Significance
A fundamental question in immunology is how the coordination of immune signals and metabolic programs regulates immune responses. The identification of metabolic pathways orchestrating the activation of lymphocytes and dendritic cells (DCs) has advanced our understanding of immune activation, but whether cell metabolism contributes to development of immune cells is unknown. Here we have genetically defined a crucial metabolic checkpoint for DC development that is mediated by the interplay between Tsc1-mTOR complex 1 signaling and Myc-dependent bioenergetic and biosynthetic programs. Dysregulation of this pathway impairs survival, proliferation, and functional differentiation of DCs, thereby highlighting the importance of metabolic programming of DC development.
Abstract
Coordination of cell metabolism and immune signals is crucial for lymphocyte priming. Emerging evidence also highlights the importance of cell metabolism for the activation of innate immunity upon pathogen challenge, but there is little evidence of how this process contributes to immune cell development. Here we show that differentiation of dendritic cells (DCs) from bone marrow precursors is associated with dynamic regulation of mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) signaling and cell metabolism. Unexpectedly, enhancing mTORC1 activity via ablation of its negative regulator tuberous sclerosis 1 (Tsc1) impaired DC development in vivo and in vitro, associated with defective cell survival and proliferation. Moreover, Tsc1 deficiency caused DC spontaneous maturation but a propensity to differentiate into other lineages, and attenuated DC-mediated effector TH1 responses. Mechanistically, Tsc1-deficient DCs exhibited increased glycolysis, mitochondrial respiration, and lipid synthesis that were partly mediated by the transcription factor Myc, highlighting a key role of Tsc1 in modulating metabolic programming of DC differentiation. Further, Tsc1 signaled through Rheb to down-regulate mTORC1 for proper DC development, whereas its effect at modulating mTOR complex 2 (mTORC2) activity was largely dispensable. Our results demonstrate that the interplay between Tsc1-Rheb-mTORC1 signaling and Myc-dependent bioenergetic and biosynthetic activities constitutes a key metabolic checkpoint to orchestrate DC development.
Cell metabolism refers to the intracellular chemical reactions that convert nutrients and endogenous molecules into energy and biomass (proteins, nucleic acids, and lipids). Emerging evidence highlights an intimate interaction between metabolism and immunity (1–3). For example, activated T cells are highly glycolytic and rely on glycolysis to generate ATP (even in the presence of high levels of oxygen), a phenomenon known as Warburg metabolism, which is unique to cancer cells and activated lymphocytes. Blocking glycolysis impairs activation and differentiation of T cells and the outcome of adaptive immune responses, thereby indicating a prerequisite role of metabolism in T-cell fate determination (4–6). Other modes of metabolism, such as lipid metabolism and fatty acid oxidation, are also important regulators of T-cell responses (7–10). Although most studies of metabolic controls of cell fate are focused on T cell-mediated adaptive immunity, we are beginning to appreciate that activation of innate immune cells is also metabolically demanding. Engagement of toll-like receptors (TLRs) expressed by dendritic cells (DCs), the specialized antigen-presenting cells for bridging innate and adaptive immunity, triggers a profound metabolic transition to aerobic glycolysis, similar to Warburg metabolism. Glucose restriction inhibits the activation and life span of TLR-stimulated DCs (11, 12). Glucose metabolism is also a limiting step in the activation of the inflammasome and TLR signaling for the production of the inflammatory cytokine IL-1β (13, 14). Despite advances in our understanding of metabolic regulation of immune cell activation, there is little evidence that cell metabolism is involved in the development of immune cells.
The evolutionarily conserved mechanistic target of rapamycin (mTOR) pathway integrates various environmental signals to regulate fundamental physiological functions such as cell growth and proliferation, autophagy, and nutrient sensing and uptake (15). Whereas the most well-established molecular function of mTOR is in protein translation, recent studies have identified an important role of mTOR in activating a metabolic gene-regulatory network via controlling the respective transcription factors in glycolysis and lipid synthesis, HIF1α and SREBP (16). mTOR exists in two complexes, mTORC1 and mTORC2, both of which contribute to T-cell activation and differentiation (17–19). In the innate immune system, mTOR and the upstream PI3K-AKT pathway have a well-established role in modulating the balance between TLR-induced production of pro- and anti-inflammatory DC cytokines, especially IL-12 and IL-10, thereby affecting DC function and immune responses (20–24). Additionally, mTOR signaling promotes the production of type I IFN from plasmacytoid DCs (pDCs) (25), and regulates other cellular events induced by TLR stimulation such as survival of activated DCs (12, 26). These results collectively illustrate an important role of mTOR signaling in the activation of both innate and adaptive immune systems.
In contrast, the function of mTOR signaling in the development of DCs is less understood, with many of the findings to date obtained via pharmacological approaches. For instance, blocking mTORC1 activity by rapamycin inhibits DC development and/or maturation, and instead endows DCs with a strong tolerogenic activity to promote T-cell tolerance (19, 27–29). However, rapamycin is not an efficient inhibitor of 4EBP1 phosphorylation downstream of mTORC1 activation (30), and may also inhibit mTORC2 activity with prolonged treatment and/or at a high dose (31, 32). To conclusively establish the roles of mTOR in DC development, we have used genetic approaches to eliminate mTOR signaling components, including tuberous sclerosis 1 (Tsc1, a modulator of mTORC1 and mTORC2 activities), Rheb (a key upstream activator of mTORC1), and Rictor (an obligatory component of mTORC2) (15), as well as the downstream effector Myc, alone or in combination, and further complemented them with pharmacological approaches. Contrary to our expectations, deletion of Tsc1 and subsequent mTORC1 activation exerted multiple negative effects on DC development, including survival, proliferation, and lineage differentiation. Loss of Tsc1 up-regulated several metabolic programs including glycolysis, mitochondrial respiration, and lipid synthesis, partly via a Myc-dependent pathway, and this metabolic programming contributed to DC survival and differentiation but not proliferation. Further, Tsc1 signaled through Rheb and mTORC1 to control DC development, whereas mTORC2 activity was largely dispensable in this process. Importantly, DC differentiation is associated with dynamic regulation of glycolytic and lipogenic programs and the accompanying mTORC1 activity. These data point to a unique checkpoint active in DC development that is mediated by the interplay between Tsc1-mTORC1–dependent immune signaling and Myc-dependent metabolic programming.
Results
Tsc1 Plays an Important Role in DC Development in Vitro.
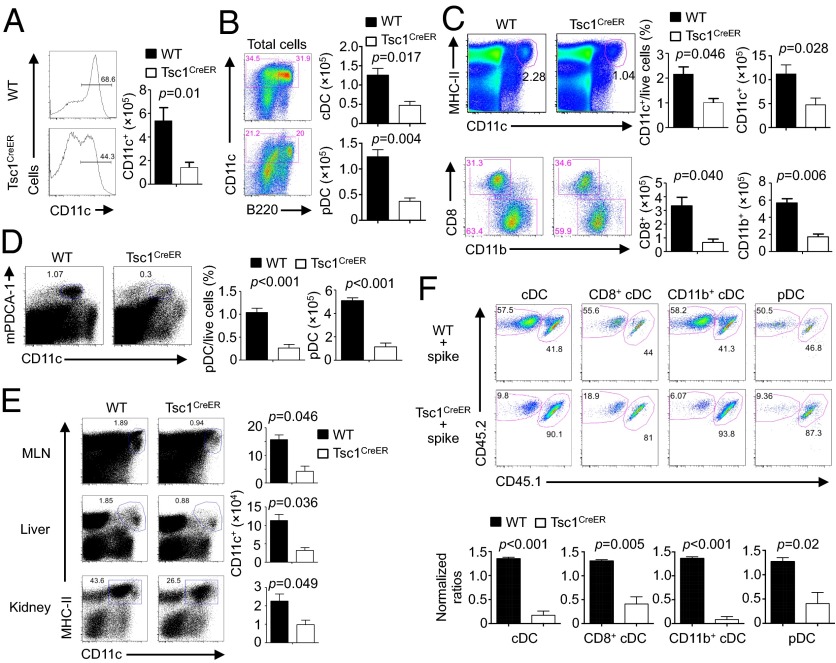
DCs originate from hematopoietic stem cells (HSCs) in the bone marrow (BM) through multiple steps of differentiation, and this can be recapitulated in the in vitro differentiation system mediated by FLT3L (33–35). To determine the importance of mTORC1 regulation in DC development, we focused on Tsc1, an upstream regulator of mTOR signaling (15), by crossing Tsc1flox/flox mice with Rosa26-Cre-ERT2 mice (a Cre-ER fusion gene was recombined into the ubiquitously expressed Rosa26 locus) to generate Tsc1flox/floxRosa26-Cre-ERT2 mice (Tsc1CreER mice). To circumvent the detrimental effects of chronic loss of Tsc1 on the function of HSCs (36, 37), we determined the requirement of Tsc1 during DC development through in vitro acute deletion. Specifically, we cultured BM cells with FLT3L in the presence of 4-hydroxytamoxifen (4-OHT), which resulted in efficient deletion of Tsc1 after 2 d of treatment (Fig. S1A). Under these conditions, Tsc1CreER BM cells showed a considerable defect to generate CD11c+ cells (Fig. 1A). Analysis of DC-specific transcripts including Zbtb46, Ccr7, Flt3, and c-Kit (35) in sorted CD11c+ cells confirmed them as DCs (Fig. S1B). The impairment in DC generation was apparent in both conventional DC (cDC; CD11c+B220–) and pDC (CD11c+B220+) subsets (Fig. 1B). Thus, Tsc1 is important for DC development in vitro.
Fig. 1.
Loss of Tsc1 disrupts DC development in vitro and in vivo. (A and B) BM cells from WT or Tsc1CreER mice were cultured with FLT3L (input, 3 × 106) in the presence of 4-hydroxytamoxifen, followed by analysis of the percentage (Left) and cell number of total CD11c+ DCs (Right) (A) and the number of cDCs (CD11c+B220–) and pDCs (CD11c+B220+) (B). (C and D) WT or Tsc1CreER mice were treated with tamoxifen for 5 d, followed by resting for 5–10 d, and the percentage and number of splenic cDCs and cDC subsets (C) or pDCs (D) were analyzed. (E) DC percentage (Left) and number (Right) in mesenteric lymph nodes (MLNs), liver, and kidney of WT or Tsc1CreER mice were analyzed as in C. (F) BM cells from tamoxifen-treated WT or Tsc1CreER CD45.2.2+ mice were mixed with cells from CD45.1.2+ mice at a 1:1 ratio and transferred into irradiated CD45.1.1+ recipients. The contributions of spike BM-derived (CD45.1.2+) and WT or Tsc1CreER donor BM-derived (CD45.2.2+) cells among total CD11c+ cDCs, CD8+ cDCs, CD11b+ cDCs, and pDCs in the reconstituted mixed chimeras were analyzed (Upper). (Lower) Normalized ratios against the percentages of CD4+ T cells. Error bars indicate SEM. Data are representative of two to four independent experiments.
We next determined the temporal requirement of Tsc1 in DC development. First, we assessed the generation of DCs from Lin–Sca-1+c-Kit+ cells (LSKs; a heterogeneous mixture of HSCs and multipotent progenitors) and macrophage and DC precursors (MDPs; one of the earliest precursor populations identified for DCs, defined as Lin–CD11c−Sca-1–CSF1R+) (38–40). Both progenitor cultures lacking Tsc1 were impaired from developing into CD11c+ DCs after FLT3L stimulation (Fig. S1 C and D), indicating the involvement of Tsc1 in DC development from multipotent and DC-committed precursors. Second, we cultured BM cells from Tsc1CreER mice with 4-OHT added at various times after the initiation of DC differentiation. Compared with wild-type (WT) control, Tsc1CreER cells were impaired from developing into CD11c+ DCs even when 4-OHT was added at 5 d after the initiation of DC differentiation (Fig. S1E). These results illustrate a direct role of Tsc1 in programming DC development.
A Cell-Autonomous Role of Tsc1 in the Generation of DCs and Precursors in Vivo.
To determine the role of Tsc1 in DC development in vivo, we treated WT and Tsc1CreER mice with tamoxifen for 5 d and then rested the mice for 5–10 d. Such treatment resulted in efficient deletion of Tsc1 in splenic DCs from Tsc1CreER mice (Fig. S1F). Compared with WT control, Tsc1CreER mice contained a reduced number of cDCs (CD11c+MHC-II+), including both CD8+ and CD11b+ subsets (Fig. 1C). The pDC (CD11clomPDCA-1+) population was also diminished (Fig. 1D). Similar defects in the cDC population were observed in mesenteric lymph nodes and nonlymphoid organs including liver and kidney (Fig. 1E). Therefore, Tsc1 plays an important role in maintaining DC populations in vivo.
To address whether the reduction of DCs in Tsc1CreER mice was an intrinsic defect, we generated mixed-BM chimeras. Specifically, the recipients (CD45.1.1+) were lethally irradiated and reconstituted with tamoxifen-treated WT or Tsc1CreER BM cells (CD45.2.2+; donor) along with WT BM cells (CD45.1.2+; spike) at a 1:1 ratio. After reconstitution, we analyzed the contributions of donor and spike cells to various immune compartments. Because CD4 T cells are relatively resistant to Tsc1 deletion (41), we used the chimerism of CD4 T cells for normalization (Fig. S1G). DCs, including various subsets, derived from Tsc1-deficient donor cells were greatly underrepresented relative to the spike cells (Fig. 1F). Thus, Tsc1 has a key cell-autonomous role in regulating DC development in vivo. Notably, other myeloid cells lacking Tsc1, such as macrophages and neutrophils, were also decreased in the chimeras (Fig. S1 H and I).
We further determined whether Tsc1 regulates DC development partly by impacting the generation of early precursor MDPs. Following tamoxifen treatment in vivo, the percentage and cell number of MDPs in Tsc1CreER mice were significantly decreased compared with those in WT mice (Fig. S2A). Furthermore, more committed precursors, including common dendritic precursors (CDPs; defined as Lin−CD11c−Sca-1−CSF1R+Flt3+CD117lo) and committed precursors of cDCs (precDCs; defined as Lin−IA/E−CD11c+Flt3+CD172αint) (33), were also reduced upon Tsc1 deletion (Fig. S2 B and C). Associated with the numerical reduction of these precursor populations was a decreased expression of GM-CSFR (CD116), although Flt3 expression was largely normal (Fig. S2D). These data indicate a requirement of Tsc1 in the generation of DC precursors, which may act in synergy with its effect at driving the differentiation from these precursors to mature DCs (Fig. S1D) to promote DC development.
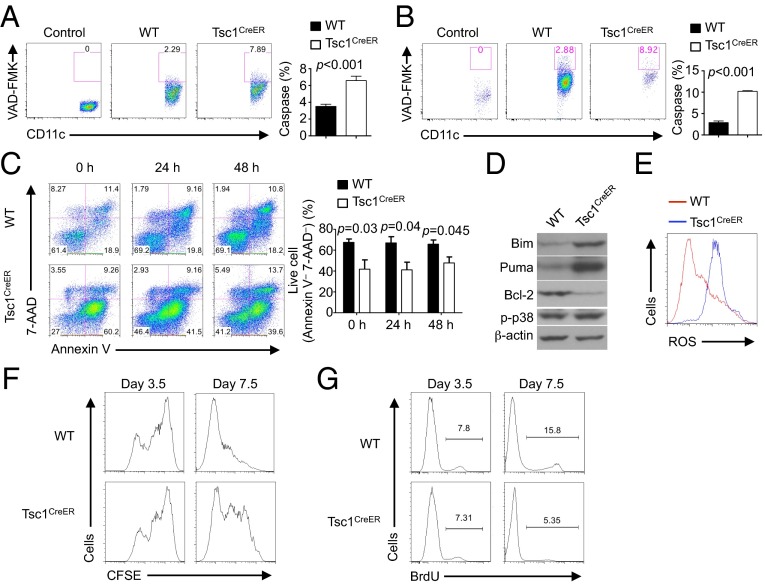
Tsc1 Promotes DC Survival and Proliferation.
The overall size of DC populations is dependent upon the rates of apoptosis and proliferation. We first evaluated the effects of Tsc1 on the survival of DCs. Caspase activity, a hallmark of apoptotic cell death, was increased in freshly isolated splenic DCs of tamoxifen-treated Tsc1CreER mice and of the chimeras reconstituted with Tsc1CreER BM cells (Fig. 2 A and B). In vitro FLT3L-derived DCs from 4-OHT–treated Tsc1CreER BM cells also showed significantly increased apoptosis as indicated by the loss of the live Annexin V− 7-AAD− population, and this increase was observed when 4-OHT was added at 0, 24, and 48 h after the initiation of DC differentiation (Fig. 2C). These data indicate an important role of Tsc1 in promoting the survival of DCs.
Fig. 2.
Tsc1 deficiency impairs DC survival and proliferation. (A and B) Caspase activity in freshly isolated splenic DCs from tamoxifen-treated WT or Tsc1CreER mice (A), or WT or Tsc1CreER BM-derived cells in mixed chimeras (generated as described in Fig. 1F) (B), assessed with FITC-VAD-FMK staining. “Control” indicates no staining with FITC-VAD-FMK. (Right) Percentage of caspase-positive cells. (C) BM cells from WT or Tsc1CreER mice were cultured with FLT3L, with 4-OHT added at 0, 24, and 48 h after the initiation of DC differentiation. The apoptosis of CD11c+ cells was analyzed by flow cytometry at day 7.5. (Right) Percentage of live cells (Annexin V– 7-AAD–). (D) WT or Tsc1CreER BM cells were cultured with FLT3L and 4-OHT, followed by purification of CD11c+ cells and immunoblot analysis. (E) ROS production in 4-OHT–treated WT or Tsc1CreER BM-derived CD11c+ cells. (F) BM cells from WT or Tsc1CreER mice were labeled with CFSE and cultured with FLT3L and 4-OHT for 3.5 or 7.5 d. (G) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT for 3.5 and 7.5 d and pulse-labeled with BrdU for 3 h before staining. Error bars indicate SEM. Data are representative of two or three independent experiments.
We then dissected molecular mechanisms by which Tsc1 regulates DC survival. Bcl-2 family proteins are important regulators of apoptotic cell death, and the balance of anti- and proapoptotic Bcl-2 family members is crucial to dictating the decision between survival and apoptosis (42). The expression of the proapoptotic Bim and Puma was increased in 4-OHT–treated Tsc1CreER DCs, whereas the antiapoptotic protein Bcl-2 was diminished. In contrast, the activity of p38 MAPK, another important cell death regulator, was comparable between WT and Tsc1-deficient DCs (Fig. 2D). Aside from Bcl-2 family proteins, production of reactive oxygen species (ROS) also contributes to apoptotic cell death (42). Tsc1-deficient DCs produced greatly increased levels of ROS (Fig. 2E). Therefore, the increased apoptosis of Tsc1-deficient DCs is associated with dysregulated Bcl-2 family proteins and elevated oxidative stress.
We next tested the possibility that the decreased DC population in the absence of Tsc1 was also partly ascribed to defective DC proliferation. We labeled freshly isolated BM cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) and cultured the cells with FLT3L in the presence of 4-OHT. The proliferation of Tsc1-deficient CD11c+ DCs was similar to that of WT cells when examined at day 3.5. However, the mutant cells showed diminished CFSE dilution when analyzed at day 7.5 (Fig. 2F). Furthermore, a bromodeoxyuridine (BrdU) incorporation assay showed that Tsc1-deficient CD11c+ DCs incorporated less BrdU than did WT cells at day 7.5 (Fig. 2G). Thus, Tsc1 is dispensable for initial DC proliferation but its absence compromises continuous DC proliferation.
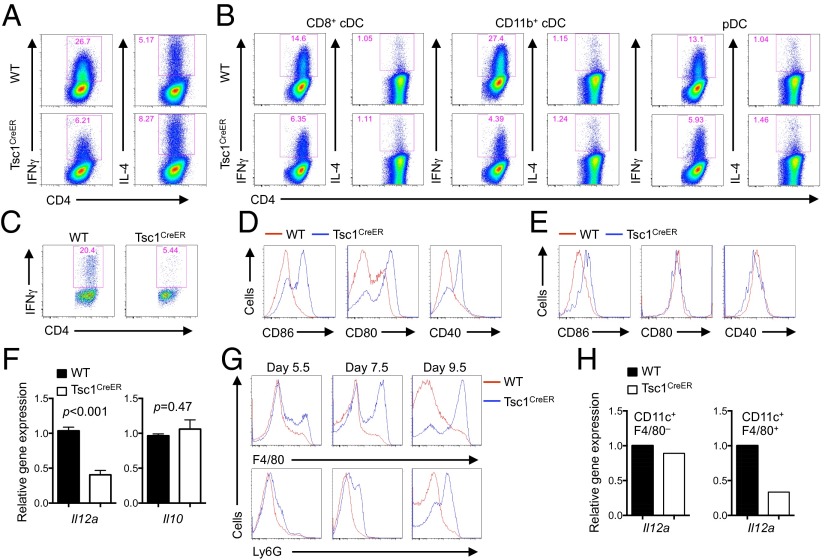
Dysregulated DC Maturation and Functional Differentiation in the Absence of Tsc1.
DCs are the most potent antigen-presenting cells for the activation and differentiation of naïve T cells. To investigate the effect of Tsc1 signaling on DC functions at mediating T-cell responses, we first cocultured WT and Tsc1-deficient DCs with naïve CD4+ T cells from OT-II mice [a T cell receptor (TCR)-transgenic model with T cells recognizing ovalbumin (OVA) amino acids 323–339] in the presence of antigen but no exogenous cytokines to mimic the physiological interaction between DCs and T cells. After 5–6 d, T cells activated with Tsc1-deficient DCs secreted decreased IFN-γ, but similar levels of IL-4, compared with those activated with WT DCs (Fig. 3A). We next used freshly isolated DC subsets from in vivo tamoxifen-treated WT and Tsc1CreER mice and cocultured them with OT-II naïve T cells. All DC subsets lacking Tsc1 showed a defective ability to drive TH1 but not TH2 differentiation (Fig. 3B). Finally, we determined the role of Tsc1 in mediating DC-dependent T-cell responses in vivo. To this end, we generated WT and Tsc1CreER BM chimeras and then transferred OVA-specific CD4+ T cells into the chimeras, followed by immunization with the cognate antigen. T cells isolated from Tsc1CreER chimera hosts produced less IFN-γ (Fig. 3C). These results demonstrate that Tsc1 signaling in DCs is required for the differentiation of antigen-specific naïve T cells into TH1 cells.
Fig. 3.
Tsc1 deficiency causes spontaneous maturation but aberrant differentiation of DCs that prevents induction of effector T-cell responses. (A) WT or Tsc1CreER BM cells were cultured with FLT3L and 4-OHT for 7–8 d and sorted for CD11c+ cells, which were then cultured with naïve OT-II T cells in the presence of OVA323–339 and LPS, followed by IFN-γ and IL-4 detection. (B) Freshly isolated splenic DC subsets from tamoxifen-treated WT or Tsc1CreER mice were cocultured with OT-II T cells and analyzed as described in A. (C) Antigen-specific T cells from OT-II TCR-transgenic mice (Thy1.1+) were transferred into complete WT and Tsc1CreER chimera mice, followed by antigen immunization in the presence of CFA. At day 7 after immunization, draining lymph node cells were isolated for IFN-γ expression analysis. (D) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT, followed by analysis of CD86, CD80, and CD40 expression on CD11c+ cells. (E) Expression of CD86, CD80, and CD40 on splenic DCs from WT or Tsc1CreER BM-derived cells in mixed chimeras (generated as described in Fig. 1F). (F and G) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT, followed by analysis of CD11c+ cells for cytokine mRNA (F) and F4/80 and Ly6G expression (G). (H) WT or Tsc1CreER BM cells were cultured with FLT3L and 4-OHT, and CD11c+F4/80– cells and CD11c+F4/80+ cells were sorted for RNA analysis. Error bars indicate SEM. Data are representative of two to four independent experiments.
To explore the mechanistic basis, we analyzed the effects of Tsc1 deficiency on DC maturation, cytokine production, and lineage differentiation. First, we examined the expression of various surface markers associated with DC maturation, including CD86, CD80, and CD40. Surprisingly, Tsc1-deficient DCs showed more elevated expression of CD86, CD80, and CD40 than WT cells did (Fig. 3D). Moreover, CD86 was up-regulated on DCs derived from Tsc1-deficient donor cells in the mixed chimeras, although CD80 and CD40 expression in vivo was largely normal (Fig. 3E). Thus, Tsc1 deficiency resulted in spontaneous maturation of DCs, with a more pronounced effect on CD86 up-regulation. However, this was unlikely to cause diminished effector T-cell responses, as these DC molecules generally promote effector responses (43). Second, we examined the expression of proinflammatory cytokines, and found that Il12a (IL-12 p35) was decreased in Tsc1-deficient DCs, whereas the anti-inflammatory cytokine Il10 was comparable between WT and Tsc1-deficient DCs (Fig. 3F). Given the importance of IL-12 for mediating TH1 differentiation (44), the impaired cytokine production from Tsc1-deficient DCs, together with the survival defect and numerical reduction, likely contributed to decreased TH1 responses in responding T cells.
Third, we determined the effects of Tsc1 deficiency on the integrity of DC differentiation. Although CD11c+ DCs developed in the absence of Tsc1 expressed normal levels of DC-specific transcripts as described above (Fig. S1B), they aberrantly acquired expression of macrophage and neutrophil markers F4/80 and Ly6G, respectively. This defect was exacerbated with more extended duration of culture (Fig. 3G). To determine whether this altered differentiation was functionally relevant to DC cytokine production, we purified F4/80+CD11c+ cells and measured their cytokine expression levels. Loss of Tsc1 resulted in a profound down-regulation of Il12a in these populations. In contrast, WT and Tsc1-deficient F4/80–CD11c+ cells expressed comparable levels of Il12a (Fig. 3H). Therefore, Tsc1 maintains the integrity of DC differentiation, the loss of which results in aberrant up-regulation of other lineage markers and impaired DC cytokine expression.
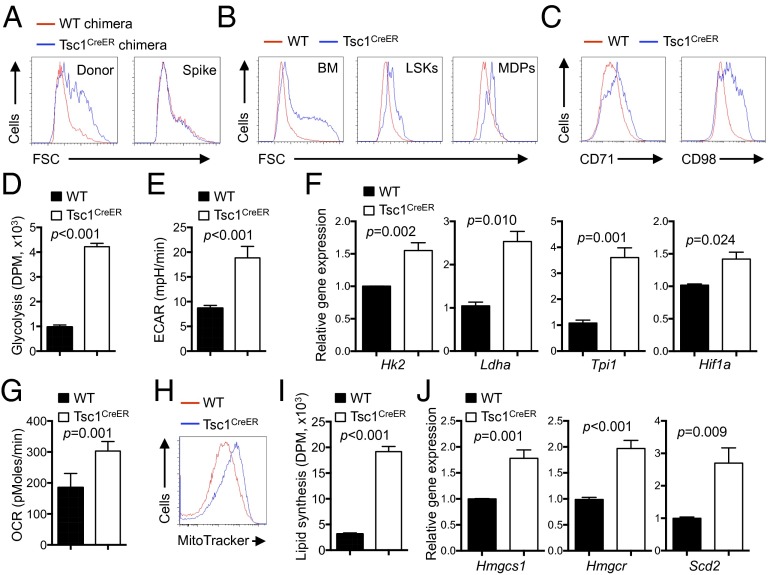
Tsc1 Coordinates Cell Growth and Metabolism in DCs.
We next explored the molecular mechanism underlying Tsc1-mediated control of DC development. We noticed that DCs developed in the absence of Tsc1 exhibited an increased cell size. This alteration was evident in BM chimeras (Fig. 4A), indicative of an intrinsic defect. Additionally, cell size was increased in Tsc1-deficient DCs developed from total BM cells, LSKs, or MDPs in vitro (Fig. 4B), and this was also observed when Tsc1 deletion was initiated at 3–5 d after the start of culture (Fig. S3A). Further, the mutant DCs contained more protein and RNA content on a per-cell basis (Fig. S3B). These results indicated that Tsc1-deficient DCs underwent more cell growth. Cell growth is dependent on the regulated expression of amino acid transporters that include CD98 as a key component, and of CD71, the transferrin receptor that mediates iron uptake. Flow cytometry analysis of surface expression of CD98 and CD71 revealed elevated levels of CD98 and CD71 upon loss of Tsc1 (Fig. 4C). This effect was also observed in DCs derived from Tsc1-deficient MDPs (Fig. S4A), indicating an important role of Tsc1 in cell-growth regulation during DC development.
Fig. 4.
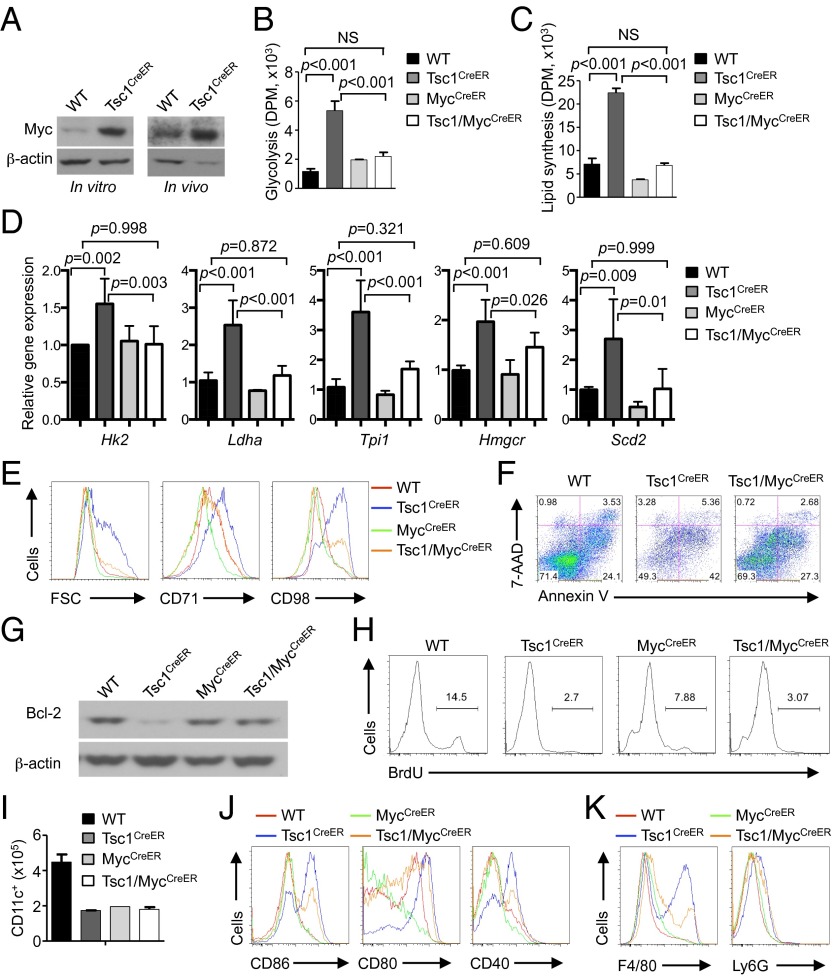
Loss of Tsc1 up-regulates cell growth and multiple metabolic programs including glycolysis, mitochondrial respiration, and lipid biosynthesis. (A) Cell size of splenic DCs from spike BM and WT or Tsc1CreER donor BM-derived cells in the mixed chimeras (generated as described in Fig. 1F). FSC, forward scatter. (B) Size of CD11c+ cells derived from 4-OHT–treated WT or Tsc1CreER total BM cells (Left), LSKs (Center), or MDPs (Right). (C–J) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT, followed by analysis of CD71 and CD98 expression (C), glycolytic activity through measurement of the generation of 3H2O from [3-3H]glucose (DPM, disintegrations per minute) (D), extracellular acidification rate (mpH, milli-pH units) (E), mRNA expression of glycolysis-related genes (F), oxygen consumption rate (G), mitochondria mass as determined by MitoTracker Green (H), de novo lipogenesis assay (I), and mRNA expression of lipogenic genes (J). Error bars indicate SEM. Data are representative of two to five independent experiments.
Cell growth is coupled with cell metabolism, which consists of bioenergetic and biosynthetic activities (1, 17). Two major modes of bioenergetics are glycolysis and mitochondrial respiration. To determine the role of Tsc1 in DC metabolism, we first measured the glycolytic activity of WT and Tsc1-deficient cells by the generation of 3H-labeled H2O from [3-3H]glucose. Compared with FLT3L-derived WT DCs, those lacking Tsc1 displayed up-regulated glycolysis (Fig. 4D). This was verified by the elevated extracellular acidification rate (ECAR) as measured by the Seahorse extracellular flux analyzer (Fig. 4E). Further, mRNA expression of glycolytic enzymes, including Hk2, Ldha, and Tpi1, as well as Hif1a, the transcription factor that regulates glucose metabolism, were all elevated in DCs developed from Tsc1-deficient BM cells (Fig. 4F) or MDPs (Fig. S4B). Moreover, oxygen consumption rate (OCR), indicative of mitochondrial respiration activity, was increased in Tsc1-deficient cells (Fig. 4G), and this was associated with increased mitochondrial mass (Fig. 4H). Therefore, Tsc1-deficient cells exhibit increased glycolysis and mitochondrial respiration.
Having established an important role of Tsc1 in DC bioenergetics, we next determined whether it also contributes to cell biosynthetic activity. In particular, de novo synthesis of lipids (cholesterol and fatty acids) has been shown to depend upon mTOR activity (45). However, the role of Tsc1 in this process remains unclear because it can either promote or inhibit lipid synthesis in a context-dependent manner (16, 46, 47). We therefore measured the incorporation of [1-14C]acetate into chloroform/methanol-soluble lipids in DCs with normal or absent Tsc1 expression; [1-14C]acetate was used here to bypass the requirement of Tsc1 in glycolysis or mitochondrial activity, thereby allowing us to directly evaluate the role of Tsc1 in de novo lipid biosynthesis rate (46). Compared with WT DCs, those lacking Tsc1 showed a markedly increased lipid synthesis rate (Fig. 4I). Furthermore, expression of many genes in cholesterol and fatty acid metabolism, including Hmgcs1, Hmgcr, and Scd2, was greatly elevated in DCs derived from Tsc1-deficient BM cells (Fig. 4J) or MDPs (Fig. S4C). Altogether, these results indicate that Tsc1 has a key role in negatively controlling cell growth, bioenergetics, and lipid biosynthesis, thereby coupling cell growth and metabolism.
The Tsc1–Myc Axis Orchestrates Metabolic Programming of DCs.
The transcription factor Myc is an established regulator of cell metabolism, especially glycolytic activity, and plays a key role in metabolic reprogramming of activated T cells (4). We hypothesized that the increased metabolic activity of Tsc1-deficient DCs is partly dependent upon Myc. Indeed, Myc expression was up-regulated in Tsc1-deficient BM-derived CD11c+ DCs or freshly isolated DCs following in vivo tamoxifen treatment (Fig. 5A). To determine the functional relevance of Myc in Tsc1-mediated control of DC metabolism, we generated Tsc1flox/floxMycflox/floxRosa26-Cre-ERT2 (Tsc1/MycCreER) mice. Remarkably, loss of Myc nearly completely blocked elevated glycolysis and de novo lipid synthesis in Tsc1-deficient DCs, even though Myc deficiency alone did not cause major alteration of DC metabolism (Fig. 5 B and C). This rescue effect was associated with the restoration of glycolytic and lipogenic gene expression in DCs deficient in both Tsc1 and Myc (Fig. 5D). Moreover, the increased cell size and CD71 and CD98 expression in Tsc1-deficient DCs were partly rescued by the simultaneous loss of Myc (Fig. 5E). Therefore, the aberrant induction of Myc in Tsc1-deficient DCs accounted, at least partially, for the dysregulated growth and metabolism.
Fig. 5.
Tsc1 controls metabolic programming of DCs through inhibition of Myc expression. (A) Immunoblot analysis of Myc expression in BM-derived CD11c+ cells from WT or Tsc1CreER mice cultured with FLT3L and 4-OHT (Left) or freshly isolated splenic CD11c+ cells from tamoxifen-treated WT or Tsc1CreER mice (Right). (B–D) BM cells from WT, Tsc1CreER, MycCreER, or Tsc1/MycCreER mice were cultured with FLT3L and 4-OHT, followed by analyses of metabolic parameters including glycolysis (B), de novo lipogenesis (C), and mRNA expression of glycolytic and lipogenic genes (D). (E–K) BM cells from the indicated mice were cultured with FLT3L and 4-OHT, followed by analysis of CD11c+ cells for cell size and CD71 and CD98 expression (E), Annexin V and 7-AAD staining (F), expression of Bcl-2 (G), cell proliferation as determined by BrdU incorporation (H), DC production (input BM cells, 3 × 106) (I), expression of maturation markers (J), and expression of F4/80 and Ly6G (K). NS, not significant. Error bars indicate SEM. Data are representative of two or three independent experiments.
We next evaluated the extent to which Myc-dependent metabolism contributes to DC development. Importantly, the exacerbated apoptosis in Tsc1-deficient DCs was partly blocked by Myc deficiency (Fig. 5F). This blocking was associated with the partial rescue of Bcl-2 (Fig. 5G) and Bim dysregulation (Fig. S5A), although ROS production was not corrected (Fig. S5B). In contrast, Myc deletion alone diminished DC proliferation, and DCs developed in the absence of both molecules were still defective in proliferation and total DC production (Fig. 5 H and I). Thus, aberrant induction of Myc contributes to defective survival but not proliferation in Tsc1-deficient DCs. Further, Myc deletion partially blocked the spontaneous maturation of DCs lacking Tsc1, as the increased expression of the maturation markers CD86, CD80, and CD40 in these cells was down-regulated by the deletion of Myc (Fig. 5J). Analysis of BM chimeras further showed that Myc deficiency partially blocked the aberrant up-regulation of CD86 on Tsc1-deficient DCs in vivo (Fig. S5C). Finally, the increased F4/80 and Ly6G expression in the absence of Tsc1 was partly rescued by simultaneous loss of Tsc1 and Myc (Fig. 5K). Taken together, Myc plays a key role in mediating the dysregulated metabolism and contributes to the impaired survival and maturation of Tsc1-deficient DCs. However, other effects, such as the defective proliferation and in vivo differentiation in Tsc1-deficient cells, occur independent of Myc.
Rapamycin-Sensitive mTORC1 Signaling Contributes to Tsc1-Deficient DC Defects.
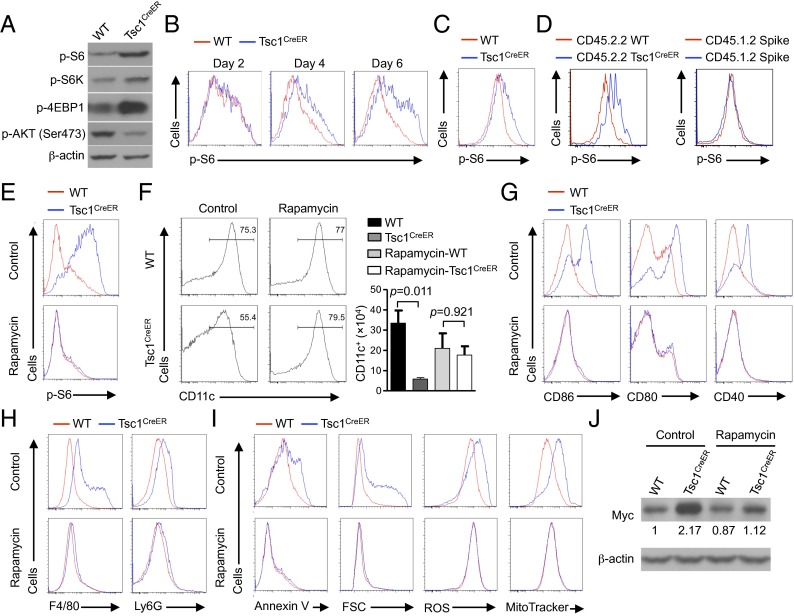
We investigated the signaling mechanisms by which Tsc1 controls DC development. Although Tsc1 is known to negatively regulate mTORC1 (15), it also regulates mTORC2 activity (48). Indeed, the defective survival of Tsc1-deficient T cells is insensitive to rapamycin treatment in vitro (49), and has been suggested to be ascribed to diminished mTORC2 activity (50). To determine the biochemical functions of Tsc1 in DCs, we first used immunoblotting to examine canonical targets of mTORC1, namely the phosphorylation of S6, S6K1, and 4EBP1 (15). DCs developed in the absence of Tsc1 showed increased activities of these mTORC1 targets (Fig. 6A). In contrast, the phosphorylation of Akt (Ser473), which is a target of mTORC2, was diminished in the mutant cells (Fig. 6A). Thus, Tsc1 serves as a negative regulator of mTORC1 in DCs but promotes mTORC2 activity. To further examine the role of Tsc1 in restraining mTORC1 activity at the single-cell level, we used flow cytometry to measure phosphorylation of S6 at different time points of DC differentiation. Increased phosphorylation of S6 was observed at 4 d after the initiation of 4-OHT treatment (Fig. 6B), which was largely in agreement with the timing of Tsc1 RNA deletion (Fig. S1A). Moreover, in vivo tamoxifen-treated Tsc1CreER splenic DCs had higher levels of S6 activation than WT controls (Fig. 6C). Similarly, DCs derived from Tsc1CreER BM donor cells in mixed chimeras also had increased p-S6 than those derived from WT BM or spike BM cells (Fig. 6D). These results indicate that Tsc1 plays an important role in inhibiting mTORC1 activation in DCs in vitro and in vivo.
Fig. 6.
Rapamycin-sensitive mTORC1 signaling contributes to Tsc1-deficient DC defects. (A) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT, followed by immunoblot analysis of p-S6, p-S6K, p-4EBP1, and p-AKT (Ser473) in purified CD11c+ cells. (B) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT, followed by flow cytometry analysis of p-S6 at the indicated time points of DC differentiation. (C and D) Phosphorylation of S6 was determined in splenic DCs from tamoxifen-treated WT or Tsc1CreER mice (C) or from WT or Tsc1CreER donor BM-derived cells (Left) and spike BM cells (Right) in the mixed chimeras (generated as described in Fig. 1F) (D). (E–J) BM cells from WT or Tsc1CreER mice were cultured with FLT3L and 4-OHT in the presence of rapamycin or control, followed by analyses of phosphorylation of S6 (E), CD11c+ DC percentage (Left) and number (Right) (input BM cells, 2 × 106) (F), maturation marker expression (G), F4/80 and Ly6G expression (H), Annexin V staining, cell size, ROS production, and mitochondrial mass (I), and Myc expression (the numbers below the lanes indicate the band intensity relative to that of the loading control β-actin) (J). Error bars indicate SEM. Data are representative of two to four independent experiments.
To determine whether elevated mTORC1 activation contributes to the defects in Tsc1-deficient DCs, we treated BM cells in vitro with the mTORC1 inhibitor rapamycin. As expected, rapamycin blocked the elevated mTORC1 activity in Tsc1-deficient DCs (Fig. 6E). Whereas rapamycin treatment had a modest effect in reducing DC production from WT cells, it largely restored the percentage and total number of Tsc1-deficient DCs (Fig. 6F). Rapamycin also blocked excessive expression of CD86, CD80, and CD40 (Fig. 6G), and rescued the aberrant induction of F4/80 and Ly6G in the mutant cells (Fig. 6H). At the cellular and molecular levels, rapamycin blocked the up-regulation of apoptosis, cell size, ROS production, and mitochondrial mass (Fig. 6I), as well as Myc expression (Fig. 6J), as observed in Tsc1-deficient DCs. Taken together, the increased mTORC1 activity is required to drive the developmental and metabolic defects of Tsc1-deficient DCs.
One caveat remains, however, because the extended treatment of rapamycin can also inhibit mTORC2 activity (31, 32). To determine the effects of mTORC2 on DC development, we used a similar strategy as Tsc1 deletion and generated Rictorflox/floxRosa26-Cre-ERT2 mice (RictorCreER), which allowed us to acutely delete Rictor, the mTORC2-defining component. BM cells with deletion of Rictor in vitro developed normally into DCs, although total DC number trended lower (Fig. S6A). This finding is in agreement with a recent study showing a dispensable role of Rictor in GM-CSF–derived DC development (29). The mutant DCs showed comparable cell death, cell size, and expression of DC maturation markers as WT cells (Fig. S6 B–D). Therefore, deficiency of Rictor did not alter DC development, suggesting that loss of mTORC2 activity in Tsc1-deficient DCs is insufficient to drive the disrupted immune homeostasis. These results collectively indicate that aberrant activation of mTORC1 largely accounts for impaired DC development.
Tsc1 Signals Through Rheb to Control mTORC1 and DC Development.
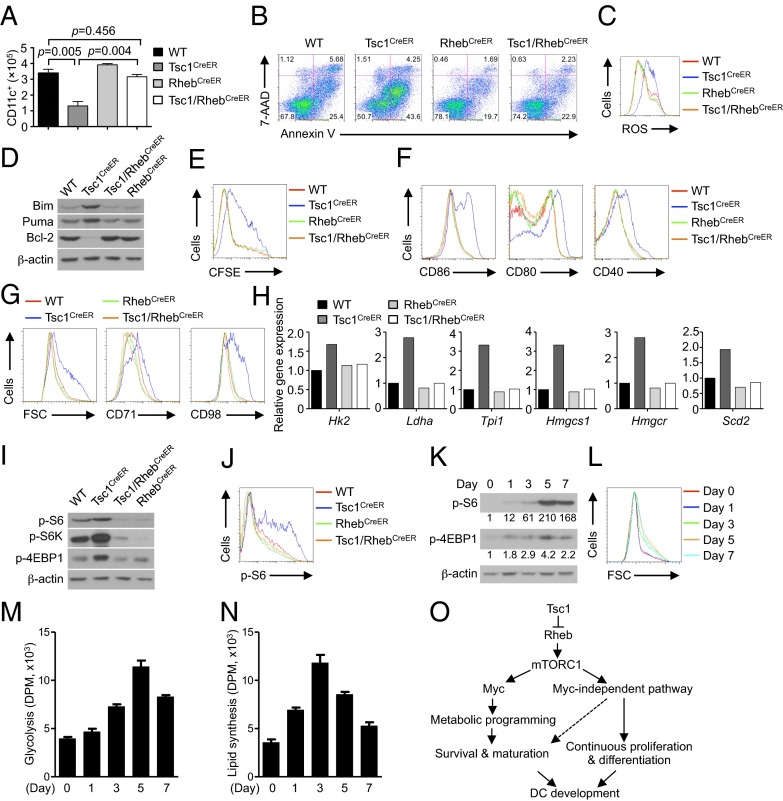
One of the important inputs to mTORC1 is mediated by the small G protein Rheb, although Rheb-independent pathways have also been identified (17). For example, the kinase AMPK can directly phosphorylate Raptor to mediate a metabolic checkpoint (51). Having established a role for Tsc1 in DC development via mTORC1 inhibition, we next determined whether this effect is mediated by Rheb. To this end, we used the Rosa26-Cre-ERT2 system and generated mice lacking both Tsc1 and Rheb (Tsc1/RhebCreER). We cultured BM cells from the mutant mice and proper controls with FLT3L and examined DC generation following acute deletion in vitro. The diminished generation of DCs in the absence of Tsc1 was restored to WT levels by the simultaneous loss of Rheb (Fig. 7A). We then determined the effects of Rheb deficiency on the altered apoptosis and proliferation of Tsc1-deficient DCs. The increased apoptosis of Tsc1-deficient DCs was nearly completely restored in Tsc1/Rheb-deficient DCs (Fig. 7B), associated with the reversal of ROS overproduction and dysregulated expression of Bcl-2 family proteins (Fig. 7 C and D). Additionally, the defective proliferation of Tsc1-deficient DCs was similarly rescued by the loss of Rheb (Fig. 7E). Furthermore, the altered maturation markers as observed in Tsc1-deficient DCs were restored to WT levels in Tsc1/Rheb-deficient cells (Fig. 7F). These results indicate that Tsc1 signals to mTORC1 inhibition in a strictly Rheb-dependent manner. Surprisingly, deficiency of Rheb alone did not have a strong effect on DC development, survival, or proliferation (Fig. 7 A–F). Thus, whereas an excessive Rheb-mTORC1 activity is detrimental to DC generation, loss of function of this pathway appears to be compatible with DC development.
Fig. 7.
Tsc1 signals through Rheb to control mTORC1 activation in DC development. (A–D) BM cells from WT, Tsc1CreER, RhebCreER, or Tsc1/RhebCreER mice were cultured with FLT3L and 4-OHT, followed by analysis of CD11c+ DC number (input BM cells, 2 × 106) (A), Annexin V and 7-AAD staining (B), ROS production (C), and immunoblot analysis of Bim, Puma, and Bcl-2 in purified CD11c+ DCs (D). (E) BM cells from WT or Tsc1CreER mice were labeled with CFSE and cultured with FLT3L and 4-OHT, followed by analysis of CFSE dilution at day 7.5. (F–J) BM cells were cultured with FLT3L and 4-OHT, followed by analysis of CD11c+ DCs for maturation marker expression (F), cell size and CD71 and CD98 expression (G), mRNA analysis of glycolytic and lipogenic genes (H), immunoblot of p-S6, p-S6K, and p-4EBP1 (I), and intracellular staining of p-S6 (J). (K–N) WT BM cells were cultured with FLT3L for the indicated times, followed by analysis of p-S6 by immunoblot (the numbers below the lanes indicate the band intensity relative to that of the loading control β-actin) (K), cell size (L), glycolysis (M), and de novo lipogenesis (N). Error bars indicate SEM. Data are representative of two or three independent experiments. (O) The interplay between Tsc1-Rheb-mTORC1 signaling and Myc-dependent metabolic programming in DC development.
Given the importance of cell growth and metabolism in DC development, we further measured the role of the Tsc1–Rheb axis in this process. The increased cell growth and nutrient receptor expression in Tsc1-deficient DCs were blocked by the simultaneous loss of Rheb (Fig. 7G). Further, the elevated mRNA expression of glycolytic (Hk2, Ldha, and Tpi1) and lipogenic genes (Hmgcs1, Hmgcr, and Scd2), as observed in Tsc1-deficient DCs, was completely restored in Tsc1/Rheb-deficient cells (Fig. 7H). Finally, Rheb deficiency restored the aberrant induction of p-S6 and p-4EBP1 observed in Tsc1-deficient cells (Fig. 7 I and J).
We next explored how mTORC1 activity is regulated during DC development. We cultured BM cells with FLT3L and examined phosphorylation of S6 and 4EBP1 at different time points. Phosphorylation of these molecules reached a high level at day 5 and then abated (Fig. 7K). Importantly, the cell size and glycolytic and lipogenic activities followed a similar biphasic pattern of regulation, with the highest levels observed at days 3–5 (Fig. 7 L–N). Therefore, mTORC1 activity and mTORC1-dependent metabolism are dynamically regulated during DC development, whereas uncontrolled mTORC1 activation impairs DC development (Fig. 7O).
Discussion
A fundamental question in immunology is how the development of DCs is regulated by cellular and molecular processes. The recent identification of distinct DC precursors and transcription factors orchestrating differentiation of DCs and various subsets has revolutionized our understanding of DC biology (33–35). In contrast, we have limited information on how intracellular signaling networks program DC development. Moreover, whereas emerging evidence highlights a role of cell metabolism in the activation of innate and adaptive immunity (1–3), its involvement in the development of immune cells, and in particular DCs, remains undefined. Here we describe that the interplay between Tsc1-mTORC1 signaling and Myc-dependent metabolism orchestrates a previously unappreciated metabolic checkpoint to control DC development. Surprisingly, loss of Tsc1 and the ensuing mTORC1 up-regulation disrupt development of DCs by impairing their survival, proliferation, and differentiation. These defects impair TH1 effector responses, despite the elevated expression of maturation markers. Mechanistically, Tsc1 controls DC development in part by actively repressing Myc-mediated metabolic programs, especially glycolysis and lipid synthesis. This process requires Tsc1 to signal through Rheb to inhibit mTORC1 activity. Our results therefore identify Tsc1-Rheb-mTORC1 as a central pathway to program DC development by mediating the interplay between immune signaling and Myc-mediated metabolic programs.
Emerging evidence from mouse genetic models highlights a role of mTOR in DC development and activation. Specifically, deletion of Pten facilitates FLT3L but not GM-CSF–driven DC development in vitro and the expansion of CD8+ DCs in vivo (27). Further, deficiency of Tsc1 has been recently shown to impair TLR-induced activation and maturation of DCs derived from GM-CSF, although Tsc1 loss does not affect DC development in response to GM-CSF (52) or DC terminal differentiation or maintenance (27). In contrast, our results have identified a detrimental effect of Tsc1 deficiency on the generation of DCs in vivo and in response to FLT3L stimulation in vitro, thereby highlighting an intrinsic role of Tsc1 in DC development. The simplest interpretation is that the mTOR regulators exert pathway- and context-specific regulation of DC biology. For instance, Pten is an inhibitor of both mTORC1 and mTORC2 activities by repressing upstream PI3K activity (17). Pten also has functions independent of PI3K/mTOR, such as acting in the nucleus to maintain chromosomal stability (53). In contrast, loss of Tsc1 up-regulates mTORC1 but diminishes mTORC2 activity, although the reduction of mTORC2 activity alone (as achieved by Rictor deletion) is not sufficient to alter DC development. Consistent with a dominant role of mTORC1 in this process, the defect in Tsc1-deficient DCs can be rescued by rapamycin and, more importantly, by Rheb deficiency. Furthermore, deletion of Pten but not Tsc1 using CD11c-Cre mice affects homeostasis of DCs (27). These results collectively support an important but complex role of mTORC1 signaling in DC development.
Aside from a reduction in overall DC populations, DCs that were able to develop in the absence of Tsc1 (as indicated by the normal expression of DC-specific transcripts) aberrantly up-regulate markers for macrophages and neutrophils. These results indicate that Tsc1 deficiency results in a loss of DC developmental integrity by failing to repress diversion into alternative lineages. Further, these mutant DCs undergo spontaneous maturation, as shown by the excessive induction of costimulatory molecules, especially CD86, that was obvious in Tsc1-deficient DCs in vivo and in vitro. This finding is in agreement with the observation that rapamycin treatment during DC development reduces DC maturation (19, 28, 29), thereby providing genetic evidence for a role of mTORC1 in promoting DC maturation. Despite the increased CD86 expression, DCs developed in the absence of Tsc1 were unable to effectively mediate the differentiation of TH1 cells, and such a functional loss is associated with aberrant lineage development. Therefore, the aberrant differentiation of Tsc1-deficient DCs, likely acting in synergy with the survival defect and numerical reduction, contributes to the loss of DC functional fitness, further highlighting the importance of active control of mTORC1 activity for DC development and function.
We further link Tsc1-mTORC1 functions to metabolic programming of DC development. Remarkably, loss of Tsc1 results in extensive changes of multiple metabolic programs. First, DCs developed in the absence of Tsc1 up-regulate glycolysis and expression of key glycolytic enzymes. Second, the mutant cells show increased mitochondrial metabolism as shown by increased rate of mitochondrial respiration, excessive ROS production, and elevated mitochondrial mass. Third, lipid synthesis is markedly increased as well, associated with up-regulated genes in both fatty acid and cholesterol metabolic pathways. Further, the dysregulated metabolism in Tsc1-deficient cells is contingent upon Rheb function, thereby highlighting an important role of Tsc1-Rheb-mTORC1 to coordinately regulate metabolic programs during DC development. However, DC development proceeds normally in the absence of Rheb, suggesting the presence of a Rheb-independent pathway [e.g., the compensation from the homolog Rhebl1/Rheb2 (54) or a distinct GTPase that is yet to be identified] to mediate mTORC1 activation during early DC development. Importantly, expression of the key metabolic transcription factor Myc is enhanced in Tsc1-deficient cells, and deletion of Myc has a marked effect to block the dysregulated metabolism and also partly rectifies abnormal DC survival and maturation. These results point to a unique metabolic checkpoint mediated by the interplay between Tsc1-Rheb-mTORC1 signaling and Myc-dependent bioenergetic and biosynthetic activities that actively orchestrates DC development. Our study has established a key metabolic checkpoint in immune cell development, and this is reminiscent of the recent identification of an Lkb1-dependent but mTORC1-independent metabolic control mechanism required for HSC development (55–57).
Despite the general role of mTORC1 and metabolism in promoting cell proliferation, we had an unexpected finding that DCs developed in the absence of Tsc1 were impaired in continuous proliferation whereas the initial proliferation was normal. Additionally, this effect was independent of Myc. In fact, deficiency of Myc alone reduced DC proliferation. Although the precise mechanism remains to be established, we noticed that deletion of another mTOR inhibitor, Pten, depletes HSCs by inducing the expression of p53 and other tumor suppressors (58). We speculate that the aberrant cell growth and metabolism due to mTORC1 hyperactivation in the absence of Tsc1 induces metabolic stress that, once accumulated, leads to cell-cycle arrest and apoptosis induction, possibly via a tumor suppressor response. Future work is warranted to identify the molecular components involved.
In summary, we have identified a unique metabolic checkpoint crucial for DC development (Fig. 7O). Although emerging evidence highlights a role of glucose metabolism in the activation of innate immunity, especially in response to TLR stimulation (11–14), whether cell metabolism contributes to DC development remains unclear. Similarly, whereas the role of mTOR signaling, including the Tsc1/Tsc2 pathway, in TLR responses in macrophages, DCs, and other cells is beginning to be recognized (20, 52, 59), its involvement in the development of DCs has not been appreciated. Given the evolutionarily conserved function of mTOR signaling, we speculate that mTORC1-dependent metabolic programming of DC development represents a previously unappreciated paradigm of metabolic control of immune cell development that can be applied to the generation of other myeloid cells and additional immune lineages.
Materials and Methods
Mice and BM Chimeras.
C57BL/6, CD45.1, Thy1.1, and OT-II mice were purchased from The Jackson Laboratory. Tsc1flox/flox, Mycflox/flox, Rhebflox/flox, and Rictorflox/flox mice were bred with Rosa26-Cre-ERT2 mice, and had been backcrossed to the C57BL/6 background for at least eight generations. WT controls were Cre+ mice in the same genetic background to account for Cre effects. For in vivo tamoxifen treatment, WT or Tsc1CreER mice were injected i.p. with 2 mg tamoxifen (Sigma) per mouse for 5 consecutive days and rested for 5–10 d before analyses. For mixed-BM experiments, BM cells from in vivo tamoxifen-treated WT or Tsc1CreER (CD45.2.2+) mice were mixed with cells from CD45.1.2+ mice at a 1:1 ratio and transferred into lethally irradiated (11 Gy) CD45.1.1+ mice, as described previously (60). For complete BM experiments, BM cells from tamoxifen-treated WT or Tsc1CreER CD45.2.2+ mice were transferred into lethally irradiated CD45.1.1+ mice. Animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Cell Purification and Cultures.
Mouse spleens were digested with Collagenase D (Roche), and DCs (CD11c+TCR–CD19–DX5–), CD8+ cDCs (CD11c+CD8+CD11b–TCR–CD19–DX5–), CD11b+ cDCs (CD11c+CD11b+CD8–TCR–CD19–DX5–), or pDCs (CD11c+mPDCA-1+TCR–CD19–DX5–) were sorted on a Reflection (iCyt). DCs from nonlymphoid organs were isolated as described (61). Lymphocytes were sorted for naïve T cells (CD4+CD62LhiCD44loCD25–). For DC culture, BM cells (2–4 × 106) were cultured in RPMI-1640 medium containing 10% (vol/vol) FBS and mouse FLT3L (200 ng/mL), in the presence of 4-hydroxytamoxifen (0.5 µM) when indicated. Unless otherwise noted, FLT3L-stimulated cells were analyzed at days 7–8 for DC generation or at days 5–6 for metabolic or molecular mechanisms. For labeling with CFSE (Life Technologies), BM cells were incubated in RPMI-1640 medium with 5% (vol/vol) FBS and 4 μM CFSE at 37 °C for 25 min, followed by extensive washes. The BM precursor populations were isolated as follows: LSK cells were defined as Lin– (CD3, CD45R, Ter119, CD11b, and Gr1) Sca-1hic-Kithi, and MDPs as Lin–Sca-1–CSF1R+. Rapamycin (50 nM) was added from day 0 of culture. For DC and T-cell cocultures, BM cells were cultured with FLT3L and 4-OHT for 7 d and sorted for CD11c+ DCs, which were then mixed with naïve OT-II T cells at a 1:10 ratio in the presence of OVA323–339 peptide and 100 ng/mL LPS; alternatively, freshly isolated DCs from spleen were used for cocultures with T cells. After 5–6 d of culture, live T cells were collected and stimulated with PMA (phorbol 12-myristate 13-acetate) and ionomycin for intracellular cytokine staining.
Antigen Challenge.
Antigen-specific T cells from OT-II TCR-transgenic mice (Thy1.1+) were sorted and transferred into complete WT or Tsc1CreER chimera mice. Twenty-four hours later, the mice were injected s.c. with OVA323–339 (100 μg) in the presence of complete Freund’s adjuvant (CFA; Difco). At day 7 after immunization, draining lymph node cells were isolated and stimulated with PMA and ionomycin for intracellular cytokine staining.
Flow Cytometry.
Flow cytometry was performed as described previously (61, 62). For intracellular cytokine detection, cells were stimulated for 5 h with PMA and ionomycin in the presence of monensin before staining, according to the manufacturer’s instructions (BD Biosciences). For caspase activity detection, cells were stained with FITC-VAD-FMK according to the manufacturer’s instructions (Promega). ROS was measured by incubation with CM-H2DCFDA (10 μM; Invitrogen) at 37 °C for 30 min after staining of surface markers. To stain mitochondria, lymphocytes were incubated with MitoTracker Green (20 nM; Invitrogen) at 37 °C for 20 min after staining of surface markers. BrdU labeling was performed according to the manufacturer’s instructions (BD Biosciences) (41). For detection of phosphorylated signaling proteins, cells were fixed with Phosflow Lyse/Fix Buffer, followed by permeabilization with Phosflow Perm Buffer III (BD Biosciences) and staining with antibodies to S6 phosphorylated at Ser235 and Ser236 (D57.2.2E; Cell Signaling Technology). Bim staining was done according to the manufacturer’s instructions (Cell Signaling Technology). Flow cytometry data were acquired on an LSR II or LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (Tree Star). Precursor populations were gated as follows: MDPs, Lin– (CD3–, CD45R–, Ter119–, CD11b–, and Gr1–) CD11c−Sca-1–CSF1R+; CDPs, Lin−CD11c−Sca-1−CSF1R+Flt3+CD117lo; and precDCs, Lin−MHC-II−CD11c+Flt3+CD172αint.
De Novo Lipogenesis, Glycolysis, and Bioenergetics Assays.
Bone marrow cultures were incubated with 4 μCi/mL [1-14C]acetic acid (PerkinElmer) for 4 h. Following incubation, cells were collected, washed twice with PBS, and lysed in 0.5% (vol/vol) Triton X-100. Lipids were extracted by the addition of a chloroform and methanol mixture (2:1, vol/vol) with vortexing, followed by the addition of water with vortexing. After centrifugation (350 × g, 15 min), the lipid-containing phase (at the bottom) was separated and 14C incorporation was measured using a Beckman LS6500 scintillation counter. Glycolytic flux was determined by measuring the detritiation of [3-3H]glucose, as we have previously described (5). The bioenergetic activities of the ECAR and OCR pathways were measured using the Seahorse XF24-3 Extracellular Flux Analyzer per the manufacturer’s instructions (Seahorse Bioscience).
RNA and Protein Analyses.
Real-time PCR analysis was performed with primers and probe sets from Applied Biosystems, or using Power SYBR Green Master Mix from Life Technologies. The cycling threshold value of the endogenous control gene (HPRT or actin) was subtracted from the cycling threshold value of each target gene to generate the change in cycling threshold (ΔCT). The relative expression of each target gene is expressed as the fold change relative to that of WT samples (2−ΔΔCT), as described (61, 62), and the values of biological replicates are presented. Immunoblotting was performed as described (61, 62) using the following antibodies: p-S6, p70 S6K (p-S6K), p-AKT (Ser473), p-4EBP1, Myc (all from Cell Signaling Technology), Bcl-2 (Santa Cruz), Bim (Abcam), Puma, and β-actin (both from Sigma). Protein concentration was measured by the BCA Protein Assay Kit (Thermo Scientific Pierce), and RNA concentration was measured with a NanoDrop spectrophotometer.
Statistical Analysis.
P values were calculated using Student t test or ANOVA (GraphPad Prism). P values of less than 0.05 were considered significant. All error bars in the graphs represent SEM.
Supplementary Material
Acknowledgments
We acknowledge D. Green for Mycflox mice, C. Cloer and N. Brydon for animal colony management, and the St. Jude Immunology FACS core facility for cell sorting. This work was supported by the National Institutes of Health (Grants R21 AI094089, R01 NS064599, and R01 AI101407; to H.C.), a research grant from the National Multiple Sclerosis Society (RG 4691-B-2; to H.C.), and the American Lebanese Syrian Associated Charities (H.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308905110/-/DCSupplemental.
References
- 1.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13(10):907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 2.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38(4):633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensinger SJ, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134(1):97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14(5):489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everts B, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120(7):1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115(23):4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masters SL, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat Immunol. 2010;11(10):897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9(5):324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29(4):565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Ohtani M, et al. Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood. 2008;112(3):635–643. doi: 10.1182/blood-2008-02-137430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz F, et al. Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur J Immunol. 2008;38(11):2981–2992. doi: 10.1002/eji.200838761. [DOI] [PubMed] [Google Scholar]
- 23.Haidinger M, et al. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J Immunol. 2010;185(7):3919–3931. doi: 10.4049/jimmunol.1000296. [DOI] [PubMed] [Google Scholar]
- 24.Ohtani M, et al. Cutting edge: mTORC1 in intestinal CD11c+ CD11b+ dendritic cells regulates intestinal homeostasis by promoting IL-10 production. J Immunol. 2012;188(10):4736–4740. doi: 10.4049/jimmunol.1200069. [DOI] [PubMed] [Google Scholar]
- 25.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9(10):1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amiel E, et al. Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. J Immunol. 2012;189(5):2151–2158. doi: 10.4049/jimmunol.1103741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sathaliyawala T, et al. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 2010;33(4):597–606. doi: 10.1016/j.immuni.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnquist HR, et al. mTOR and GSK-3 shape the CD4+ T-cell stimulatory and differentiation capacity of myeloid DCs after exposure to LPS. Blood. 2010;115(23):4758–4769. doi: 10.1182/blood-2009-10-251488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosborough BR, et al. Murine dendritic cell rapamycin-resistant and rictor-independent mTOR controls IL-10, B7-H1, and regulatory T-cell induction. Blood. 2013;121(18):3619–3630. doi: 10.1182/blood-2012-08-448290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105(45):17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 32.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234(1):45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 35.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205(10):2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan B, et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci USA. 2008;105(49):19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fogg DK, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 39.Varol C, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204(1):171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waskow C, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9(6):676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12(9):888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M, Wang J. Programmed cell death of dendritic cells in immune regulation. Immunol Rev. 2010;236(1):11–27. doi: 10.1111/j.1600-065X.2010.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joffre O, Nolte MA, Spörri R, Reis e Sousa C. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol Rev. 2009;227(1):234–247. doi: 10.1111/j.1600-065X.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8(3):224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14(1):21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenerson HL, Yeh MM, Yeung RS. Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation. PLoS One. 2011;6(3):e18075. doi: 10.1371/journal.pone.0018075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28(12):4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien TF, et al. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41(11):3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, et al. TSC1/2 signaling complex is essential for peripheral naïve CD8+ T cell survival and homeostasis in mice. PLoS One. 2012;7(2):e30592. doi: 10.1371/journal.pone.0030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan H, et al. Critical role of the tumor suppressor tuberous sclerosis complex 1 in dendritic cell activation of CD4 T cells by promoting MHC class II expression via IRF4 and CIITA. J Immunol. 2013;191(2):699–707. doi: 10.4049/jimmunol.1201443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 54.Zou J, et al. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20(1):97–108. doi: 10.1016/j.devcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468(7324):701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468(7324):659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7(5):593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan H, O’Brien TF, Zhang P, Zhong XP. The role of tuberous sclerosis complex 1 in regulating innate immunity. J Immunol. 2012;188(8):3658–3666. doi: 10.4049/jimmunol.1102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang G, Wang Y, Shi LZ, Kanneganti TD, Chi H. Signaling by the phosphatase MKP-1 in dendritic cells imprints distinct effector and regulatory T cell fates. Immunity. 2011;35(1):45–58. doi: 10.1016/j.immuni.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, et al. Transforming growth factor beta-activated kinase 1 (TAK1)-dependent checkpoint in the survival of dendritic cells promotes immune homeostasis and function. Proc Natl Acad Sci USA. 2012;109(6):E343–E352. doi: 10.1073/pnas.1115635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang G, et al. Signaling via the kinase p38α programs dendritic cells to drive TH17 differentiation and autoimmune inflammation. Nat Immunol. 2012;13(2):152–161. doi: 10.1038/ni.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.