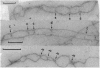
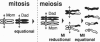Abstract
A physical connection between homologs is required for reductional segregation at the first division of meiosis. This connection is usually provided by one or a few well-spaced crossovers. A speculative overview of processes leading to formation of these crossovers is presented.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett M. D. The time and duration of meiosis. Philos Trans R Soc Lond B Biol Sci. 1977 Mar 21;277(955):201–226. doi: 10.1098/rstb.1977.0012. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992 May 1;69(3):439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E., Laemmli U. K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988 Dec 23;55(6):937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. Chiasma function. Cell. 1994 Jul 1;77(7):957–962. doi: 10.1016/0092-8674(94)90434-0. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma. 1979;75(3):259–292. doi: 10.1007/BF00293472. [DOI] [PubMed] [Google Scholar]
- Dawe R. K., Sedat J. W., Agard D. A., Cande W. Z. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994 Mar 11;76(5):901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- De Massy B., Baudat F., Nicolas A. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11929–11933. doi: 10.1073/pnas.91.25.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas L. T. Meiosis VII: "detorsive bending" as a basis for geometric shapes of late prophase bivalents. Genetica. 1970;41(2):231–256. doi: 10.1007/BF00958909. [DOI] [PubMed] [Google Scholar]
- Dresser M. E., Giroux C. N. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol. 1988 Mar;106(3):567–573. doi: 10.1083/jcb.106.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. The synaptonemal complex and the distribution of meiotic recombination events. Trends Genet. 1995 Jun;11(6):206–208. doi: 10.1016/s0168-9525(00)89046-0. [DOI] [PubMed] [Google Scholar]
- Foss E., Lande R., Stahl F. W., Steinberg C. M. Chiasma interference as a function of genetic distance. Genetics. 1993 Mar;133(3):681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson L. A., Stahl F. W. Initiation of meiotic recombination is independent of interhomologue interactions. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11934–11937. doi: 10.1073/pnas.91.25.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Abián J. F., Clarke D. J., Mullinger A. M., Downes C. S., Johnson R. T. A postprophase topoisomerase II-dependent chromatid core separation step in the formation of metaphase chromosomes. J Cell Biol. 1995 Oct;131(1):7–17. doi: 10.1083/jcb.131.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981 Sep;25(3):591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- Goldway M., Sherman A., Zenvirth D., Arbel T., Simchen G. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double strand breaks. Genetics. 1993 Feb;133(2):159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993 Jan;13(1):373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994 May;125(3):517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J. J., McKim K. S., Boyd J. B., Hawley R. S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995 Sep 8;82(5):815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Havekes F. W., de Jong J. H., Heyting C., Ramanna M. S. Synapsis and chiasma formation in four meiotic mutants of tomato (Lycopersicon esculentum). Chromosome Res. 1994 Jul;2(4):315–325. doi: 10.1007/BF01552725. [DOI] [PubMed] [Google Scholar]
- Henderson S. A. The time and place of meiotic crossing-over. Annu Rev Genet. 1970;4:295–324. doi: 10.1146/annurev.ge.04.120170.001455. [DOI] [PubMed] [Google Scholar]
- Heyting C., Dietrich A. J. Synaptosomal complexes and the organization of chromatin during meiotic prophase. Cell Biol Int Rep. 1992 Aug;16(8):749–760. doi: 10.1016/s0309-1651(05)80019-0. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Clark S. H., Chovnick A. The effect of DNA sequence polymorphisms on intragenic recombination in the rosy locus of Drosophila melanogaster. Genetics. 1991 Nov;129(3):779–781. doi: 10.1093/genetics/129.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C. Coming undone: how to untangle a chromosome. Cell. 1994 Jul 1;77(7):955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Jang J. K., Messina L., Erdman M. B., Arbel T., Hawley R. S. Induction of metaphase arrest in Drosophila oocytes by chiasma-based kinetochore tension. Science. 1995 Jun 30;268(5219):1917–1919. doi: 10.1126/science.7604267. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995 Apr;139(4):1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. H. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992 Oct;132(2):387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Kleckner N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996 May;1(5):475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- King J. S., Mortimer R. K. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990 Dec;126(4):1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Padmore R., Bishop D. K. Meiotic chromosome metabolism: one view. Cold Spring Harb Symp Quant Biol. 1991;56:729–743. doi: 10.1101/sqb.1991.056.01.082. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Weiner B. M. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Kohli J., Bähler J. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia. 1994 Mar 15;50(3):295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- Lande R., Stahl F. W. Chiasma interference and the distribution of exchanges in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1993;58:543–552. doi: 10.1101/sqb.1993.058.01.061. [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein F., Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994 Jun;125(6):1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOLE-BAJER J. Cine-micrographic analysis of C-mitosis in endosperm. Chromosoma. 1958;9(4):332–358. doi: 10.1007/BF02568085. [DOI] [PubMed] [Google Scholar]
- Maguire M. P. Is the synaptonemal complex a disjunction machine? J Hered. 1995 Sep-Oct;86(5):330–340. doi: 10.1093/oxfordjournals.jhered.a111600. [DOI] [PubMed] [Google Scholar]
- McCarroll R. M., Esposito R. E. SPO13 negatively regulates the progression of mitotic and meiotic nuclear division in Saccharomyces cerevisiae. Genetics. 1994 Sep;138(1):47–60. doi: 10.1093/genetics/138.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W. Y., Orr-Weaver T. L. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Moens P. B., Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma. 1995 Nov;104(3):175–182. doi: 10.1007/BF00352182. [DOI] [PubMed] [Google Scholar]
- Molnar M., Bähler J., Sipiczki M., Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995 Sep;141(1):61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møens P. B., Pearlman R. E. Chromatin organization at meiosis. Bioessays. 1988 Nov;9(5):151–153. doi: 10.1002/bies.950090503. [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., Scherthan H., Rockmill B., Bhargava J., Roeder G. S. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995 Sep;141(1):75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B. Chromosome distribution: experiments on cell hybrids and in vitro. Philos Trans R Soc Lond B Biol Sci. 1977 Mar 21;277(955):267–276. doi: 10.1098/rstb.1977.0017. [DOI] [PubMed] [Google Scholar]
- Ohta K., Shibata T., Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994 Dec 1;13(23):5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L. Meiosis in Drosophila: seeing is believing. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10443–10449. doi: 10.1073/pnas.92.23.10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991 Sep 20;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Pukkila P. J. Meiotic sister chromatid recombination. Adv Genet. 1995;33:41–62. doi: 10.1016/s0065-2660(08)60330-2. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Goldsmith M. R., Hamkalo B. A. Chromosome organization during male meiosis in Bombyx mori. Chromosoma. 1981;82(3):341–351. doi: 10.1007/BF00285760. [DOI] [PubMed] [Google Scholar]
- Raymond W. E., Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993 Aug 11;21(16):3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Sym M., Scherthan H., Roeder G. S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995 Nov 1;9(21):2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Roeder G. S. Chromosome synapsis and genetic recombination: their roles in meiotic chromosome segregation. Trends Genet. 1990 Dec;6(12):385–389. doi: 10.1016/0168-9525(90)90297-j. [DOI] [PubMed] [Google Scholar]
- Roeder G. S. Sex and the single cell: meiosis in yeast. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol Cell Biol. 1993 Jun;13(6):3445–3455. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Bähler J., Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994 Oct;127(2):273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmekel K., Daneholt B. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 1995 Jun;5(6):239–242. doi: 10.1016/s0962-8924(00)89017-0. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995 Dec 1;83(5):783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994 Jan 14;76(1):51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Sears D. D., Hieter P., Simchen G. An implanted recombination hot spot stimulates recombination and enhances sister chromatid cohesion of heterologous YACs during yeast meiosis. Genetics. 1994 Dec;138(4):1055–1065. doi: 10.1093/genetics/138.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples G. J., Leach D. R. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995 Sep;17(6):1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- Sherman J. D., Herickhoff L. A., Stack S. M. Silver staining two types of meiotic nodules. Genome. 1992 Dec;35(6):907–915. doi: 10.1139/g92-140. [DOI] [PubMed] [Google Scholar]
- Simchen G., Hugerat Y. What determines whether chromosomes segregate reductionally or equationally in meiosis? Bioessays. 1993 Jan;15(1):1–8. doi: 10.1002/bies.950150102. [DOI] [PubMed] [Google Scholar]
- Solari A. J. Synaptic behaviour and recombination nodules in the human XY pair. Genetica. 1988 Sep 30;77(2):149–158. doi: 10.1007/BF00057766. [DOI] [PubMed] [Google Scholar]
- Solari A. J., Tandler C. J. Presence of a centromeric filament during meiosis. Genome. 1991 Dec;34(6):888–894. doi: 10.1139/g91-136. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Xu L., Cao L., Kleckner N. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suja J. A., Antonio C., Rufas J. S. Involvement of chromatid cohesiveness at the centromere and chromosome arms in meiotic chromosome segregation: a cytological approach. Chromosoma. 1992 Jun;101(8):493–501. doi: 10.1007/BF00352472. [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J. A., Roeder G. S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993 Feb 12;72(3):365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G. S. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994 Oct 21;79(2):283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Weiner B. M., Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994 Jul 1;77(7):977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Worth L., Jr, Clark S., Radman M., Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994 Jan 28;263(5146):515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- Xu L., Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995 Oct 16;14(20):5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth D., Arbel T., Sherman A., Goldway M., Klein S., Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992 Sep;11(9):3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. Development of the synaptonemal complex and the "recombination nodules" during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma. 1977 Jun 23;61(4):289–316. doi: 10.1007/BF00288615. [DOI] [PubMed] [Google Scholar]
- Zickler D., Moreau P. J., Huynh A. D., Slezec A. M. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992 Sep;132(1):135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen S. W., Holm P. B. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]