Abstract
The objective of this study is to examine the cumulative effect of the less studied genetic variants in PLEKHA1/ARMS2/HTRA1 on age-related macular degeneration (AMD). We performed an extensive literature search for studies on the association between AMD and the less studied genetic variants in PLEKHA1/ARMS2/HTRA1. Multiple meta-analyses were performed to evaluate the association between individual genetic variants and AMD. A gene-cluster analysis was used to investigate the cumulative effect of these less studied genetic variants on AMD. A total of 23 studies from 20 published papers met the eligibility criteria and were included in our analyses. Several genetic variants in the gene cluster are significantly associated with AMD in our meta-analyses or in individual studies. Gene-cluster analysis reveals a strong cumulative association between these genetic variants in this gene cluster and AMD (p<10−5). However, two previously suspected SNPs in ARMS2, including rs2736911, the SNP having the largest number of studies in our meta-analyses; and rs3793917, the SNP with the largest sample size, were not significantly associated with AMD (both p’s>0.12). Sensitivity analyses reveal significant association of AMD with rs2736911 in Chinese but not in Caucasian, with c.372_815del443ins54 in Caucasian but not in Chinese, and with rs1049331 in both ethnic groups. These less studied genetic variants have a significant cumulative effect on wet AMD. Our study provides evidence of the joint contribution of genetic variants in PLEKHA1/ARMS2/HTRA1 to AMD risk, in addition to the two widely studied genetic variants whose association with AMD was well established.
Keywords: Macular degeneration, Polymorphism, Meta-analysis, Gene-cluster analysis
Introduction
Age-related macular degeneration (AMD) is a degenerative disorder in the macular region and a primary cause of irreversible blindness in developed countries [1] and the third leading cause worldwide [2]. In the United States, approximately 1.75 million (1.47%) individuals 40 years or older have AMD, with a projected number of persons having AMD increasing by more than 50% to around 2.95 million in 2020 [3]. In developing countries in Asia, the prevalence of AMD was estimated to be around 7.0% among individuals 40 years and older [4]. Worldwide, AMD affects about 50 million individuals [1]. AMD has been estimated to account for over 54% of visual impairment and approximately 23% of blindness among Caucasians [5]. In addition to having a devastating effect on patients’ lives, AMD also causes substantial economic burden both to the patients, their families and to the society.
Early stage AMD is characterized by the presence of medium-size drusen and/or retinal pigmentary abnormalities, and advanced AMD is typically classified into non-neovascular (dry, atrophic or nonexudative) and neovascular (wet or exudative). Although wet/neovascular AMD accounts for only about 10–15% of AMD cases, it is responsible for more than 80% of cases of severe vision loss or legal blindness [6].
Previous studies identified several risk factors for AMD, with age being the highest risk factor.[7] Cigarette smoking is a preventable risk factor that is consistently associated with AMD.[8] Several other risk factors, including obesity [9], hypertension [10] and White race [11], have also been implicated in the increased risk of AMD.
Over the past few years, numerous studies have also been conducted to search for susceptibility genes for AMD. Specifically, CFH, located on 1q32 and the gene cluster PLEKHA1/ARMS2/HTRA1, located on 10q26, have been identified as major AMD-susceptibility genes. The Tyr402His variant (rs1061170) in CFH, where a tyrosine is replaced by a histidine, has been reported to confer major risk for AMD [12–15]. The protective genotypes of rs1061170 (CT or TT) can prevent activation of inflammatory cascades in retinal pigment epithelium (RPE) cell and macrophages by binding to oxidized phospholipids (oxPLs), resulting in reduction of abnormal angiogenesis [16]. The Ala69Ser variant (rs10490924) in ARMS2 (or LOC387715), where alanine is replaced by serine, was found be significantly associated with AMD, with GG and GT carriers having approximately 2.7-fold and 8.2-fold increased risk of developing AMD, respectively [17]. This genetic variation has been hypothesized to alter the function of LOC387715/ARMS2 protein, probably by influencing its conformation and interaction with other genes [18]. The SNP rs11200638 in HTRA1, located within the putative HTRA1 promoter, was found to significantly increase the risk of AMD [19,20], with AG and AA carriers having 2.24 and 8.67 times of risk of developing AMD, respectively [21]. Genetic variation in rs11200638 can increase the expression of HTRA1 mRNA and protein [20,19], leading to an altered elastogenesis in Bruch’s membrane (BM) through fibulin 5 cleavage [22]. The SNP is in almost complete linkage disequilibrium (LD) with rs10490924 [20].
In addition to the two widely studied SNPs in PLEKHA1/ARMS2/HTRA1, the association between AMD and many other less studied genetic variants in this gene cluster has also been explored in many studies, with conflicting results. In this study, we performed meta-analyses of these less studied genetic variants in the gene cluster, and conducted a gene-cluster analysis to examine the cumulative effect of these genetic variants in the three genes on AMD risk.
Methods
Search strategy and study selection
We did an extensive literature search in MEDLINE in June 2011 on the association of genetic variants in PLEKHA1/ARMS2/HTRA1 with AMD. Search terms included “PLEKHA1,” “LOC387715,” “ARMS2,” “HTRA1,” “age related macular degeneration,” and “AMD.” Studies were included in our analysis if they met the following criteria: 1) studies on human subjects; 2) outcomes of interest include age-rated macular degeneration; and 3) report of genotype data of individual SNPs in PLEKHA1/ARMS2/HTRA1 of participants with and without AMD (or provided odds ratios and their variances). All potentially relevant publications were retrieved and evaluated for inclusion. References of all relevant publications were also hand-searched for additional studies missed by the database search. Only studies published in the English language were included. Two authors (WY and JY) performed the search independently. Disagreement over eligibility of a study was resolved by discussion until a consensus was reached.
Data extraction
Two reviewers (SD and JY) independently extracted the following data according to a pre-specified protocol: first author’s name, year of publication, characteristics (sample size, mean age, percentage of male and race/country of participants) of the study populations, genotype data for subjects with and without AMD (or odds ratio and the corresponding variances for the SNPs), and the genetic model used (additive, allelic, dominant or recessive). Discrepancies were resolved by discussion. Extracted data were entered into a computerized spreadsheet for analysis.
Statistical analysis
Odds ratio (OR) was used as a measure of the association between the genetic variants in the gene cluster and AMD. We used random-effects models to calculate OR and the corresponding 95% confidence interval (CI) for the meta-analysis when there was significant heterogeneity between the studies; otherwise, fixed-effect models were used [23]. The Z-test was used to calculate the p-value of the overall effect for the meta-analysis. We used forest plots to graphically present the calculated pooled ORs and their 95% CIs. Each study was represented by a square in the plot, and the area of the square is proportional to the weight of the study. The overall effect from the meta-analysis is represented by a diamond, with its width representing the 95% CI for the estimate. In the random-effects meta-analysis, we used the inverse of the variance of each study as the weight for the study. We used Q-test for assessment of between-study heterogeneity. Publication bias was assessed using Egger’s regression test.
In order to assess the cumulative association between AMD and the less studied genetic variants in the gene cluster, we conducted a gene-cluster analysis using the p-values of each genetic variant in PLEKHA1/ARMS2/HTRA1 calculated from meta-analyses and/or from individual publications. We used three popular p-value combination methods to assess this association: Fisher’s method [24], Simes method [25], and truncated product method (TPM) [26]. A detailed description of the three methods was reported elsewhere [27]. To deal with differences in sample size when assessing the association of each individual SNP, we calculated un-weighted and weighted TPM. Un-weighted TPM disregards the difference in sample size, and weighted TPM employs the sample size as its weight, allowing studies with larger sample size to play a larger role in the calculation [26]. Because these p-values are most likely dependent, we used 100,000 simulations to estimate the p-value for the two TPMs.
Sensitivity Analysis
To examine whether there is difference between races in the association between genetic variants in the gene cluster and AMD, we analyzed the association in Chinese and Caucasian separately. In addition, we analyzed the association between these genetic variants and wet AMD which is responsible for the majority of severe vision loss or legal blindness.
Meta-analysis was performed using Stata 11.2 (StataCorp LP, College Station, TX). All the other analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC) and Matlab 7.10.0.499 (The MathWorks, Inc., Natick, MA).
Results
Literature search and eligible studies
The flow diagram showing the selection of studies to be included in our analysis adheres to the QUOROM statement and is shown in Fig 1 [28]. Using our pre-defined search strategy, we identified a total of 220 potential publications through our initial search. After screening the abstracts of these studies, 72 were excluded either because they were irrelevant, not about human subjects, or not published in English. The remaining 148 studies were retrieved for more detailed evaluations, which excluded an additional 128 studies because they were irrelevant, there were not sufficient data, the outcome of interest was not AMD, or they were meta-analyses or review studies. This left 20 potentially relevant publications (with 23 studies) to be included in our analysis. A further review of the references of these studies and review papers identified one more study. Further exploration of the data from these studies excluded one more study with insufficient data. A total of 23 studies from 20 published papers met the eligibility criteria and were included in our analyses [29–45,18,46,17].
Figure 1.
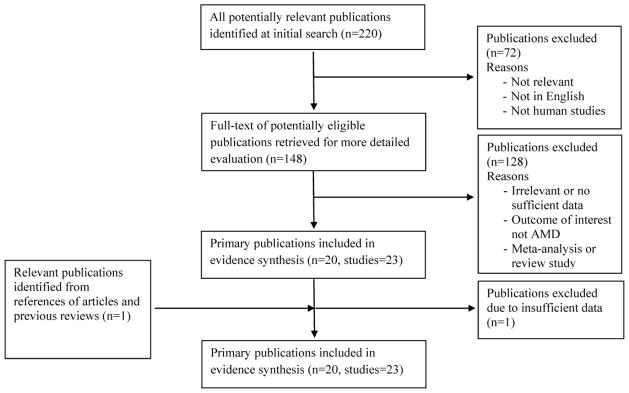
Flow diagram of studies included in the systematic review.
All qualified publications were published since 2005 and had sample sizes ranging from 130 to 3,307 (Table 1). Prevalence of AMD ranged from 19% to 77%. Of these 23 studies, eight studies reported association results for rs2736911, five studies for rs3750848 and four studies for c.372_815del443ins54, rs2014307, rs2672587, and rs3793917. The combined study population included 2,832 participants in the meta-analysis of rs2736911, 2,847 of rs3750848, 2,288 of c.372_815del443ins54, 2,114 of rs2014307, 2,685 of rs2672587, and 5,680 of rs3793917. Meta-analyses for other genetic variants in the gene cluster are based on fewer studies (Table 2). In addition to the 38 genetic variants included in the meta-analyses, the association between AMD and 56 additional genetic variants in the gene cluster was reported in individual studies (or calculated based on individual studies). The results, together with results obtained from our mea-analyses, were included in our gene-cluster analysis.
Table 1.
Basic characteristics of all studies.
| Study (author, year) | Race/Country | AMD | Control | ||||
|---|---|---|---|---|---|---|---|
| n | Age (Mean±SD) | Male (%) | n | Age (Mean±SD) | Male (%) | ||
| Liang et al., 2012 | Chinese | 156 | 75.9±7.4 | 52.6 | 248 | 73.6±7.4 | 44.8 |
| Teper et al., 2012 | Poland | 90 | NA | NA | 40 | NA | NA |
| Gotoh et al., 2010 | Japanese | 84 | 76.2±8.6 | 73.8 | 276 | 46.6±6.7 | NA |
| Richardson et al., 2010 | Australia | 402 | 72.7±7.4 | 36.6 | 119 | 71.8±7.7 | 44.5 |
| Chen et al., 2010 | America | 2157 | 78.6 | 38.2 | 1150 | 74.1 | 44.1 |
| Wang et al., 2010 | Caucasian | 819 | 76.1±7.9 | 34.7 | 329 | 69.5±9.0 | 41.0 |
| Goto et al., 2009 | Japanese | 100 | 74.6±8.8 | 73.0 | 190 | 72.2±8.5 | 45.3 |
| Yang et al. (a), 2010 | America | 705 | 83.3 | 45.8 | 650 | 75.3 | 38.5 |
| Yang et al. (b), 2010 | America | 442 | 80.5 | 45.0 | 434 | 78.1 | 47.5 |
| Yang et al. (c), 2010 | Chinese | 138 | 68.1 | 52.2 | 591 | 66.5 | 54.0 |
| Ricci et al., 2009 | Caucasian | 159 | 75.0±7.8 | 45.9 | 286 | 69.2±11.1 | 44.1 |
| Gotoh et al., 2009 | Japanese | 56 | 76.2±9.1 | 64.3 | 77 | 73.4±9.0 | 45.5 |
| Bergeron-Sawitzke et al., 2009 | Primarily Caucasian | 421 | 64.8 | 45.1 | 215 | 66.5 | 47.0 |
| Zhang et al., 2008 | Caucasian | 134 | 71.3±8.3 | 45.5 | 134 | 72.8±9.0 | 39.6 |
| Fritsche et al., 2008 | Germany | 794 | 76.8±6.6 | 35.6 | 612 | 76.2±5.3 | 37.9 |
| Kaur et al., 2008 | India | 250 | 68.8±3.1 | NA | 250 | 64.4±4.8 | NA |
| Tam et al., 2008 | Chinese | 163 | 75.5±7.5 | 54.0 | 183 | 75.3±6.5 | 49.7 |
| DeAngelis et al., 2008 | Caucasian | 134 | 71.3±8.3 | 45.5 | 134 | 72.8±9.0 | 39.6 |
| Leveziel et al., 2007 | France | 200 | 72.3±3.8 | 22.5 | 116 | 72.0±3.8 | 39.7 |
| Kanda et al., 2007 | Caucasian | 466 | NA | NA | 280 | NA | NA |
| Fisher et al., 2007 | Russian | 155 | 72.6±7.6 | NA | 151 | 71.1±7.3 | NA |
| Rivera et al. (a), 2005* | Germany | 794 | 76.8±6.6 | 35.6 | 612 | 76.2±5.3 | 37.9 |
| Rivera et al. (b), 2005 | Germany | 373 | 75.0±7.5 | 35.1 | 335 | 68.3±8.1 | 44.5 |
Used the data from the same participants as in Fritsche et al., 2008.
Multiple studies with a paper are marked using (a), (b), …
AMD: age-related macular degeneration; SD: standard deviation; NA: not available.
Table 2.
Meta-analyses of the association between AMD and the less studied genetic variants in PLEKHA1/ARMS2/HTRA1a.
| SNP | Gene | # of studies | AMD | Control | OR (95% CI) | P |
|---|---|---|---|---|---|---|
| c.372_815del443ins54 | ARMS2 | 4 | 1128 | 1160 | 1.07 (0.48–2.35) | 0.874 |
| rs10490923 | ARMS2 | 2 | 682 | 554 | 1.31 (1.04–1.65) | 0.023 |
| rs10664316 | ARMS2 | 3 | 342 | 455 | 0.85 (0.63–1.17) | 0.323 |
| rs2736911 | ARMS2 | 8 | 1471 | 1361 | 0.77 (0.55–1.07) | 0.122 |
| rs2736912 | ARMS2 | 2 | 212 | 325 | 0.49 (0.36–0.69) | <0.0001 |
| rs36212731 | ARMS2 | 2 | 207 | 324 | 1.11 (0.97–1.26) | 0.125 |
| rs36212732 | ARMS2 | 2 | 204 | 324 | 1.10 (0.96–1.25) | 0.18 |
| rs36212733 | ARMS2 | 2 | 204 | 324 | 1.10 (0.96–1.25) | 0.18 |
| rs3750846 | ARMS2 | 2 | 205 | 324 | 1.09 (0.96–1.24) | 0.205 |
| rs3750847 | ARMS2 | 3 | 655 | 601 | 1.09 (1.01–1.18) | 0.032 |
| rs3750848 | ARMS2 | 5 | 1513 | 1334 | 1.27 (0.93–1.74) | 0.136 |
| rs2014307 | ARMS2-HTRA1 | 4 | 1106 | 1008 | 0.91 (0.65–1.26) | 0.562 |
| -502C>T | HTRA1 | 2 | 393 | 367 | 1.05 (0.88–1.25) | 0.617 |
| rs1049331 | HTRA1 | 2 | 297 | 317 | 1.29 (1.14–1.46) | <0.0001 |
| rs11200644 | HTRA1 | 2 | 182 | 465 | 0.41 (0.12–1.41) | 0.157 |
| rs2248799 | HTRA1 | 2 | 1214 | 827 | 1.28 (0.94–1.75) | 0.119 |
| rs2250804 | HTRA1 | 2 | 180 | 461 | 0.63 (0.36–1.12) | 0.113 |
| rs2253755 | HTRA1 | 2 | 1214 | 827 | 1.36 (0.98–1.89) | 0.066 |
| rs2268345 | HTRA1 | 2 | 1211 | 775 | 1.22 (1.15–1.30) | <0.0001 |
| rs2268356 | HTRA1 | 3 | 617 | 685 | 0.73 (0.36–1.46) | 0.37 |
| rs2300431 | HTRA1 | 2 | 1201 | 777 | 1.26 (1.18–1.35) | <0.0001 |
| rs2672587 | HTRA1 | 4 | 1397 | 1288 | 0.93 (0.51–1.68) | 0.8 |
| rs2672591 | HTRA1 | 2 | 843 | 823 | 0.63 (0.11–3.61) | 0.602 |
| rs2672598 | HTRA1 | 3 | 525 | 500 | 1.22 (1.03–1.45) | 0.022 |
| rs2736914 | HTRA1 | 2 | 1214 | 827 | 1.22 (1.14–1.29) | <0.0001 |
| rs3793917 | HTRA1 | 4 | 3433 | 2247 | 1.49 (0.78–2.87) | 0.231 |
| rs4752699 | HTRA1 | 2 | 539 | 554 | 0.54 (0.13–2.17) | 0.382 |
| rs4752700 | HTRA1 | 2 | 524 | 495 | 0.67 (0.18–2.55) | 0.56 |
| rs7093894 | HTRA1 | 2 | 184 | 465 | 0.46 (0.15–1.37) | 0.162 |
| rs714816 | HTRA1 | 2 | 1208 | 774 | 1.43 (1.25–1.64) | <0.0001 |
| rs763720 | HTRA1 | 2 | 842 | 824 | 0.88 (0.24–3.18) | 0.84 |
| rs932275 | HTRA1 | 2 | 1200 | 774 | 1.67 (0.92–3.03) | 0.093 |
| rs1045216 | PLEKHA1 | 3 | 1286 | 1045 | 0.99 (0.54–1.79) | 0.965 |
| rs2292625 | PLEKHA1 | 2 | 1209 | 821 | 1.22 (1.15–1.30) | <0.0001 |
| rs2421016 | PLEKHA1 | 2 | 915 | 700 | 1.22 (0.90–1.64) | 0.201 |
| rs4146894 | PLEKHA1 | 3 | 1333 | 893 | 1.38 (1.29–1.48) | <0.0001 |
| rs4405249 | PLEKHA1 | 2 | 1210 | 822 | 1.23 (1.15–1.30) | <0.0001 |
| rs10510110 | PLEKHA1- ARMS2 | 2 | 1222 | 771 | 1.38 (1.28–1.49) | <0.0001 |
Assuming an additive model.
AMD: age-related macular degeneration.
Assessment of publication bias
Egger’s test was used to assess publication bias. There was modest publication bias for the meta-analysis of rs2736911 (p=0.099) and no publication bias for the meta-analysis of rs3750848, c.372_815del443ins54, rs2014307, rs2672587 or rs3793917 (all p>0.15). Assessment of publication bias for the meta-analysis of other SNPs is not very meaningful due to the low number of studies included in the corresponding meta-analysis.
Association of individual SNPs with AMD
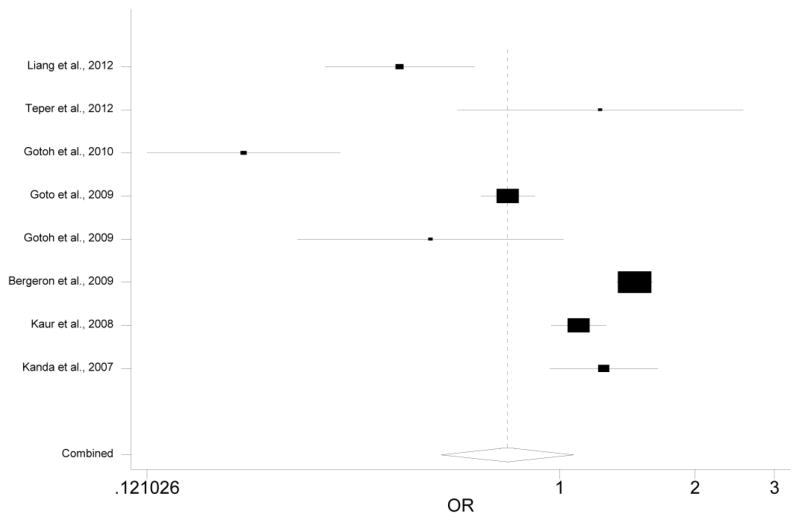
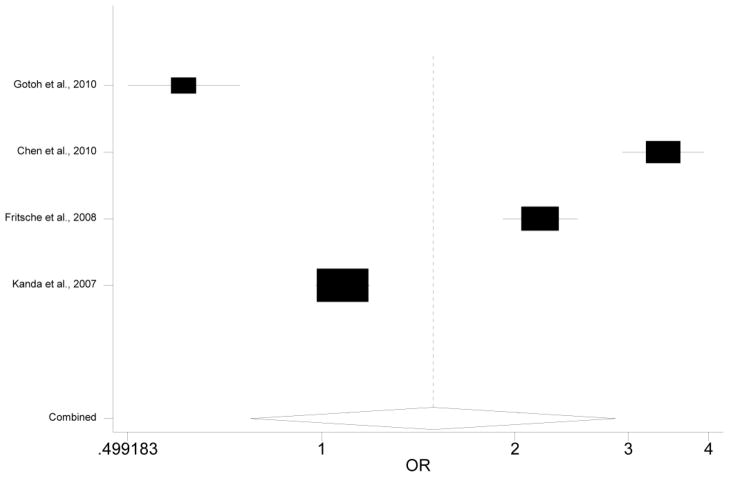
We calculated the association between the less studied genetic variants in PLEKHA1/ARMS2/HTRA1 and AMD assuming four different genetic models (additive, allelic, dominant and recessive). Due to space limit, we present the results using an additive model. Results obtained using other models can be found in the supplementary file. Both the meta-analyses and the individual association studies reveal a few SNPs that are significantly associated with AMD (Table 2 and Table 3). However, rs2736911, the SNP having the largest number of studies in our meta-analysis, is not strongly associated with AMD (OR=0.77, 95% CI: 0.55–1.07, p=0.122; Fig 2, Table 4). Similarly, rs3793917, the SNP with the largest sample size in our meta-analysis, is not significantly associated with AMD (OR=1.49, 95% CI 0.78–2.87, p=0.231, Fig 3).
Table 3.
Association between AMD and less studied genetic variants in PLEKHA1/ARMS2/HTRA1 from single studiesa.
| SNP | Gene | Study | AMD | Control | OR (95% CI) | P |
|---|---|---|---|---|---|---|
| -9G>A | ARMS2 | Liang et al., 2012 | 153 | 248 | 2.00 (0.18–22.05) | 0.571 |
| EU427528(310–311) | ARMS2 | Gotoh et al., 2009 | 56 | 77 | 1.16 (0.88–1.52) | 0.298 |
| IVS1+664A>G | ARMS2 | Liang et al., 2012 | 153 | 248 | 0.50 (0.05–5.51) | 0.571 |
| IVS1-746A>G | ARMS2 | Liang et al., 2012 | 154 | 247 | 1.00 (0.06–15.99) | 1 |
| rs61544945 | ARMS2 | Liang et al., 2012 | 151 | 248 | 1.09 (0.93–1.26) | 0.285 |
| * | ARMS2-HTRA1 | Gotoh et al., 2009 | 56 | 76 | 0.52 (0.26–1.02) | 0.057 |
| § | ARMS2-HTRA1 | Gotoh et al., 2009 | 56 | 77 | 0.56 (0.19–1.66) | 0.292 |
| c.324+838C>T | ARMS2-HTRA1 | Liang et al., 2012 | 149 | 247 | 0.43 (0.29–0.64) | <0.0001 |
| c.324+965T>C | ARMS2-HTRA1 | Liang et al., 2012 | 156 | 248 | 0.31 (0.15–0.66) | 0.002 |
| -497C>T | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.95 (0.85–1.05) | 0.328 |
| 34delCinsTCCT | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.96 (0.86–1.07) | 0.476 |
| 59C>T | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.96 (0.85–1.08) | 0.505 |
| IVS2+100C>T | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.95 (0.85–1.06) | 0.342 |
| IVS2+317C>T | HTRA1 | Tam et al., 2008 | 163 | 183 | 2.00 (0.18–22.05) | 0.571 |
| rs11538140 | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.95 (0.85–1.05) | 0.304 |
| rs12267142 | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.95 (0.85–1.06) | 0.383 |
| rs12571363 | HTRA1 | Kanda et al., 2007 | 453 | 230 | 1.54 (1.06–2.23) | 0.023 |
| rs2239586 | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.97 (0.84–1.12) | 0.684 |
| rs2239587 | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.97 (0.84–1.12) | 0.684 |
| rs2239588 | HTRA1 | Kanda et al., 2007 | 438 | 229 | 1.23 (0.84–1.81) | 0.284 |
| rs2268346 | HTRA1 | Kanda et al., 2007 | 448 | 276 | 1.27 (1.18–1.38) | <0.0001 |
| rs2272599 | HTRA1 | Tam et al., 2008 | 163 | 183 | 1.00 (0.83–1.22) | 0.961 |
| rs2293870 | HTRA1 | Tam et al., 2008 | 163 | 183 | 1.24 (1.07–1.45) | 0.006 |
| rs2672582 | HTRA1 | Tam et al., 2008 | 163 | 183 | 0.99 (0.81–1.20) | 0.921 |
| rs2672583 | HTRA1 | Tam et al., 2008 | 163 | 183 | 1.04 (0.86–1.27) | 0.658 |
| rs2672585 | HTRA1 | Tam et al., 2008 | 163 | 183 | 1.00 (0.82–1.22) | 1 |
| rs2672588 | HTRA1 | Kanda et al., 2007 | 451 | 278 | 1.07 (0.89–1.29) | 0.472 |
| rs2672589 | HTRA1 | Gotoh et al., 2010 | 83 | 275 | 0.56 (0.48–0.65) | <0.0001 |
| rs2736917 | HTRA1 | Kanda et al., 2007 | 440 | 271 | 1.14 (0.92–1.43) | 0.236 |
| rs736960 | HTRA1 | Kanda et al., 2007 | 450 | 228 | 1.49 (1.04–2.13) | 0.028 |
| rs760336 | HTRA1 | Fritsche et al., 2008 | 760 | 549 | 1.33 (1.22–1.44) | <0.0001 |
| rs878107 | HTRA1 | Gotoh et al., 2010 | 83 | 276 | 0.33 (0.25–0.45) | <0.0001 |
| rs909290 | HTRA1 | Kanda et al., 2007 | 440 | 205 | 1.61 (1.10–2.35) | 0.014 |
| rs10082476 | PLEKHA1 | Kanda et al., 2007 | 442 | 268 | 1.20 (0.97–1.47) | 0.086 |
| rs10887149 | PLEKHA1 | Kanda et al., 2007 | 456 | 276 | 1.22 (1.03–1.45) | 0.019 |
| rs11200621 | PLEKHA1 | Kanda et al., 2007 | 454 | 278 | 1.29 (1.19–1.41) | <0.0001 |
| rs17564097 | PLEKHA1 | Fritsche et al., 2008 | 760 | 549 | 1.22 (1.15–1.30) | <0.0001 |
| rs2280141 | PLEKHA1 | Gotoh et al., 2010 | 82 | 275 | 0.53 (0.44–0.63) | <0.0001 |
| rs2421022 | PLEKHA1 | Fritsche et al., 2008 | 760 | 549 | 1.31 (1.22–1.41) | <0.0001 |
| rs4598609 | PLEKHA1 | Kanda et al., 2007 | 452 | 275 | 1.17 (0.75–1.82) | 0.499 |
| rs4612730 | PLEKHA1 | Fritsche et al., 2008 | 760 | 549 | 1.41 (1.29–1.53) | <0.0001 |
| rs7097701 | PLEKHA1 | Fritsche et al., 2008 | 760 | 549 | 1.41 (1.29–1.53) | <0.0001 |
| rs7902176 | PLEKHA1 | Kanda et al., 2007 | 457 | 226 | 1.38 (1.17–1.62) | <0.0001 |
| rs1474526 | PLEKHA1- ARMS2 | Kanda et al., 2007 | 454 | 278 | 1.10 (0.78–1.55) | 0.599 |
| rs2292627 | PLEKHA1- ARMS2 | Gotoh et al., 2010 | 83 | 274 | 0.58 (0.47–0.72) | <0.0001 |
| rs2421027 | PLEKHA1- ARMS2 | Kanda et al., 2007 | 452 | 273 | 1.03 (0.74–1.42) | 0.869 |
| rs2736930 | PLEKHA1- ARMS2 | Fritsche et al., 2008 | 760 | 549 | 1.23 (1.16–1.31) | <0.0001 |
| rs4752695 | PLEKHA1- ARMS2 | Gotoh et al., 2010 | 84 | 273 | 0.18 (0.11–0.29) | <0.0001 |
| rs6585829 | PLEKHA1- ARMS2 | Fritsche et al., 2008 | 760 | 549 | 1.22 (1.15–1.30) | <0.0001 |
| rs11200572 | near PLEKHA1 | Kanda et al., 2007 | 436 | 269 | 1.24 (1.13–1.35) | <0.0001 |
| rs11200580 | near PLEKHA1 | Kanda et al., 2007 | 453 | 277 | 1.30 (1.18–1.44) | <0.0001 |
| rs1998345 | near PLEKHA1 | Kanda et al., 2007 | 440 | 229 | 1.43 (1.28–1.60) | <0.0001 |
| rs2038596 | near PLEKHA1 | Kanda et al., 2007 | 456 | 278 | 1.17 (0.96–1.43) | 0.119 |
| rs7074542 | near PLEKHA1 | Kanda et al., 2007 | 455 | 278 | 1.28 (1.07–1.52) | 0.006 |
| rs7904674 | near PLEKHA1 | Kanda et al., 2007 | 449 | 275 | 1.34 (1.10–1.64) | 0.004 |
| rs984668 | near PLEKHA1 | Kanda et al., 2007 | 392 | 245 | 1.32 (1.18–1.47) | <0.0001 |
Assuming an additive model.
bp 124207277(NT_030059.12).
bp 124207404 (NT_030059.12).
AMD: age-related macular degeneration.
Figure 2. Forest plot for meta-analysis of rs2736911.
Each study was represented by a square whose area was proportional to the weight of the study. The overall effect from meta-analysis is represented by a diamond whose width represents the 95% CI for the estimated OR.
Table 4.
Meta-analysis of the association between AMD and rs2736911a.
| Study | Case | Control | OR (95% CI) | P-value |
|---|---|---|---|---|
| Liang et al., 2012 | 151 | 248 | 0.44 (0.30–0.65) | 2.76×10−5 |
| Teper et al., 2012 | 90 | 40 | 1.23 (0.59–2.56) | 0.578 |
| Gotoh et al., 2010 | 83 | 274 | 0.20 (0.12–0.33) | 1.53×10−10 |
| Goto et al., 2009 | 100 | 188 | 0.77 (0.67–0.88) | 1.82×10−4 |
| Gotoh et al., 2009 | 56 | 77 | 0.52 (0.26–1.02) | 0.057 |
| Bergeron et al., 2009 | 421 | 215 | 1.47 (1.34–1.61) | 4.44×10−16 |
| Kaur et al., 2008 | 112 | 94 | 1.10 (0.96–1.27) | 0.175 |
| Kanda et al., 2007 | 458 | 225 | 1.25 (0.95–1.65) | 0.110 |
| Total | 1,471 | 1,361 | 0.77 (0.55–1.07) | 0.122 |
Assuming an additive model.
AMD: age-related macular degeneration.
Figure 3. Forest plot for meta-analysis of rs3793917.
Each study was represented by a square whose area was proportional to the weight of the study. The overall effect from meta-analysis is represented by a diamond whose width represents the 95% CI for the estimated OR.
Gene-cluster analysis
To examine the cumulative association of these genetic variants with AMD, we performed a gene-cluster analysis using the p-values obtained above (Table 5). Additionally, we examined whether the association varies in meta-studies only (including only studies that are covered in meta-analyses) and in individual-studies only (including only studies that are covered in individual genetic variant analyses). Our gene-cluster analysis indicates a strong association between the genetic variants in this gene cluster and AMD (all p’s<10−5), and the association holds in both meta-studies only and individual-studies only.
Table 5.
Gene-cluster analysis for cumulative association of the less studied genetic variants in PLEKHA1/ARMS2/HTRA1 with AMD.
| Data | Fisher | Simes | TPMa | |
|---|---|---|---|---|
| Meta-studies only | <10−6 | 3.80×10−6 | Un-weighted | <10−5 |
| Weighted | <10−5 | |||
| Single-studies only | <10−6 | 2.80×10−6 | Un-weighted | <10−5 |
| Weighted | <10−5 | |||
| All | <10−6 | 3.13×10−6 | Un-weighted | <10−5 |
| Weighted | <10−5 | |||
Based on 100,000 simulations
Sensitivity analysis
Sensitivity analysis reveals strong association between AMD and rs2736911 in Chinese (p=2.77×10−5) but the association is not significant in Caucasian (p=0.11). On the contrary, the association between AMD and c.372_815del443ins54 is highly significant among Caucasians (1.04×10−17) but not in Chinese (p=0.35). In both ethnic groups, AMD is strongly associated with genetic polymorphism rs1049331 in HTRA1 (both p’s <5×10−3) and the gene cluster (both p’s≤6.94×10−4).
Additionally we examined the association between genetic variants in the gene cluster and wet AMD. The meta-analysis indicates strong association between wet AMD and rs1049331 and rs2736912 (both p’s≤7.56×10−5). Two other genetic variants (rs2736911 and rs2672598) are also significantly associated with wet AMD (both p’s ≤0.03). Another SNP (rs2268356) is marginally associated with wet AMD in meta-analysis (p=0.055). Our gene-cluster analysis indicates a significant cumulative effect of the genetic variants in this gene cluster on wet AMD risk (all p’s<10−5), and the association holds in both meta-studies only (p≤9.0×10−5) and individual-studies only (all p’s<10−5).
Discussion
In this study, we did an extensive literature search for publications on the association of less studied genetic variants in PLEKHA1/ARMS2/HTRA1. Our analysis reveals that several genetic variants are significantly associated with AMD, and gene-cluster analysis indicates that the gene cluster has significant cumulative association with AMD, implying that, in addition to the two widely studied SNPs (rs10490924 and rs11200638) in this gene cluster, other genetic variants in this gene cluster also contribute to AMD risk. To the best of our knowledge, this is the first meta-analysis on a number of less studied genetic variants in PLEKHA1/ARMS2/HTRA1, and the first gene-cluster analysis on the cumulative effect of genetic variants in the gene cluster.
The effect of multiple less studied genetic variants in PLEKHA1/ARMS2/HTRA1 on AMD risk has been studied in many studies. The SNP rs2736911 is a non-synonymous coding SNP in LOC387715/ARMS2 leading to a premature stop codon (R38X). A recent study found that a common disease haplotype, comprising rs2736911, rs10490924, c.372_815del443ins54 and rs11200638, can lead to transcriptional upregulation of HTRA1 [36]. The result, however, could not be replicated in a subsequent study and is still controversial [47]. Our meta-analysis with 2,832 participants from eight studies found no significant association between this SNP and AMD risk under an additive model (p=0.122; Table 4). Under the additive model, the results from individual studies are inconsistent, with some studies showing strong association and others indicating insignificant results (Table 4). However, we found that this SNP is significantly associated with AMD under the dominant model (p<0.0001, Supplementary Table 2). We could not perform haplotype analysis due to unavailability of data, and hence could not analyze haplotype effect in influencing AMD risk; this warrants further investigation.
Another SNP rs3793917 is highly correlated with several other SNPs in ARMS2-HTRA1.[41] Our meta-analyses of 5,680 participants failed to find a significant association between this SNP and AMD under all genetic models. Another interesting genetic variant, c.372_815del443ins54, is an indel located within the 3′-UTR of the ARMS2 gene, where a deletion removes the polyadenylation signal sequence at position *395_400 and the insertion introduces a 54-bp AU-rich element. Expression of ARMS2 was reported to be non-detectable in homozygous carriers of the indel variant [41]. In our meta-analysis, this SNP is significantly associated with AMD only when assuming a recessive model (Supplementary Table 3). Sensitivity analyses show significant association between this indel and AMD in Caucasians but not in Chinese. We also found that the LD structures spanning rs10490924-rs11200638 are very different between Han Chinese and Caucasians (http://www.hapmap.org) [48], confirming the difference in genetic background between the two ethnic groups.
Our analysis identified several SNPs that are significantly associated with AMD. The gene-cluster analysis indicates that these less studied genetic variants in PLEKHA1/ARMS2/HTRA1 as a whole are significantly associated with AMD (p<10−5; Table 5), implying that these less studied genetic variants also contribute, directly or indirectly and individually or jointly, to the pathology of AMD. Information is scarce about these less studied genetic variants showing significant association in our analysis, and little is known about their relationship with the extensively studied genetic variants whose association with AMD are well-established (e.g., rs1049024 and rs11200638). One study reported that a haplotype block consisting of five SNPs including rs1049331 and rs11200638 in HTRA1 significantly predisposes individuals to AMD [43]. Another haplotype consisting of rs2736911 and rs10490924 was found to be significantly associated with PCV but not with exudative AMD [29]. Identifying the true causal genetic variant is challenging due to the high LD across the genetic variants in this gene cluster [49]. Further studies are warranted on the underlying mechanism leading to the association, particularly on the genetic variants showing significant association in this study.
Our study has some limitations. Due to the unavailability of relevant data, our meta-analysis did not adjust by age, sex or smoking status. Future studies are needed to validate our results especially large consortium studies which control for such confounding factors. Second, some meta-analyses were based on few studies, and the gene-cluster analysis used some results from individual studies. Third, sensitivity analyses by race are limited because race information was unavailable in many studies. Forth, the definition of AMD was not consistent across the 23 studies for the meta-and gene-based analyses. Fifth, due to limited number of studies included in some of meta-analyses, we could not test publication bias for them. This might lead to bias in the resulting data, and subsequently influence the validity of the gene-based analysis. Finally, there are other types of genetic variations that are not included in our study, such as copy number variation which was recently reported to be associated with AMD [50,51].
In summary, we conducted an extensive literature search for publications on the effect of less studied genetic variants in PLEKHA1/ARMS2/HTRA1 on risk of AMD, and performed meta- and gene-cluster analyses to evaluate the cumulative effect of multiple genetic variants within the gene cluster. We identified several significant SNPs and found significant cumulative effects of these genetic variants. Our results suggest that in addition to the two widely studied SNPs in this gene cluster, these less studied genetic variants also contribute to AMD risk. The genetic variants included for analysis this paper, particularly those showing significant association, might influence AMD susceptibility (e.g., by altering protein sequence directly or through LD with a nearby non-synonymous genetic variant) or help prioritize nearby genetic variants or genes for future study. Further studies are warranted to validate our findings and explore the potential mechanisms underlying the association, particularly studies with larger sample size that re-sequence the gene cluster to rigorously evaluate rare variants while taking into account potential interaction with ethnicity. Generation of murine models bearing these corresponding SNPs will also provide insights into the validation and mechanism of the association demonstrated in our study.
Supplementary Material
Acknowledgments
Financial Support: This research was supported by Award Number P50DA010075-16 from the National Institute on Drug Abuse (NIDA) and NIH/NCI R01 CA168676. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIDA or the National Institutes of Health. The sponsor or funding organization had no role in the design or conduct of this research.
Footnotes
Conflict of interest: no conflicting relationship exists for any author.
References
- 1.Pascolini D, Mariotti SP, Pokharel GP, Pararajasegaram R, Etya’ale D, Negrel AD, Resnikoff S. 2002 global update of available data on visual impairment: a compilation of population-based prevalence studies. Ophthalmic Epidemiol. 2004;11 (2):67–115. doi: 10.1076/opep.11.2.67.28158. [DOI] [PubMed] [Google Scholar]
- 2.Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82 (11):844–851. S0042-96862004001100009. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122 (4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 4.Cheung CM, Tai ES, Kawasaki R, Tay WT, Lee JL, Hamzah H, Wong TY. Prevalence of and risk factors for age-related macular degeneration in a multiethnic Asian cohort. Arch Ophthalmol. 2012;130 (4):480–486. doi: 10.1001/archophthalmol.2011.376. [DOI] [PubMed] [Google Scholar]
- 5.Congdon N, O’Colmain B, Klaver CC, Klein R, Munoz B, Friedman DS, Kempen J, Taylor HR, Mitchell P. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122 (4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 6.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358 (24):2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 7.VanNewkirk MR, Nanjan MB, Wang JJ, Mitchell P, Taylor HR, McCarty CA. The prevalence of age-related maculopathy: the visual impairment project. Ophthalmology. 2000;107 (8):1593–1600. doi: 10.1016/s0161-6420(00)00175-5. [DOI] [PubMed] [Google Scholar]
- 8.Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, Kelly SP. Smoking and age-related macular degeneration: a review of association. Eye (Lond) 2005;19 (9):935–944. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- 9.Adams MK, Simpson JA, Aung KZ, Makeyeva GA, Giles GG, English DR, Hopper J, Guymer RH, Baird PN, Robman LD. Abdominal obesity and age-related macular degeneration. Am J Epidemiol. 2011;173 (11):1246–1255. doi: 10.1093/aje/kwr005. [DOI] [PubMed] [Google Scholar]
- 10.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137 (3):486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 11.Bressler SB, Munoz B, Solomon SD, West SK. Racial differences in the prevalence of age-related macular degeneration: the Salisbury Eye Evaluation (SEE) Project. Arch Ophthalmol. 2008;126 (2):241–245. doi: 10.1001/archophthalmol.2007.53. [DOI] [PubMed] [Google Scholar]
- 12.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308 (5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakkinstian A, Han P, McEvoy M, Smith W, Hoh J, Magnusson K, Zhang K, Attia J. Systematic review and meta-analysis of the association between complement factor H Y402H polymorphisms and age-related macular degeneration. Hum Mol Genet. 2006;15 (18):2784–2790. doi: 10.1093/hmg/ddl220. [DOI] [PubMed] [Google Scholar]
- 14.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308 (5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 15.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308 (5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 16.Shaw PX, Zhang L, Zhang M, Du H, Zhao L, Lee C, Grob S, Lim SL, Hughes G, Lee J, Bedell M, Nelson MH, Lu F, Krupa M, Luo J, Ouyang H, Tu Z, Su Z, Zhu J, Wei X, Feng Z, Duan Y, Yang Z, Ferreyra H, Bartsch DU, Kozak I, Lin F, Sun H, Feng H, Zhang K. Complement factor H genotypes impact risk of age-related macular degeneration by interaction with oxidized phospholipids. Proc Natl Acad Sci U S A. 2012;109 (34):13757–13762. doi: 10.1073/pnas.1121309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14 (21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 18.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104 (41):16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314 (5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 20.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314 (5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 21.Tong Y, Liao J, Zhang Y, Zhou J, Zhang H, Mao M. LOC387715/HTRA1 gene polymorphisms and susceptibility to age-related macular degeneration: A HuGE review and meta-analysis. Mol Vis. 2010;16:1958–1981. [PMC free article] [PubMed] [Google Scholar]
- 22.Vierkotten S, Muether PS, Fauser S. Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch’s membrane via cleavage of extracellular matrix components. PLoS One. 2011;6 (8):e22959. doi: 10.1371/journal.pone.0022959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borenstein M. Introduction to meta-analysis. John Wiley & Sons; Chichester, U.K: 2009. [Google Scholar]
- 24.Fisher RA. Biological monographs and manuals. V. 5. Oliver and Boyd; Edinburgh: 1932. Statistical methods for research workers. [Google Scholar]
- 25.Simes RJ. An Improved Bonferroni Procedure for Multiple Tests of Significance. Biometrika. 1986;73 (3):751–754. [Google Scholar]
- 26.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22 (2):170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- 27.Sheng X, Yang J. Truncated product methods for panel unit root tests. Oxfor Bulletin of Economic and Statistics. 2012 doi: 10.1111/j.1468-0084.2012.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moher D, Cook DJ, Eastwood S, Olkin I, Rennie D, Stroup DF. Improving the quality of reports of meta-analyses of randomised controlled trials: the QUOROM statement. Quality of Reporting of Meta-analyses. Lancet. 1999;354 (9193):1896–1900. doi: 10.1016/s0140-6736(99)04149-5. [DOI] [PubMed] [Google Scholar]
- 29.Liang XY, Lai TY, Liu DT, Fan AH, Chen LJ, Tam PO, Chiang SW, Ng TK, Lam DS, Pang CP. Differentiation of exudative age-related macular degeneration and polypoidal choroidal vasculopathy in the ARMS2/HTRA1 locus. Invest Ophthalmol Vis Sci. 2012;53 (6):3175–3182. doi: 10.1167/iovs.11-8135. [DOI] [PubMed] [Google Scholar]
- 30.Teper SJ, Nowinska A, Wylegala E. A69S and R38X ARMS2 and Y402H CFH gene polymorphisms as risk factors for neovascular age-related macular degeneration in Poland - a brief report. Med Sci Monit. 2012;18(2):PR1–3. doi: 10.12659/MSM.882447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotoh N, Yamashiro K, Nakanishi H, Saito M, Iida T, Yoshimura N. Haplotype analysis of the ARMS2/HTRA1 region in Japanese patients with typical neovascular age-related macular degeneration or polypoidal choroidal vasculopathy. Jpn J Ophthalmol. 2010;54 (6):609–614. doi: 10.1007/s10384-010-0865-2. [DOI] [PubMed] [Google Scholar]
- 32.Richardson AJ, Islam FM, Aung KZ, Guymer RH, Baird PN. An intergenic region between the tagSNP rs3793917 and rs11200638 in the HTRA1 gene indicates association with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51 (10):4932–4936. doi: 10.1167/iovs.09-5114. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, Tan PL, Conley YP, Kanda A, Kopplin L, Li Y, Augustaitis KJ, Karoukis AJ, Scott WK, Agarwal A, Kovach JL, Schwartz SG, Postel EA, Brooks M, Baratz KH, Brown WL, Brucker AJ, Orlin A, Brown G, Ho A, Regillo C, Donoso L, Tian L, Kaderli B, Hadley D, Hagstrom SA, Peachey NS, Klein R, Klein BE, Gotoh N, Yamashiro K, Ferris F, III, Fagerness JA, Reynolds R, Farrer LA, Kim IK, Miller JW, Corton M, Carracedo A, Sanchez-Salorio M, Pugh EW, Doheny KF, Brion M, Deangelis MM, Weeks DE, Zack DJ, Chew EY, Heckenlively JR, Yoshimura N, Iyengar SK, Francis PJ, Katsanis N, Seddon JM, Haines JL, Gorin MB, Abecasis GR, Swaroop A. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107 (16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Spencer KL, Scott WK, Whitehead P, Court BL, Ayala-Haedo J, Mayo P, Schwartz SG, Kovach JL, Gallins P, Polk M, Agarwal A, Postel EA, Haines JL, Pericak-Vance MA. Analysis of the indel at the ARMS2 3′ UTR in age-related macular degeneration. Hum Genet. 2010;127 (5):595–602. doi: 10.1007/s00439-010-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goto A, Akahori M, Okamoto H, Minami M, Terauchi N, Haruhata Y, Obazawa M, Noda T, Honda M, Mizota A, Tanaka M, Hayashi T, Tanito M, Ogata N, Iwata T. Genetic analysis of typical wet-type age-related macular degeneration and polypoidal choroidal vasculopathy in Japanese population. J Ocul Biol Dis Infor. 2009;2 (4):164–175. doi: 10.1007/s12177-009-9047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Tong Z, Chen Y, Zeng J, Lu F, Sun X, Zhao C, Wang K, Davey L, Chen H, London N, Muramatsu D, Salasar F, Carmona R, Kasuga D, Wang X, Bedell M, Dixie M, Zhao P, Yang R, Gibbs D, Liu X, Li Y, Li C, Campochiaro B, Constantine R, Zack DJ, Campochiaro P, Fu Y, Li DY, Katsanis N, Zhang K. Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS Genet. 2010;6 (2):e1000836. doi: 10.1371/journal.pgen.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricci F, Zampatti S, D’Abbruzzi F, Missiroli F, Martone C, Lepre T, Pietrangeli I, Sinibaldi C, Peconi C, Novelli G, Giardina E. Typing of ARMS2 and CFH in age-related macular degeneration: case-control study and assessment of frequency in the Italian population. Arch Ophthalmol. 2009;127 (10):1368–1372. doi: 10.1001/archophthalmol.2009.237. [DOI] [PubMed] [Google Scholar]
- 38.Gotoh N, Nakanishi H, Hayashi H, Yamada R, Otani A, Tsujikawa A, Yamashiro K, Tamura H, Saito M, Saito K, Iida T, Matsuda F, Yoshimura N. ARMS2 (LOC387715) variants in Japanese patients with exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Am J Ophthalmol. 2009;147 (6):1037–1041. 1041 e1031–1032. doi: 10.1016/j.ajo.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Bergeron-Sawitzke J, Gold B, Olsh A, Schlotterbeck S, Lemon K, Visvanathan K, Allikmets R, Dean M. Multilocus analysis of age-related macular degeneration. Eur J Hum Genet. 2009;17 (9):1190–1199. doi: 10.1038/ejhg.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Morrison MA, Dewan A, Adams S, Andreoli M, Huynh N, Regan M, Brown A, Miller JW, Kim IK, Hoh J, Deangelis MM. The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med Genet. 2008;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40 (7):892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 42.Kaur I, Katta S, Hussain A, Hussain N, Mathai A, Narayanan R, Reddy RK, Majji AB, Das T, Chakrabarti S. Variants in the 10q26 gene cluster (LOC387715 and HTRA1) exhibit enhanced risk of age-related macular degeneration along with CFH in Indian patients. Invest Ophthalmol Vis Sci. 2008;49 (5):1771–1776. doi: 10.1167/iovs.07-0560. [DOI] [PubMed] [Google Scholar]
- 43.Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, DeWan A, Hoh J, Lam DS, Pang CP. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci. 2008;49 (6):2357–2365. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deangelis MM, Ji F, Adams S, Morrison MA, Harring AJ, Sweeney MO, Capone A, Jr, Miller JW, Dryja TP, Ott J, Kim IK. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115 (7):1209–1215. e1207. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leveziel N, Souied EH, Richard F, Barbu V, Zourdani A, Morineau G, Zerbib J, Coscas G, Soubrane G, Benlian P. PLEKHA1-LOC387715-HTRA1 polymorphisms and exudative age-related macular degeneration in the French population. Mol Vis. 2007;13:2153–2159. [PubMed] [Google Scholar]
- 46.Fisher SA, Rivera A, Fritsche LG, Babadjanova G, Petrov S, Weber BH. Assessment of the contribution of CFH and chromosome 10q26 AMD susceptibility loci in a Russian population isolate. Br J Ophthalmol. 2007;91 (5):576–578. doi: 10.1136/bjo.2006.105577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanda A, Stambolian D, Chen W, Curcio CA, Abecasis GR, Swaroop A. Age-related macular degeneration-associated variants at chromosome 10q26 do not significantly alter ARMS2 and HTRA1 transcript levels in the human retina. Mol Vis. 2010;16:1317–1323. [PMC free article] [PubMed] [Google Scholar]
- 48.The International HapMap Project. Nature. 2003;426 (6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y, Huang L, Li X, Zhou P, Zeng W, Zhang C. Genetic and functional dissection of ARMS2 in age-related macular degeneration and polypoidal choroidal vasculopathy. PLoS One. 2013;8 (1):e53665. doi: 10.1371/journal.pone.0053665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu MM, Agron E, Chew E, Meyerle C, Ferris FL, 3rd, Chan CC, Tuo J. Copy number variations in candidate genes in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52 (6):3129–3135. doi: 10.1167/iovs.10-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JH, Lee S, Yu HG, Kim JI, Seo JS. Copy number variation of age-related macular degeneration relevant genes in the Korean population. PLoS One. 2012;7 (2):e31243. doi: 10.1371/journal.pone.0031243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.