Significance
Cancer immune evasion is well described. In some cases, this may be overcome by enhancing T-cell responses. We show that despite the presence of antitumor T cells, immunotherapeutic antibodies are ineffective in a murine pancreatic cancer model recapitulating the human disease. Removing the carcinoma-associated fibroblast (CAF) expressing fibroblast activation protein (FAP) from tumors permitted immune control of tumor growth and uncovered the efficacy of these immunotherapeutic antibodies. FAP+ CAFs are the only tumoral source of chemokine (C-X-C motif) ligand 12 (CXCL12), and administering AMD3100, an inhibitor of chemokine (C-X-C motif) receptor 4, a CXCL12 receptor, also revealed the antitumor effects of an immunotherapeutic antibody and greatly diminished cancer cells. These findings may have wide clinical relevance because FAP+ cells are found in almost all human adenocarcinomas.
Keywords: KPC mouse, tumor stroma, T cell exclusion, tumor immunogenicity
Abstract
An autochthonous model of pancreatic ductal adenocarcinoma (PDA) permitted the analysis of why immunotherapy is ineffective in this human disease. Despite finding that PDA-bearing mice had cancer cell-specific CD8+ T cells, the mice, like human patients with PDA, did not respond to two immunological checkpoint antagonists that promote the function of T cells: anti-cytotoxic T-lymphocyte-associated protein 4 (α-CTLA-4) and α-programmed cell death 1 ligand 1 (α-PD-L1). Immune control of PDA growth was achieved, however, by depleting carcinoma-associated fibroblasts (CAFs) that express fibroblast activation protein (FAP). The depletion of the FAP+ stromal cell also uncovered the antitumor effects of α-CTLA-4 and α-PD-L1, indicating that its immune suppressive activity accounts for the failure of these T-cell checkpoint antagonists. Three findings suggested that chemokine (C-X-C motif) ligand 12 (CXCL12) explained the overriding immunosuppression by the FAP+ cell: T cells were absent from regions of the tumor containing cancer cells, cancer cells were coated with the chemokine, CXCL12, and the FAP+ CAF was the principal source of CXCL12 in the tumor. Administering AMD3100, a CXCL12 receptor chemokine (C-X-C motif) receptor 4 inhibitor, induced rapid T-cell accumulation among cancer cells and acted synergistically with α-PD-L1 to greatly diminish cancer cells, which were identified by their loss of heterozygosity of Trp53 gene. The residual tumor was composed only of premalignant epithelial cells and inflammatory cells. Thus, a single protein, CXCL12, from a single stromal cell type, the FAP+ CAF, may direct tumor immune evasion in a model of human PDA.
Immunotherapy of cancer has made recent progress by focusing on overcoming T-cell immunological checkpoints with blocking monoclonal antibodies to cytotoxic T-lymphocyte associated protein-4 (CTLA-4) and the programmed cell death 1/programmed cell death 1 ligand 1 (PD-1/PD-L1) receptor/ligand pair (1–7). Many patients, however, did not respond to these immunological checkpoint antagonists for reasons that are not understood. In particular, patients with pancreatic ductal adenocarcinoma (PDA), the fourth most common cause of cancer-related deaths in the United States, had no objective responses to anti (α)-CTLA-4 (7) or α-PD-L1 monoclonal antibodies (5).
A mesenchymal tumoral stromal cell that is present in almost all human adenocarcinomas (8) and is identified by its expression of the membrane protein, fibroblast activation protein (FAP), was shown recently to mediate immunosuppression in a transplanted murine tumor model (9). Because FAP+ stromal cells are present in human PDA (8), we wished to investigate whether the immunosuppressive activity of the murine FAP+ stromal cell might be involved in the resistance of this cancer to immunotherapy. We were able to carry out this analysis because of the availability of the autochthonous LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre (KPC) model of PDA (10). We find that this PDA model replicates the resistance of human PDA to checkpoint antagonists, despite the presence of systemic anti-PDA immunity. The failure of immune surveillance is attributable to local immunosuppression mediated by the FAP+ stromal cell, which comprises essentially all carcinoma-associated fibroblasts (CAFs). Immunosuppression manifests as exclusion of T cells from regions of the tumor containing cancer cells and involves the production of chemokine (C-X-C motif) ligand 12 (CXCL12) by FAP+ CAFs. Inhibiting chemokine (C-X-C motif) receptor 4 (CXCR4), a CXCL12 receptor, promotes T-cell accumulation and synergizes with the checkpoint antagonist, α-PD-L1, to cause cancer regression.
Results and Discussion
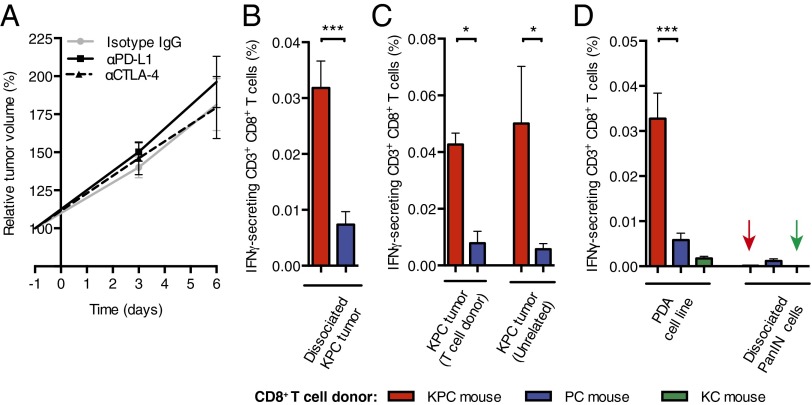
In the KPC model, Cre-mediated expression of Trp53R172H and KrasG12D is targeted to the pancreas, causing the development of invasive and metastatic carcinoma that recapitulates many aspects of human PDA, including the loss of heterozygosity (LOH) of Trp53 in cancer cells but not in premalignant pancreatic intraepithelial neoplasia (PanIN) (10). KPC mice with appropriately sized tumors demonstrate consistent tumor growth, which permits robust analyses of experimental interventions (SI Appendix, Fig. S1). We examined whether blocking immunological checkpoints with α-CTLA-4 and α-PD-L1 would promote immune control of the tumor. Administering these antibodies over 6 d to mice bearing PDA did not diminish the ∼80% increase in tumor volume observed in mice receiving control IgG (Fig. 1A). We determined whether this could be explained by the absence of an immune response to the PDA. Splenic CD8+ T cells from PDA-bearing and non–tumor-bearing mice were stimulated with tumor cells, and IFN-γ–secreting CD8+ T cells reporting antigenic stimulation were detected in an enzyme-linked immunosorbent spot (ELISpot) assay. Tumor cells induced significantly more ELISpots among CD8+ T cells from tumor-bearing mice than from non–tumor-bearing mice (Fig. 1B). The frequency of IFN-γ–secreting CD8+ T cells was the same when the stimulating tumor cells were from the T-cell donor or from another PDA-bearing mouse (Fig. 1C). An established PDA cell line also was stimulatory, whereas dissociated cells from pancreata of LSL-KrasG12D/+;Pdx-1-Cre mice with premalignant PanIN lesions expressing only KrasG12D or from young KPC mice before the development of cancer, were not (Fig. 1D and SI Appendix, Fig. S2). Therefore, PDA-bearing mice have a spontaneous adaptive immune response to antigens that are shared by cancer cells from different PDA tumors, and the ineffectiveness of α-CTLA-4 and α-PD-L1 suggests an additional immunosuppressive mechanism.
Fig. 1.
Immunological characteristics of murine PDA. (A) Increase in PDA volume (mean ± SEM) following treatment of mice with α-PD-L1 (n = 6), α-CTLA-4 (n = 6), or control (n = 4) antibodies was measured by ultrasound. (B–D) Induction of IFN-γ secretion by splenic CD8+ T cells from various donor types following stimulation by different sources of pancreatic cells was measured by ELISpot assay (n ≥ 8 in B and D; Mann–Whitney test, n = 4 in C). *P < 0.05; ***P < 0.001.
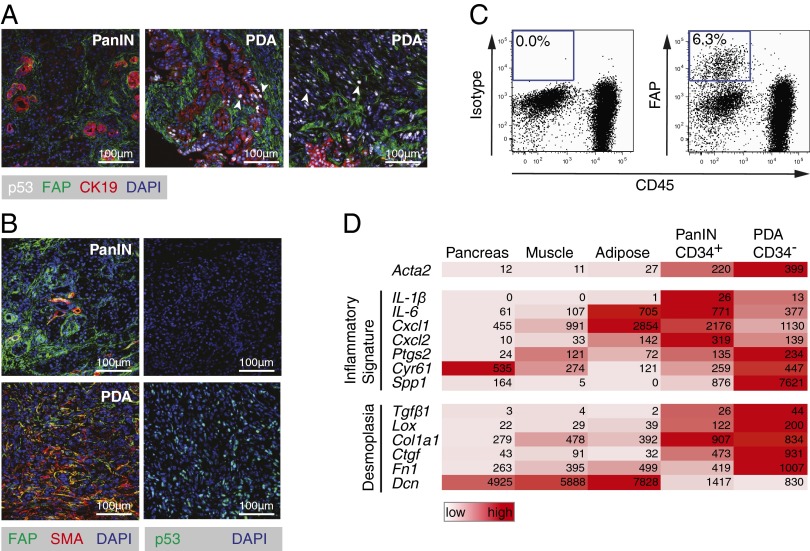
We considered the possibility that the FAP+ stromal cell mediates this immunosuppression. FAP+ cells were present in PanIN and both cytokeratin-19+ (CK19+) and CK19− PDA lesions (Fig. 2A). FAP+ cells in PanIN expressed CD34 (SI Appendix, Fig. S3 A and B) but rarely α-smooth muscle actin (αSMA) (Fig. 2B), whereas FAP+ cells among PDA cells were CD34− (SI Appendix, Fig. S3B) and αSMA+ (Fig. 2B). All FAP+ cells were PDGF receptor-α+ (SI Appendix, Fig. S3A) and CD45− (Fig. 2C), confirming their mesenchymal origin, and their frequency among dispersed tumor cells was 3.7% [95% confidence interval (CI): 1.1–6.3%; n = 8]. The expression of FAP by 92% of the αSMA+ fibroblasts (95% CI: 87.5–97.1%; n = 5) suggested that they are CAFs. This suggestion was supported by the transcriptomes of CD34+FAP+ (PanIN-associated) and CD34−FAP+ (PDA-associated) cells (Fig. 2D), which exhibited the “inflammatory gene signature” of CAFs (11). FAP+ cells from three normal tissues also displayed the signature and were clustered together by a principal component analysis of their transcriptomes (SI Appendix, Fig. S3C), indicating that FAP may identify a stromal cell lineage. The increased expression by tumoral FAP+ cells of genes encoding extracellular matrix proteins verifies their role in the desmoplasia of PDA, whereas their lower expression of decorin may relate to similar observations in human cancer (Fig. 2D).
Fig. 2.
Characterization of FAP+ stromal cells in murine PDA. (A) Tissue sections from PDA tumors were stained for FAP, CK19, and p53 and then analyzed by immunofluorescent (IF) microscopy. White arrows indicate examples of p53+ LOH cells. (B) Tissue sections from PDA tumors were stained for αSMA and FAP and analyzed by IF microscopy. (C) Single-cell suspensions of PDA tumor cells were analyzed by flow cytometry. (D) Heat map presents the reads per kilobase of transcript per million mapped reads of RNA-seq analyses of FACS-purified FAP+ cells.
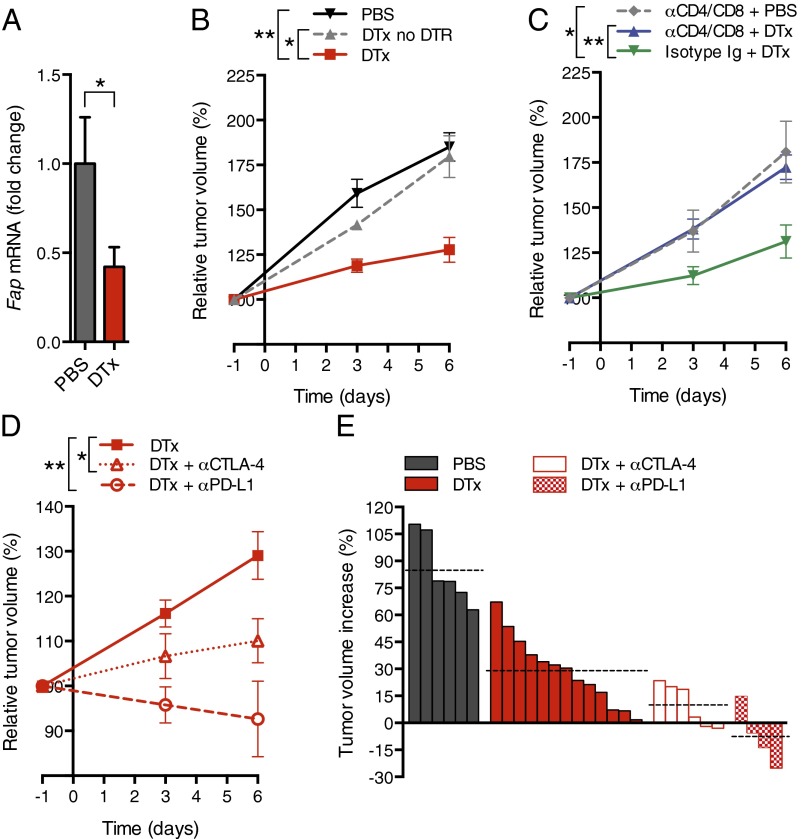
We introduced into the KPC line the bacterial artificial chromosome (BAC) transgene containing a modified Fap gene that drives the expression of the human diphtheria toxin receptor (DTR) selectively in cells that are FAP+ (9). Administering diphtheria toxin (DTx) to PDA-bearing BAC transgenic mice depleted ∼55% the tumoral FAP+ cell content (Fig. 3A). DTx appeared to deplete FAP+ cells throughout the tumor. Depleting FAP+ cells slowed PDA growth (Fig. 3B), but not when CD4+ and CD8+ T cells were removed (Fig. 3C). Combining depletion of FAP+ cells with administration of α-CTLA-4 or α-PD-L1 further diminished tumor growth (Fig. 3 D and E), indicating that the FAP+ cell contributes to the resistance of murine PDA to these checkpoint antagonists. The absence of an increase in IFN-γ–secreting CD8+ T cells from the spleens of DTx- and α-PD-L1–treated mice indicates that immune control was not accomplished by enhanced priming of cancer-specific CD8+ T cells (SI Appendix, Fig. S2).
Fig. 3.
Conditional depletion of FAP+ stromal cells and immune control of PDA. (A) KPCD mice with PDA received DTx or PBS; after 6 d, tumoral Fap mRNA was measured by quantitative RT-PCR (qRT-PCR) (PBS, n = 5; DTx, n = 7). (B) KPC mice with or without the DTR BAC transgene received DTx or PBS, and tumor volumes were measured by ultrasound (PBS, n = 6; DTx, n = 8; DTx to non-DTR transgenic, n = 4). (C) KPCD mice with PDA received CD4- and CD8-depleting antibodies or control IgG before and during treatment with DTx or PBS. Tumor volumes were measured (α-CD4/8 + PBS, n = 3; α-CD4/8 + DTx, n = 5; isotype IgG + DTx, n = 5). (D) KPCD mice with PDA received α-CTLA-4 or α-PD-L1 during treatment with DTx or PBS, and tumor volumes were measured [DTx, n = 13 (representing all DTx-treated KPCD mice presented in B and C); α-CTLA-4 + DTx, n = 6; α-PD-L1 + DTx, n = 4]. (E) Waterfall plots demonstrate the final tumor volume changes in individual mice. Dashed lines indicate the mean tumor volume for each treatment group. *P < 0.05; **P < 0.01.
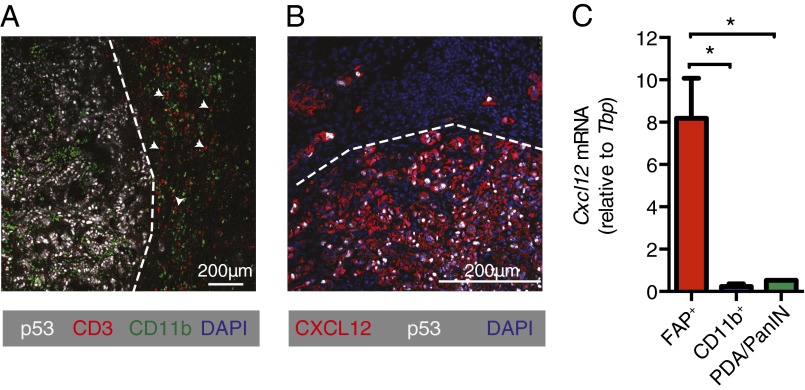
Therapy involving the depletion of FAP+ cells is precluded by their essential roles in normal tissues (12), and a therapeutic target that accounts for their immunosuppression had to be identified. We noted a paucity of CD3+ T cells in the vicinity of cancer cells (Fig. 4A), a characteristic of human PDA that is associated with FAP+ cells (SI Appendix, Fig. S4) and other carcinomas (13, 14). This T-cell trafficking problem directed attention to the chemokine, CXCL12, which localizes to cancer cells in both human (15) (SI Appendix, Fig. S4) and murine (Fig. 4B) PDA. We identified the source of CXCL12 as the tumoral FAP+ cell (Fig. 4C), as has been previously reported for CAFs (16). FAP+ cells in the s.c. Lewis lung carcinoma (LL2) model also are the tumoral source of CXCL12 (SI Appendix, Fig. S5A). CXCR4 is unlikely to mediate the uptake of the chemokine because cancer cell expression of CXCR4 is low (SI Appendix, Fig. S6). We hypothesize that high mobility group box 1 (HMGB1), which is overexpressed and secreted by metabolically stressed cancer cells (17), captures CXCL12 by forming a high-affinity heterocomplex (18), thus explaining the paradoxical localization of CXCL12 on cancer cells despite their not producing CXCL12 (Fig. 4C).
Fig. 4.
FAP+ cell-derived CXCL12 and T-cell exclusion. (A) Tissue sections from PDA tumors were stained for CD3, p53, and CD11b and then analyzed by IF microscopy. White arrows demonstrate examples of CD3+ cells. (B) Tissue sections from PDA tumors were stained for CXCL12 and p53 and analyzed by IF microscopy. (C) Single-cell suspensions of PDA tumors (n = 3) were stained with antibodies for FAP, CD45, and CD11b. FAP+ cells, CD11b+ cells, and PDA/PanIN cells (FAP−CD45−CD11b−) were FACS-purified. Their levels of Cxcl12 mRNA were assessed by qRT-PCR. *P < 0.05.
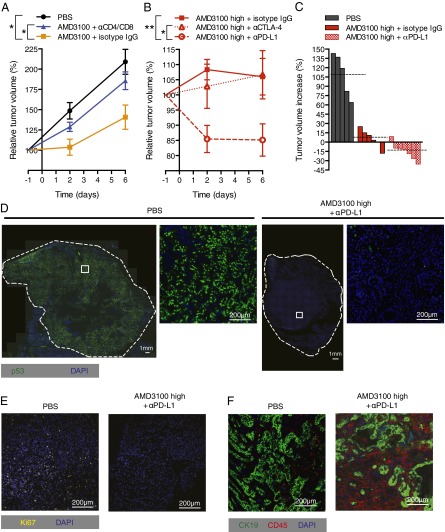
To assess the role of CXCL12 in tumoral immunosuppression, we administered AMD3100, a specific CXCR4 inhibitor that is licensed for clinical use (19), to mice bearing PDA in the presence or absence of depleting antibodies to CD4+ and CD8+ T cells. Tumor growth was slowed by AMD3100 in a T cell-dependent manner (Fig. 5A). AMD3100 also induced T cell-dependent control of s.c. immunogenic LL2 tumors (SI Appendix, Fig. S5 B and C). We administered a higher dose of AMD3100 that almost completely arrested PDA growth and combined it with immunological checkpoint antagonists (Fig. 5 B and C). The combination of AMD3100 with α-PD-L1 led to a significant 15% decline in PDA volume by 48 h and no further decrease over the following 4 d. α-CTLA-4 did not augment the antitumor effect of AMD3100.
Fig. 5.
Inhibition of CXCR4 by AMD3100 and immune elimination of PDA cells. (A) PDA-bearing mice, some of which had been pretreated with depleting CD4 and CD8 antibodies or control IgG, received PBS or AMD3100 by continuous infusion, and tumor volumes were measured by ultrasound (PBS, n = 5; AMD3100 + isotype IgG, n = 6; AMD3100 + α-CD4/8, n = 4). (B) PBS or AMD3100 (high dose) was given to PDA-bearing mice that were treated with α-CTLA-4, α-PD-L1, or control IgG (AMD3100 high + α-CTLA-4, n = 4; AMD3100 high + α-PD-L1, n = 7; AMD3100 high + isotype IgG, n = 6). (C) Waterfall plots present the final changes in tumor volumes in individual mice from A and B. Dashed lines indicate the mean tumor volume for each treatment group. (D) Tissue sections from PDA tumors taken from mice treated with PBS or AMD3100 high + α-PD-L1 for 6 d were stained for p53, and the entire cross-sections were imaged. White squares show the regions corresponding to the magnified images. Tissue sections from PDA tumors of mice treated with AMD3100 high + α-PD-L1 or PBS for 6 d were stained for Ki67 (E) or CD45 and CK19 (F) and analyzed by IF microscopy. *P < 0.05; **P < 0.01.
Rapid, therapy-induced, clinically relevant reductions in cancer cell number are often not associated with comparable decreases in tumor volume (20). To evaluate more definitively the response of the PDA to immunotherapy, we examined tumors after 6 d of treatment for the presence of cancer cells. These can be unambiguously identified because their loss of the wild type p53 allele results in the appearance and stabilization of the protein product of the mutant p53R172H allele (10). These p53+ PDA cells were abundant in tumors from mice that had received PBS or α-PD-L1 but were greatly diminished in tumors from mice that had received AMD3100 alone or with α-PD-L1 (Fig. 5D and SI Appendix, Fig. S7). The marked decrease in Ki67+ cells reflects this diminution in proliferating cancer cells (Fig. 5E and SI Appendix, Fig. S4). Serial sections demonstrate that the residual tumors were composed of CK19+ epithelial cells and CD45+ inflammatory cells (Fig. 5F). The selective elimination of p53+ LOH cells is consistent with the finding that the CD8+ T-cell response is cancer cell-specific (Fig. 1D).
To determine if this immunotherapeutic effect was caused by enhanced T-cell accumulation among cancer cells, we treated mice for 24 h with AMD3100 and α-PD-L1. α-PD-L1 alone had no effect, whereas AMD3100 increased the accumulation of T cells (Fig. 6 and SI Appendix, Fig. S9A). The combination of α-PD-L1 with AMD3100 amplified this effect and led to an apparent decrease in the frequency of p53+ LOH cancer cells (Fig. 6 and SI Appendix, Fig. S9A). Local T-cell proliferation could not account for the increase in tumoral T cells, because there were few Ki67+ CD3+ cells in AMD3100- and α-PD-L1–treated tumors (mean of 1.58%; range: 0.75–2.99%; n = 3) (SI Appendix, Fig. S9B). Furthermore, because Foxp3+ cells also were increased in tumors of mice given AMD3100 and α-PD-L1 (SI Appendix, Fig. S9C), we conclude that regulatory T (Treg) cells are not critically involved once immunosuppression by CXCR4/CXCL12 and PD-1/PD-L1 is overcome.
Fig. 6.
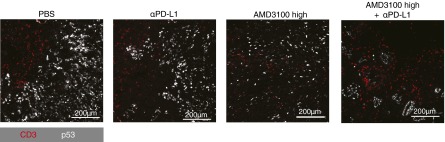
Accumulation of CD3+ T cells in cancer cell-containing regions of PDA induced by AMD3100 and α-PD-L1. Tissue sections from PDA tumors taken from mice that had been treated for 24 h with PBS, α-PD-L1, AMD3100 high, or AMD3100 high + α-PD-L1 were stained for CD3 and p53 and analyzed by IF microscopy.
These studies reveal a hierarchy of immunosuppression in murine PDA, with that mediated by the FAP+ CAF being dominant over two T-cell checkpoints. Depleting the FAP+ stromal cell or inhibiting the interaction of its chemokine, CXCL12, with CXCR4 uncovers the antitumor activity of α-CTLA-4 and α-PD-L1. We do not know whether CXCR4-mediated exclusion of T cells reflects T-cell apoptosis, as occurs with HIV gp120 (21), or a chemorepulsive effect of CXCL12 (22). A direct effect of AMD3100 on cancer cells, however, is excluded by their not expressing CXCR4 (SI Appendix, Fig. S6). The absence of a synergistic interaction between AMD3100 and α-CTLA-4 may indicate that inhibiting CXCR4 so strongly promotes the accumulation of T cells among cancer cells such that any augmented priming by α-CTLA-4 is superfluous. We excluded the possibility that other mechanisms may have accounted for the absence of an effect of α-CTLA-4 in combination with AMD3100. The α-CTLA-4 clone, 9H10, that was used in this experiment depletes Treg cells (23) and was effective when combined with a suboptimal reduction of FAP+ CAFs in PDA-bearing mice (Fig. 3A). Furthermore, macrophages that are capable of engaging in Fc receptor-mediated Treg cell clearance (23) were abundant in tumors treated with AMD3100 (SI Appendix, Fig. S10). Finally, the finding that Treg cells even accumulated in tumors along with other CD3+ cells during treatment with AMD3100 and α-PD-L1 (SI Appendix, Fig. S9C) suggests that the inhibitory function of Treg cells only plays a minor role once the immune suppressive effect of CXCL12 is neutralized. Hence, it is plausible that immune control of PDA may be achieved without incurring the severe adverse effects of α-CTLA-4 that have been observed in human patients with melanoma, especially when coadministered with another T-cell checkpoint antagonist (6).
Because T cells are also excluded from other carcinomas (14, 15) in which FAP+ stromal cells are abundant (8) and CXCL12 is associated with cancer cells (24), our findings may be widely relevant to tumor immunotherapy. Finally, this study strongly supports the use of autochthonous, genetically engineered models of cancer to study the mechanisms of escape from immune surveillance. These models have permitted the demonstration that cancers may maintain the expression of antigens despite the occurrence of systemic immune responses (25) and that metastatic lesions may be immunologically controlled despite outgrowth of primary tumors (26), observations that focus attention on the need to understand fully the rules of tumoral immunosuppression.
Materials and Methods
Mice.
All experiments were performed in accordance with institutional guidelines and were approved by the UK Home Office and the animal ethics committee of Cancer Research UK Cambridge Institute and the University of Cambridge. The generation of KPC and FAP-DTR BAC transgenic mice has been described previously (9, 10). These strains were crossed to generate KPCD (LSL-KrasG12D/+;LSL-Tp53R172H/+;Pdx-1-Cre;FAP-DTR) mice. Tumors that were 6–9 mm in diameter were selected, and volumes were monitored by high-resolution ultrasound as detailed in SI Appendix. KPC (±DTR transgene) mice were treated every 48 h with 25 ng/g of DTx (List Biologicals) in PBS, 160 μg of α-PD-L1 (10F.9G2; Biolegend), 100 μg of α-CTLA-4 (9H10; Biolegend), or isotype control antibody by i.p. injection. AMD3100 (Sigma–Aldrich) was administered by means of an AZLET osmotic pump (Charles River) at 30 mg/mL or 90 mg/mL (high dose). For T-cell depletion, mice received 300 μg each of α-CD4 (GK1.5; Biolegend) and α-CD8α (53-6.7; Biolegend) or isotype control antibodies for 3 consecutive days before treatment and on days 2 and 5 of treatment via i.p. injection. For s.c. tumors, 2 × 105 LL2/ovalbumin (OVA) cells were injected into the flanks of C57BL/6 and mouse strain Rag2−/− mice. Tumor volumes were calculated from caliper measurements using a standard formula listed in SI Appendix. AMD3100 (30 mg/mL) treatment via the AZLET osmotic pump commenced when tumors reached at least 62 mm3.
Cell Lines.
The generation of the LL2 cell line expressing chicken OVA (LL2/OVA) has been previously reported (9). Pancreatic cancer cell lines were derived from tumors arising in KPC mice (TB32964, K8484).
ELISpot Assays.
Single-cell suspensions of CD8+ splenocytes and of whole tumors depleted of CD3ε+ T cells were purified by magnetic-activated cell separation as described in SI Appendix. T cells were challenged with stimulator cells in a 12-h IFN-γ release ELISpot assay. IFN-γ–secreting CD8+ T-cell frequency was calculated from a dose–response curve.
Immunofluorescence.
Frozen tissue sections were prepared and imaged by confocal microscopy as described in SI Appendix. Slides of p53+ cells and Treg cells were analyzed on the ARIOL XT (Leica Biosystems) system.
Immunohistochemistry.
Slides were prepared as described in SI Appendix and imaged on the ARIOL XT system.
Flow Cytometry/Cell Sorting.
Single-cell suspensions were prepared by tissue digestion, stained, and analyzed as described in SI Appendix.
RNA Analysis.
RNA was extracted from whole-tumor samples and sorted cells as described in SI Appendix. For RT-PCR, we used Taqman assays, as detailed in SI Appendix. Data were normalized to endogenous Tbp. For analysis of Fap fold change, data were further normalized to the mean of the control group.
RNA-Sequencing and Computational Methods.
Cell sorting, RNA extraction, and sequencing were performed as previously described (12). Computational analysis is described in SI Appendix. The RNA-sequencing data generated in this investigation were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database and can be accessed using GEO accession no. GSE42605.
Statistical Analyses.
For multiple comparisons, ANOVA with Bonferroni’s post hoc test was applied. Significance was otherwise determined using the Student t test, unless specified in individual figure legends. Data are presented as the mean ± SEM. Statistical comparison of growth curves was performed using a permutation-based, pairwise test (SI Appendix).
Supplementary Material
Acknowledgments
We thank Frances Connor, Paul Mackin, and Lisa Young for providing KPC mice. We also thank the staff of the Cancer Research UK Cambridge Institute core facilities. This work was supported by Cancer Research UK (D.A.T., D.T.F.), the Wellcome Trust (D.T.F., T.J.), the Ludwig Institute for Cancer Research (D.T.F.), The Anthony Cerami and Anne Dunn Foundation for World Health (D.T.F.), the Medical Research Council (D.T.F.), Addenbrooke’s Charitable Trust (D.T.F.), and GlaxoSmith Kline (D.T.F.). This work has also been supported by the National Institute for Health Research Cambridge Biomedical Research Centre. C.F. was supported by a European Molecular Biology Organization long-term fellowship and a Marie Curie Intra European Fellowship within the Seventh European Community Framework Programme. R.J.B.W. received funding from the University of Cambridge School of Clinical Medicine, Rosetrees Trust, and Frank Edward Elmore Fund.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE42605).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320318110/-/DCSupplemental.
References
- 1.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 2.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 3.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamid O, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369(2):134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royal RE, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33(8):828–833. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci USA. 1990;87(18):7235–7239. doi: 10.1073/pnas.87.18.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraman M, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330(6005):827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 10.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17(2):135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Roberts EW, et al. Depletion of stromal cells expressing fibroblast activation protein-α from skeletal muscle and bone marrow results in cachexia and anemia. J Exp Med. 2013;210(6):1137–1151. doi: 10.1084/jem.20122344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naito Y, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58(16):3491–3494. [PubMed] [Google Scholar]
- 14.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 15.Liang JJ, et al. High levels of expression of human stromal cell-derived factor-1 are associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2598–2604. doi: 10.1158/1055-9965.EPI-10-0405. [DOI] [PubMed] [Google Scholar]
- 16.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 17.Tang D, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29(38):5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiraldi M, et al. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med. 2012;209(3):551–563. doi: 10.1084/jem.20111739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schols D, Esté JA, Henson G, De Clercq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor fusin/CXCR-4. Antiviral Res. 1997;35(3):147–156. doi: 10.1016/s0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- 20.Brindle K. New approaches for imaging tumour responses to treatment. Nat Rev Cancer. 2008;8(2):94–107. doi: 10.1038/nrc2289. [DOI] [PubMed] [Google Scholar]
- 21.Herbein G, et al. Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature. 1998;395(6698):189–194. doi: 10.1038/26026. [DOI] [PubMed] [Google Scholar]
- 22.Poznansky MC, et al. Active movement of T cells away from a chemokine. Nat Med. 2000;6(5):543–548. doi: 10.1038/75022. [DOI] [PubMed] [Google Scholar]
- 23.Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210(9):1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akishima-Fukasawa Y, et al. Prognostic significance of CXCL12 expression in patients with colorectal carcinoma. Am J Clin Pathol. 2009;132(2):202–210, quiz 307. doi: 10.1309/AJCPK35VZJEWCUTL. [DOI] [PubMed] [Google Scholar]
- 25.DuPage M, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19(1):72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyles J, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120(6):2030–2039. doi: 10.1172/JCI42002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.