplification. To address this issue, we report here 17 additional cases of PMMST with immunohistochemical and molecular genetic studies focusing on KIT, BRAF, NRAS and CCND1.
Material and methods
Patients and tissue samples
A total of 17 cases of PMMST were retrospectively and prospectively included. The cases were retrieved from department of pathology of the centre hospitalier inter-communal de Créteil and Henri Mondor hospital, between 1995 and 2012. The demographic symptoms and clinical features at presentation (gender, age epistaxis, nasal obstruction, nasal mass, polyp and pain), location of the tumors (nasal cavity, septum, frontal, ethmoidal, maxillary sinus), together with the personal and familial past medical history were reviewed in all cases.
Pathologic study
The following macroscopic features were recorded: tumor size, color, necrosis and hemorrhage. Hematoxylin and eosin stained slides were reviewed and the following histologic parameters were recorded, according to the previously published series from Thompson et al.[1]: surface epithelium (present or absent), in situ component, architectural pattern of growth (solid, epithelioid, meningothelial, hemangiopericytoma-like, peritheliomatous, papillary, storiform), cell type (undifferentiated, epithelioid, small cell, plasmacytoid, rhabdoid, giant cells), melanin pigment (present or absent), inflammatory response (present or absent), topography of inflammatory response (inside or outside of tumor), index of mitotic figures, perineural invasion and vascular invasion. Tumor necrosis was noted as : 0: absent, 1 : <5%, 2 : <50%, 3 : >50%. 0 : absent, 1 : low, 2 : moderated, 3 :important, The topography (around the in situ and/or within the invasive neoplastic components) of the inflammatory reaction was evaluated together with intensity, quantified as follows : 0 : absent, 1 : low (sparsed cells), 2 : moderated (grouped cells), 3 : important (sheets of inflammatory cells),
Immunohistochemistry
For immunohistochemistry, we used the Bond-Max device (Bond, Leica Menarini). The staining was done after antigen retrieval by heat with appropriate buffer with peroxydase and diaminobenzidine. The deparaffinized sections were stained for CKit (Dako; clone A4502; 1:900 dilution), EGFR (Zymed; clone 28-0005; 1:200 dilution), PS100 (Dako; clone Z311; 1:3000 dilution), HMB45 (Dako; clone M0634; 1/100 dilution), Cyclin D1/BCL1 (Microm; clone SP4; 1:25 dilution). The expression of each marker was investigated in the two in situ and invasive melanoma components, when applicable. We assessed both the proportion of stained cells on a scale of 0 to 10 (0, 10% … 90%, 100%) and the staining intensity (low, moderate, high).
Interphase fluorescence in situ hybridization (FISH) analysis
FISH analysis was performed on 3μM TMA tissue sections using split signal FISH DNA probes for CCND1/11q13 (probe Y5414, Dako, SA, Glostrup, Denmark) according to the manufacturer’s recommendations (www.euro-fish.org). Slides were analysed with a Zeiss AxioImager Z1 fluorescence microscope equipped with microscope-specific double filters (XF53, Omega Optical, Brattleboro, VT, USA) suitable for the fluorescein isothiocyanate and Texas Red labelled split-signal probes. Slides were analysed independently by two scorers (MB, CCB) with a 100× oil immersion objective. For archiving, images were captured with 40× objective using a Hamamatsu digital camera attached to the fluorescence microscope and Visilog software (NOESIS, Les Ulis, France).
Amplification was defined as the presence of at least 6 fusion signals per nuclei.
Molecular studies
Molecular studies were performed in 15 cases. Genomic DNA was extracted fom five 10 μM sections of the paraffin blocks, by using the QIAamp DNA FFPE kit (Qiagen). Sequence analyses were done on the BRAF exons 11 and 15, NRAS exons 2 et 3 and KIT exons 11, 13, 17 and 18. Amplified products were obtained by PCR with specific primers (Table 1). The amplification procedure was done using the Taq polymerase manufacturer’s instructions (Invitrogen). To avoid DNA amplification inhibition by melanin, 10 μg bovin serum albumin were added to the PCR mixes. PCR products were purified after migration on an agarose gel using the illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare). The PCR purified products were then sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) following the manufacturer’s instructions.
Table 1.
Sequences of primers for gene mutation and amplification studies
| Primer name | Primer sequence (5′->3′) | Hybridization temperature | PCR fragment size |
|---|---|---|---|
| (M) BRAF exon 11 F | TCCCTCTCAGGCATAAGGTAA | 55 °C | 313 bp |
| (M) BRAF exon 11 R | CGAACAGTGAATATTTCCTTTGAT | 55 °C | |
| (M) BRAF exon 15 F | TCATAATGCTTGCTCTGATAGGA | 55 °C | 224 bp |
| (M) BRAF exon 15 R | GGCCAAAAATTTAATCAGTGCA | 55 °C | |
| (M) NRAS exon 2 F | GAACCAAATGGAAGGTCACA | 55 °C | 301 bp |
| (M) NRAS exon 2 R | TGGGTAAAGATGATCCGACA | 55 °C | |
| (M) NRAS exon 3 F | GGTGAAACCTGTTTGTTGGA | 55 °C | 272 bp |
| (M) NRAS exon 3 R | AACCTAAAACCAACTCTTCCCA | 55 °C | |
| (M) KIT exon 11 F | GATCTATTTTTCCCTTTCTC | 55 °C | 174 bp |
| (M) KIT exon 11 R | AGCCCCTGTTTCATACTGAC | 55 °C | |
| (M) KIT exon 13 F | CATCAGTTTGCCAGTTGTGC | 55 °C | 295 bp |
| (M) KIT exon 13 R | AGCAAGAGAGAACAACAGTCTGG | 55 °C | |
| (M) KIT exon 17 F | TCATTCAAGGCGTACTTTTG | 55 °C | 350 bp |
| (M) KIT exon 17 R | TCGAAAGTTGAAACTAAAAATCC | 55 °C | |
| (M) KIT exon 18 F | CATTTCAGCAACAGCAGCAT | 60 °C | 287 bp |
| (M) KIT exon 18 R | CAAGGAAGCAGGACACCAAT | 60 °C | |
| (A) ACTIN F | CATGGTGCATCTCTGCCTTACA | 60 °C | 76 bp |
| (A) ACTIN R | ACAGCCTGGATAGCAACGTACA | 60 °C | |
| (A) GADPH F | GCCCCCGGTTTCTATAAATTG | 60 °C | 94 bp |
| (A) GADPH R | CTGGCGACGCAAAAGAAGAT | 60 °C | |
| (A) KIT F | TCCTCAAACAGGCATAGATTTCC | 60 °C | 69 bp |
| (A) KIT R | TGTGGATAGCATGCCTTGGA | 60 °C | |
| (A) MDM2 F | AGGACATCTTATGGCCTGCTTTAC | 60 °C | 63 bp |
| (A) MDM2 R | GGGCAGGGCTTATTCCTTTT | 60 °C | |
| (A) CDK4 F | ATTGCATCGTTCACCGAGATC | 60 °C | 65 bp |
| (A) CDK4 R | CTTGACTGTTCCACCACTTGTCA | 60 °C | |
| (A) CCDN1 F | CGTGGCCTCTAAGATGAAGGA | 60 °C | 62 bp |
| (A) CCDN1 R | CGGTGTAGATGCACAGCTTCTC | 60 °C | |
| (A) MITF F | TGTCACTGATCCACTCCTTTCCT | 60 °C | 59 bp |
| (A) MITF R | TCCGGCTGCTTGTTTTGG | 60 °C |
Abbreviations : F, forward; R, reverse; bp, base pair, M, primer used for mutation studies; A, primer used for amplification studies
Gene copy number was assessed by quantitative real-time PCR by comparison with GAPDH or Actin using primers described in Table 1. PCR reactions were done in duplicate using the Power SYBR Green PCR Master Mix kit (Applied Biosystems) following the manufacturer’s instructions. Relative copy numbers were calculated using the ΔΔCt method, where Ct is the threshold cycle for amplification. For each sample, ΔCt for the gene of interest versus GAPDH or Actin was calculated as ΔCt = Ct(gene of interest) − Ct(GAPDH/Actin). The ΔCt value for each experimental test sample was calibrated to a reference pool of genomic DNA prepared from primary cells. Relative DNA copy number was calculated using the formula 2− ΔΔCt. Relative copy numbers were converted to absolute copy numbers by assigning a value of 2 (diploid) to the reference pool and multiplying the relative copy number of test samples by a factor of 2. The threshold for increased copy number was set to 5 copies of the gene of interest relative to GAPDH/Actin.
Results
Clinical features
The patients included 12 women and 5 men (sex ratio = 0.42), who ranged in age from 46 to 97 years. Most patients (13/15, 87%) had epistaxis. Three (3/15) complained of nasal obstruction. Periorbital headache and earache were reported in 2 cases. One patient presented anosmia without epistaxis (Table 2). A history of cutaneous melanoma was found in a case that reported the removal of 2 cutaneous melanomas, one at the right forearm, and another at the left cheek without available pathologic reports. No patients in this series had a dysplastic nevus syndrome or a xeroderma pigmentosum. Tumours were located in the nasal cavity in 65% of cases (11/17), sinus without nasal involvement in 18% (3/17). A local extension to the orbit was found in 2 cases. The tumor prolapsed in the nasopharynx in one case. Tumor size ranged between 1 and 5.5 cm with a mean of 3.25 cm.
Table 2.
Clinical and pathological features
| Case | Age/sex | Symptoms | Site | Endocopy | Size (cm) | Architecturea | Cytologyb | Pigmentationc | Necrosisd | Mitotic ratee | Inflammationf | Stromag |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 74/F | Epistaxis | Nasal cavity | Polyp | 2 | S | U | M | 1 | 24 | 1,P | F |
| 2 | 70/F | Epistaxis+headache | Ethmoïdo-frontal sinus | Mass | 1,5 | S+P | S+U | H | 1 | 23 | 0 | A |
| 3 | 63/F | Epistaxis+earache | Naso-ethmoido-frontal sinuses+orbit | Mass | 3 | S | U | 0 | 3 | 21 | 1 | A |
| 4 | 80/M | Anosmia+nasal obstruction | Nasal cavity +cavum | Mass | 4 | S | R+G | M | 2 | 18 | 0 | A |
| 5 | 68/F | Epistaxis+nasal obstruction | Nasal cavity +ethmoid sinus | Mass | 2,5 | S | R+G | M | 3 | 20 | 1,T | A |
| 6 | 78/M | Nasal obstruction | Ethmoido-maxillary sinus | Polyp | 1 | S | U | M | 0 | 10 | 2,T | A |
| 7 | 58/F | Epistaxis | Nasal cavity | Polyp | 2 | S | R | M | 1 | 23 | 3,T | A |
| 8 | 97/F | Epistaxis | Nasal cavity | Mass | 1 | S | P | 0 | 3 | 12 | 0 | A |
| 9 | 85/F | Epistaxis | Nasal cavity | Mass | ? | M | E | 0 | 1 | 18 | 0 | A |
| 10 | 88/F | Epistaxis | Nasal cavity | Mass | 5,5 | S | U | M | 1 | 28 | 2,P | F |
| 11 | 75/F | Epistaxis | Sphenoidal sinus+orbit | Mass | 2,5 | S | P | 0 | 0 | 12 | 0 | F |
| 12 | 80/M | Epistaxis | Nasal cavity | Polyp | ? | S | S+U | 0 | 0 | 14 | 0 | F |
| 13 | 55/F | Epistaxis | Nasal cavity+ septum | Mass | 2 | S | U | M | 1 | 36 | 0 | A |
| 14 | 66/F | Epistaxis | Nasal cavity+ethmoid sinus | Mass | NA | S | S+E | M | 0 | 15 | 1 | A |
| 15 | 64/M | NA | Nasal cavity | NA | NA | S | E | M | 2 | 10 | 1,T | A |
| 16 | 70/F | NA | Nasal cavity+maxillo-ethmoido-frontal sinuses | Mass | NA | S | P | M | 0 | 4 | 0 | A |
| 17 | 46/M | Epistaxis | Nasal cavity+ethmoid and frontal sinuses | NA | NA | S+M | S | M | 1 | 22 | 0 | A |
NOTE : The mitotic rate is indicated as the number of mitoses for 10 high power fields.
Abbreviations :
S, solid; P, péritheliomatous; M, meningothelial;
U, undifferenciated; S, spindle; R, rhabdoid; P, plasmacytoid; E, epithélioid; G, giant cells; In situ, in situ component;
melanin; H, hemosiderin;
0, absent; 1, low; 2, moderated; 3, important;
P, peritumoral; T, intra-tumoral; Intensity of the inflammatory reaction : 0, absent; 1, low; 2, moderated; 3, important;
F, Fibrosis; A, absent.
Histologic features
The majority of samples was submitted in multiple fragments and were blackish or brownish. In one case the tumor was whitish translucent. Four cases presented as a polypoid tumor, while most cases were ill defined masses. Most cases showed, at least partially, a solid architecture (16/17, 94%), while a pure peritheliomatous (n=2) or meningothelial (n=1) architecture were less frequent (Table 2). The tumor cells were often cytologically undifferentiated (7/17). One cases associated undifferentiated and spindle cells components (2/17) or mixed rhabdoid and giant cells (2/17). Cases with plasmacytoid (1/17) and epithelioid cells (1/17) were rare. Mitotic figures were identified in all cases with a mean of 18 mitotic figures per 10 high power fields (range 4–36). An in situ component was observed in 11 cases (78%). One case that presented as a polyp displayed only an in situ component. Inflammatory cells were present in only 7 cases. Lymphoid cells were predominant beneath the in situ component. A stroma reaction was absent. Only 4 cases showed fibrosis and an ulceration was seen in 9 cases. We identified metaplastic bone within the tumors in 3 cases. Melanin Pigment was revealed in the neoplastic tumor cells in 8 cases, and was pronounced beneath the in situ components in the form of melanophages aggregates.
Immunohistochemical analyses
S100 protein was expressed in most cases (14/17, 82,6%), but the proportion of stained cells, as well as the staining intensity greatly varied from on case to another, ranging from a low expression in about 20% of cells to a strong expression in the majority of neoplastic cells. HMB45 and Melan-A (Figures 1A and 1B) were expressed in all cases, but the proportion of positive cells also varied a lot, from 5% to 100% of cases. EGFR was not expressed in most cases, except for one case in the invasive component, with all neoplastic cells being positive. In two cases, the in situ component appeared to be weakly EGFR positive, with 10% to 50% of positive cells, but neoplastic stained cells were often difficult to distinguish from normal epithelial cells. c-Kit was found to be expressed in 8 samples (47%) in the invasive components (Figure 1D). In most of these samples, the staining intensity was weak or moderate (6/8, 75%). The in situ neoplastic cells expressed c-Kit in 10 cases (Figure 1C). Four of these samples did not show c-Kit protein expression within the invasive components, while two samples with a c-Kit+ invasive components had no positive cells in situ. Cyclin D1 was found to be expressed in 10 samples in the invasive component (10/16, 62.5%, Figures 1E and 1F), with a variable proportion of stained cells, ranging from 10% to 80%. The staining intensity appeared to be strong or at least moderate in most samples (7/10, 70%). In most of these cases, we also evidenced Cyclin D1 expression in the in situ components, except for two samples, that both showed a weak expression in a minority of tumoral cells in the invasive component (10%)
Figure 1. Immunohistochemical results.
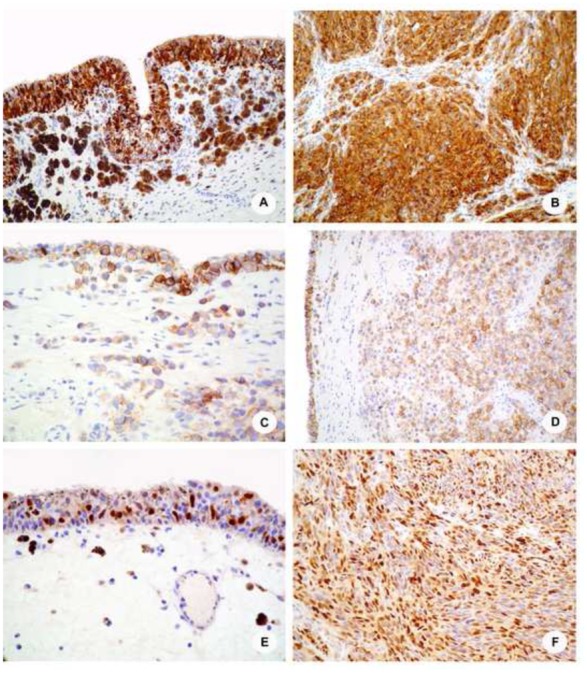
A and B, Melan-A staining in case 17 shows a significant staining of neoplastic cells in both the in situ (A), and the invasive (B) components, with a strong and diffuse expression. The invasive components is mostly composed of sheets of spindle cells; C and D, C-Kit receptor is expressed in the majority of the neoplastic, morphologically undifferenciated cells in case 10, both by in situ (C) and invasive (D) cells; E and F, Representative cyclin-D1 immunostainings showing a strong nuclear expression by in situ and invasive components of case 17 (E) and 14 (F), respectively.
Molecular and FISH studies
MDM2 gene copy numbers was within normal range in all samples using quantitative PCR, while KIT, MITF and CDK4 amplifications could be demonstrated in one case each. An amplification of CCND1 could be demonstrated in 5 samples (31%), 3 by both qPCR and FISH, 2 by qPCR alone. As seen in Figure 2, cases displaying CCND1 amplification showed a high copy number of the gene in neoplastic cells nuclei, with more than 10 to innumerable signals per nuclei (Figure 2A and B).
Figure 2. CCND1 FISH analysis using split signal FISH DNA probes.
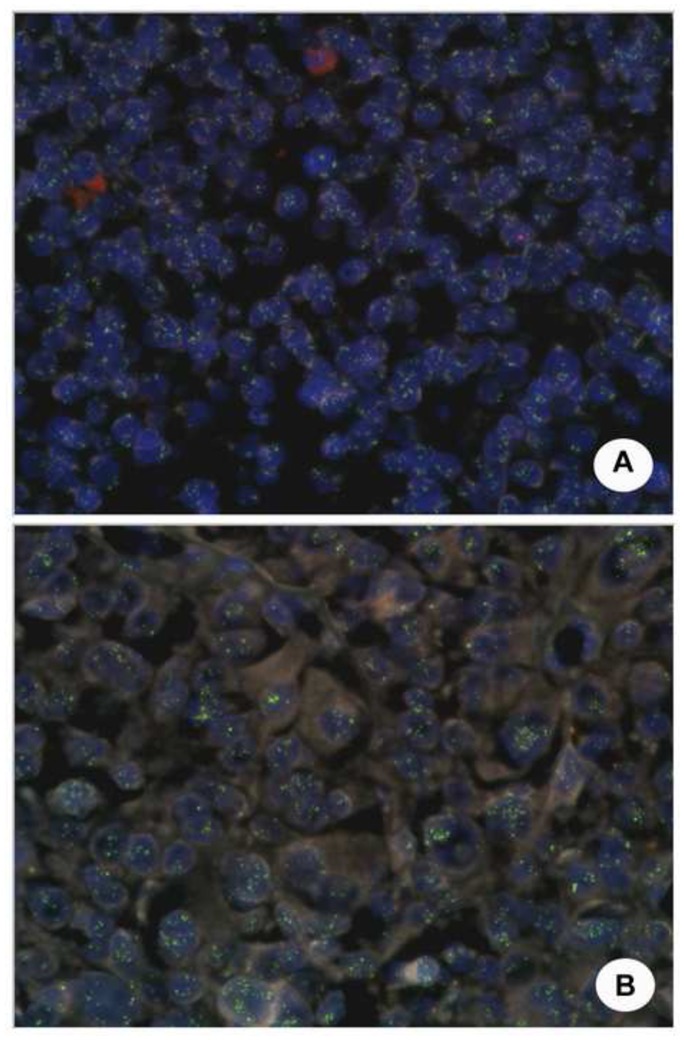
A high copy number of the CCND1 gene, with more than 10 signals per nuclei, can be evidenced in the majority (over 70% of cells) of neoplastic cells from cases 11 (A) and 4 (B).
Regarding sequence analyzes, only one case had a BRAF mutation, yielding a D594G substitution, while the classical V600E was never found. G12D (n=2) or G12A (n=1) NRAS mutations in exon 2 were demonstrated in 3 cases (18%). Finally only one sample was associated with a KIT mutation in exon 11, leading to a replacement in position 576 of a leucine by a proline (L576P) (Figure 3 and table 3).
Figure 3. Somatic mutations in sinonasal melanoma.
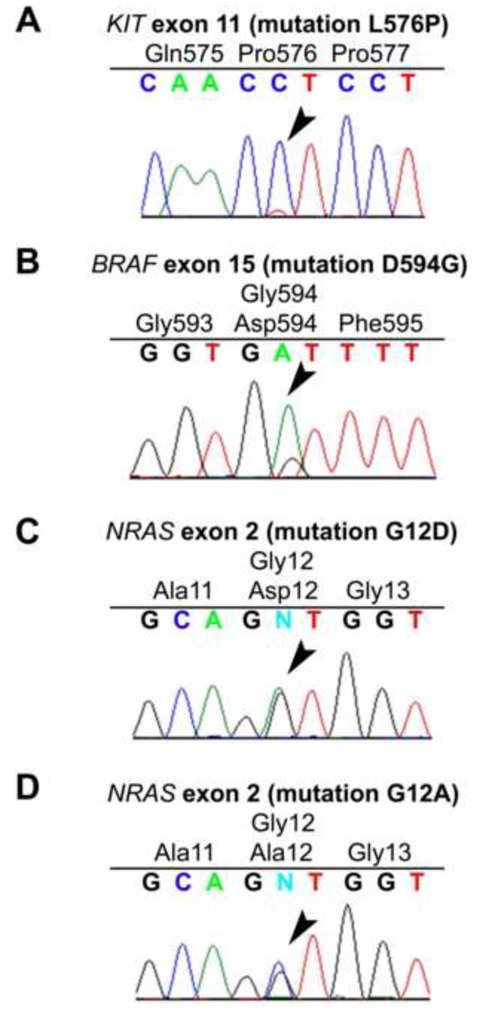
Chromatograms showing the following mutated genes: KIT (A), BRAF (B), and NRAS (C, D). Sequences are shown above the chromatograms, arrows indicate the substitutions, the wild-type amino acids are in black and the mutated one in red.
Table 3.
Mutation analyses of BRAF, NRAS and KIT
| Case | BRAF exon 15 | NRAS exon 2 | NRAS exon 3 | KIT exon 11 | KIT exon 13 | KIT exon 17 | KIT exon 18 |
|---|---|---|---|---|---|---|---|
| 1 | WT | WT | WT | L576P | WT | WT | WT |
| 2 | WT | G12D | WT | WT | WT | WT | WT |
| 3 | WT | G12A | WT | WT | WT | WT | WT |
| 4 | WT | WT | WT | WT | WT | WT | WT |
| 5 | WT | WT | WT | WT | WT | WT | WT |
| 6 | WT | WT | WT | WT | WT | WT | WT |
| 7 | WT | WT | WT | WT | WT | WT | WT |
| 8 | WT | WT | WT | WT | WT | WT | WT |
| 9 | WT | WT | WT | WT | WT | WT | WT |
| 10 | WT | WT | WT | WT | WT | WT | WT |
| 11 | WT | WT | WT | WT | WT | WT | WT |
| 14 (in situ) | WT | WT | WT | WT | WT | WT | WT |
| 14 (invasive) | WT | WT | WT | WT | WT | WT | WT |
| 15 | WT | G12D | WT | WT | WT | WT | WT |
| 16 | WT | WT | WT | WT | WT | WT | WT |
| 17 | D594G | WT | WT | WT | WT | WT | WT |
Abbreviation : WT, wild type
All the genetic aberrations described above appeared to be mutually exclusive, except for cases 9 and 15. In case 9, several genes were found to be amplified (CCND1, CDK4 and MITF), while case 15 showed both a G12D NRAS mutation and an amplification of the CCND1 gene.
Molecular and phenotypic correlations
At the protein level using immunohistochemistry, c-Kit was found to be expressed in about half cases (8/17), with a variable proportion of stained cells and various staining intensities. Interestingly, only 3 showed a high proportion (about 50% in all three) of stained cell and among them, 2 had KIT gene abnormalities, with one being amplified and the other showing an activating mutation (Table 4).
Table 4.
KIT, CCND1, CDK4, MDM2 and MITF gene copy number assessment and Ckit and cyclin D1 protein expression
| Case | KIT gene copy number | cKit protein in situ | cKit protein invasive | CCND1 gene copy number | CCND1 Amplification (FISH) | Cyclin D1 protein in situ | Cyclin D1 protein invasive | CDK4 gene copy number | MDM2 gene copy number | MITF gene copy number |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 1+ | 5+ | 1 | − | − | 1+ | 3 | 3 | 3 |
| 2 | 3 | 5+ | 0 | 2 | ND | 1++ | 2++ | 3 | 2 | 4 |
| 3 | 2 | NE | 0 | 3 | ND | NE | 0 | 3 | 2 | 4 |
| 4 | 5 | NE | 1+ | 7 | + | NE | 8+++ | 4 | 5 | 4 |
| 5 | 3 | 5++ | 0 | 9 | ND | 3++ | 3+++ | 2 | 1 | 3 |
| 6 | 3 | 3++ | 0 | 3 | ND | 2++ | 5++ | 4 | 3 | 4 |
| 7 | 1 | NE | 0 | 5 | ND | NE | 0 | 3 | 2 | 3 |
| 8 | 1 | NE | 0 | 4 | ND | NE | 0 | 2 | 2 | 5 |
| 9 | 2 | 5+ | 0 | 13 | ND | 0 | 1+ | 8 | 4 | 8 |
| 10 | 8 | 5++ | 5++ | 1 | ND | 0 | 0 | 2 | 2 | 3 |
| 11 | 2 | 5++ | 5+ | 12 | + | 0 | 0 | 3 | 2 | 3 |
| 12 | ND | 1+++ | 1+++ | ND | ND | ND | ND | ND | ND | ND |
| 14 | 2 | 9+++ | 9+++ | 3 | − | 5+++ | 7+++ | 3 | 2 | 3 |
| 15 | 4 | 0 | 0 | 7 | + | − | 1+ | 4 | 4 | 4 |
| 16 | 2 | NE | 5+ | 2 | − | NE | 5+++ | 3 | 2 | 2 |
| 17 | 1 | 10+++ | 5++ | 2 | ND | 7+++ | 8+++ | 2 | 2 | 2 |
NOTE : For immunohistochemical analyzes, the proportion of positive cells is expressed as 1 for 10%, 2 for 20%, up to 10 for 100%, and the staining intensity as + for low, ++ for moderate and +++ for high.
Abbreviations : ND, not done; NE, non evaluable.
By contrast, no strict correlation between CCND1 gene amplification and cycline D1 protein expression was observed, as only 2 of the amplified cases displayed a high proportion of stained cells, whereas 3 with no gene amplification were associated with an important proportion of stained cells (Table 4).
Discussion
Primary malignant melanoma of sinonasal tract (PMMST) is a rare form of melanoma with a poor prognosis. We here report a series of 17 cases in which we focused on oncogene mutations and amplifications. Clinical-pathological findings were similar to that reported in the large series of cases from Thompson et al. [1] In our series, we found that most cases were associated with an in situ component (78%), suggesting that PMMST follows a two step invasion process, with an initial intra-mucosal development, similar to skin melanomas. The finding of only an in situ melanoma in a patient that consulted for nasal obstruction due to a nasal polyp supports this hypothesis. Interestingly, the situ components appeared to be more pigmented than the invasive components, which may suggest that melanoma progression is associated with a loss in melanocytic differentiation. While only few cases showed a significant inflammatory infiltrate in the invasive components, in situ lesions were always associated with a coarse infiltrate. This may suggest that tumor escape mechanisms may develop and be associated with melanoma progression.
An understanding of the genetic basis of cutaneous melanoma has recently shed light on the mechanisms of melanoma genesis. Especially, somatic mutations of the BRAF, NRAS, HRAS and GNAQ genes have been shown to be associated with variable frequencies with melanoma subtypes. Due to anatomic complexity of the sinonasal tract, leading to difficult surgical removal of the primary tumor, and because they are often diagnosed at advanced stages and follow an aggressive course, PMMST are good candidates for additional treatments. To the best of our knowledge, however, only one recently published study focused on PMMST. [7] Interestingly, among the 32 cases recently published in this series, none was associated with the BRAFV600E mutation, while a D594G could be demonstrated in one sample, as in our series. The BRAFD594G mutation appears to be rare, and actually, to the best of our knowledge, was only described in a melanoma cell line.[8] Compared to the classical V600E, this mutation appears to be a low activity mutant and to signal through CRAF on the MAPK pathway. We found NRAS mutations in 18% (3/17) of our sample, a frequency similar to that previously reported.[9] However, the NRAS mutation spectrum in PMMST is different from the one found in other melanoma. Indeed, NRAS mutations in cutaneous melanoma are usually localized at codon Q61 (88%) and rarely at codons G12 (6%) and G13 (6%).[10] The high frequency of NRAS mutations at codon 61 in melanoma has been linked to the presence of mutagenic UV-induced DNA photoproducts, which have been found preferentially around codon 61 and rarely around codon 12 in UV-irradiated human cells.[11] Interestingly, the NRAS mutations we found in PMMST were all located at codon G12 and in a recent analysis of sinonasal melanoma, Turri-Zanoni M et al. also found NRAS mutations at codons G12 and G13.[7] Therefore, the NRAS mutation spectrum found in nasal melanoma is different from the one found in other melanoma but strikingly closer to the one described in tumors derived from hematopoietic and lymphoid tissue. This result suggests that the molecular mechanisms involved in NRAS mutation in PMMST are different from other melanomas, and, as expected in this anatomic localization, are unrelated to UV exposure. RAS acts as a MAPK signaling protein, upstream of RAF. Although no specific therapies aiming at RAS are available to date, it is known that inhibition of NRAS should aim both BRAF and CRAF or BRAF and PIK3CA.[12]The interaction between the MAPK pathway is actually complex. It has been shown for instance that oncogenic RAS inhibits NRAS-BRAF interaction, leading to CRAF reactivation ad cAMP phosphodiesterase hyperactivity.[13]
KIT mutations were previously shown to be preferentially associated with mucosal melanomas. They are expected at a 9–10% rate and have been shown to represent an adverse prognostic factor.[14] We actually found only one sample with a KIT mutation, while KIT mutations were found in 4 out of 32 samples in the series from Turri-Zanoni et al. Our case had a L576P mutation in exon 11, which is the most frequent KIT mutation in melanomas. We also found significant c-Kit protein expression in a subset of cases (62.5%), including the one associated with an activating mutation of the gene, a percentage however lower than the 97% of cases showing KIT protein expression in the study from Turri-Zanoni M et al. These results suggest that c-Kit is involved in PMMST cells survival and/or proliferation, but may not be as important as in other mucosal melanoma subtypes. Overall, we did not find a significant correlation between c-Kit expression and KIT mutation status, as shown for melanoma of mucous membranes from other sites,[4] further suggesting the PMMST have distinctive oncogenic properties.
We found CCND1 amplification by qPCR in a significant subset of our cases (37,5%), and confirmed it in some samples by FISH analysis. In keeping with this observation, many cases showed a nuclear expression of cyclin D1, although no strict correlation could be observed between CCND1 amplification and cyclin D1 protein expression, suggesting that cyclin D1 protein expression is regulated at many levels in melanoma. The reason why we found more frequent CCND1 amplification than Turri-Zanoni et al in their recent study is unclear, but may be due to the different probe we used or the high sensitivity of qPCR. Nevertheless, our data suggest that CCND1 is an important oncogene in sinonasal melanoma and that these tumors could be amenable for therapy targeting the cyclinD1/CDK4 pathway.
In conclusion, we confirm recently published results showing that a proportion of PMMST are associated with NRAS and less frequently KIT activating mutation, while the classical BRAFV600E is absent. In our series, c-Kit was expressed at the protein level in only half cases, and was not an indicator of KIT mutation, since only one case had a KIT mutation. All together the results further suggest that PMMST should not be sensitive to targeted therapies aiming at BRAF. PMMST appear to be heterogeneous at the molecular level and appear in our series to be associated with CCND1 amplification and NRAS mutations. Further studies are needed to determine whether cases associated with NRAS or KIT mutations or CCND1 amplifications may benefit from specific targeted therapies, together with the identification of new oncogenes in cases where no specific alterations could be identified. Also, longitudinal studies are needed to determine whether these molecular abnormalities have prognostic implications.
References
- 1.Thompson LD, Wieneke JA, Miettinen M. Sinonasal tract and nasopharyngeal melanomas: a clinicopathologic study of 115 cases with a proposed staging system. Am J Surg Pathol. 2003;27(5):594–611. doi: 10.1097/00000478-200305000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Prasad ML, Patel SG, Huvos AG, Shah JP, Busam KJ. Primary mucosal melanoma of the head and neck: a proposal for microstaging localized, Stage I (lymph node-negative) tumors. Cancer. 2004;100(8):1657–64. doi: 10.1002/cncr.20201. [DOI] [PubMed] [Google Scholar]
- 3.Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29(41):5545–55. doi: 10.1038/onc.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres-Cabala CA, Wang WL, Trent J, Yang D, Chen S, Galbincea J, et al. Correlation between KIT expression and KIT mutation in melanoma: a study of 173 cases with emphasis on the acral-lentiginous/mucosal type. Mod Pathol. 2009;22(11):1446–56. doi: 10.1038/modpathol.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J, Si L, Kong Y, Flaherty KT, Xu X, Zhu Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Oncol. 2011;29(21):2904–9. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 7.Turri-Zanoni M, Medicina D, Lombardi D, Ungari M, Balzarini P, Rossini C, et al. Sinonasal mucosal melanoma: Molecular profile and therapeutic implications from a series of 32 cases. Head Neck. 2012 doi: 10.1002/hed.23079. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28(1):85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 10.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer. 72(10):2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tormanen VT, Pfeifer GP. Mapping of UV photoproducts within ras proto-oncogenes in UV-irradiated cells: correlation with mutations in human skin cancer. Oncogene. 1992;7(9):1729–36. [PubMed] [Google Scholar]
- 12.Jaiswal BS, Janakiraman V, Kljavin NM, Eastham-Anderson J, Cupp JE, Liang Y, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4(5):e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marquette A, Andre J, Bagot M, Bensussan A, Dumaz N. ERK and PDE4 cooperate to induce RAF isoform switching in melanoma. Nat. 2011;18(5):584–91. doi: 10.1038/nsmb.2022. [DOI] [PubMed] [Google Scholar]
- 14.Kong Y, Si L, Zhu Y, Xu X, Corless CL, Flaherty KT, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res. 2011;17(7):1684–91. doi: 10.1158/1078-0432.CCR-10-2346. [DOI] [PubMed] [Google Scholar]


