Significance
Rubella virus (RV) is a human pathogen that causes serious birth defects when contracted during pregnancy. However, due to its variable shape and size, little is known about the RV structure. The RV capsid protein is an essential component of the virus and a key factor for successful replication of the virus in host cells. Here we describe the atomic structure of the RV capsid protein. This structure, along with electron microscopic data on the virus, has provided a three-dimensional picture of the virion. The capsid protein structure has also helped to identify amino acid residues that are required for virus assembly. This information can be used for the development of antiviral therapies that target the viral capsid protein.
Keywords: X-ray crystallography, cryoelectron tomography, virology
Abstract
Rubella virus (RV) is a leading cause of birth defects due to infectious agents. When contracted during pregnancy, RV infection leads to severe damage in fetuses. Despite its medical importance, compared with the related alphaviruses, very little is known about the structure of RV. The RV capsid protein is an essential structural component of virions as well as a key factor in virus–host interactions. Here we describe three crystal structures of the structural domain of the RV capsid protein. The polypeptide fold of the RV capsid protomer has not been observed previously. Combining the atomic structure of the RV capsid protein with the cryoelectron tomograms of RV particles established a low-resolution structure of the virion. Mutational studies based on this structure confirmed the role of amino acid residues in the capsid that function in the assembly of infectious virions.
Rubella virus (RV) is the only member of the Rubivirus genus which, together with alphaviruses including Chikungunya virus, Sindbis virus, and Ross River virus, make up the family Togaviridae. RV is a human pathogen that causes “German measles,” a relatively mild disease characterized by rashes and low-grade fever. However, due to its teratogenic properties, RV is a major threat to the fetus when infection occurs during the first trimester of pregnancy (1). Vaccination has been very successful in controlling RV infection; however, the virus is still endemic in many areas of the world (1, 2). RV is an enveloped virus with a 9.6-kb single-stranded, positive-sense RNA genome (1, 3). The virions have particle diameters ranging from 600 to 800 Å, with most of the spherical virions having a diameter of about 700 Å (4). RV contains three structural proteins, namely, the capsid protein (∼31 kDa) and the glycoproteins E1 (58 kDa) and E2 (42–47 kDa). The capsid protein interacts with the RNA genome and forms the nucleocapsid, which is surrounded by a lipid membrane upon which E1 and E2 are arranged. Two nonstructural proteins, p90 and p150, involved with virus replication, are also encoded by the virus (1).
Alphaviruses and RV share a similar gene order and expression strategy (3) but differ from each other in that alphaviruses are icosahedral, their nucleocapsids assemble in the cytoplasm, and virions bud from the plasma membrane (5). In contrast, RV virions are pleomorphic and the nucleocapsid assembles on Golgi membranes followed by budding of the virus into this organelle (6). The pleomorphic nature of RV virions has been a limiting factor in determining the structure of the virus particles.
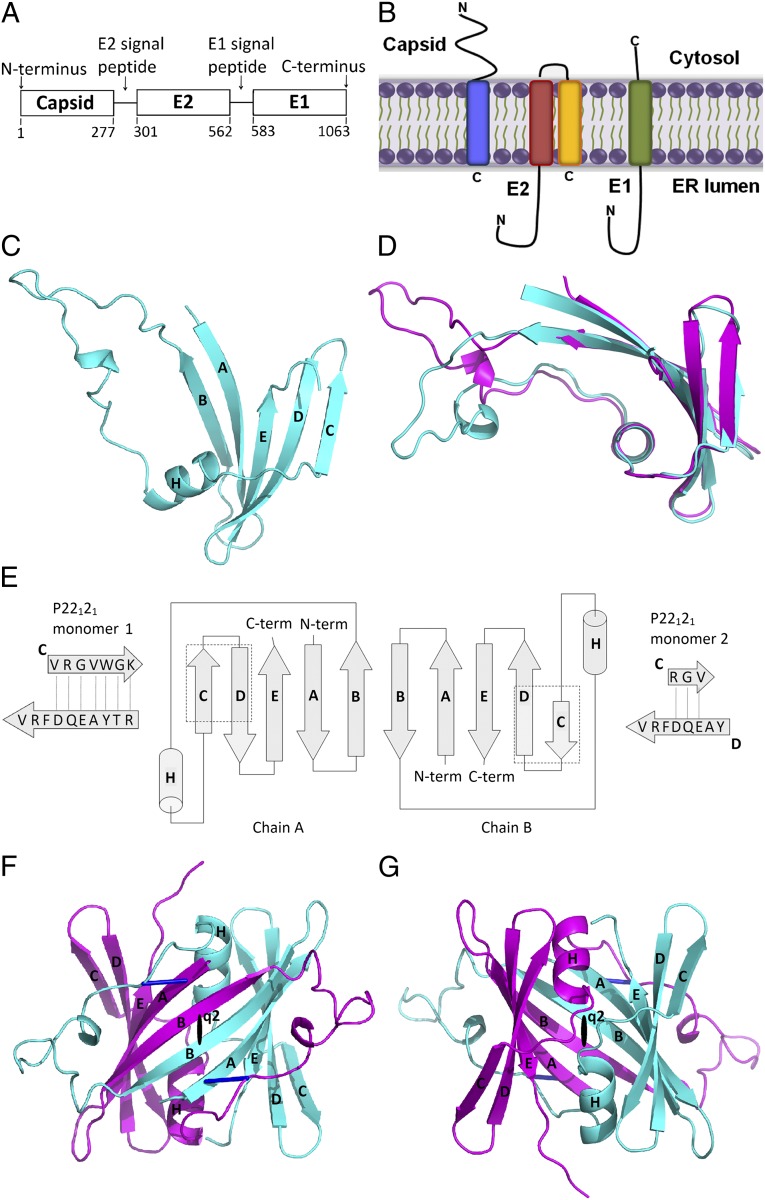
As with all Togaviruses, the structural proteins of RV are synthesized as a polyprotein precursor in association with the endoplasmic reticulum in the host cell (7) (Fig. 1A). The polyprotein is cotranslationally cleaved by the host-cell signal peptidase but, unlike alphavirus capsid proteins, the RV capsid protein remains attached to the cytoplasmic side of the membrane by virtue of the hydrophobic E2 signal peptide (8) (Fig. 1B). Similar to capsid proteins of alphaviruses (9) and small icosahedral RNA plant viruses (10), about 100 amino-terminal residues of RV capsid protein are enriched in basic amino acids. These basic residues are generally associated with the RNA viral genome and are disordered, implying that there is no unique structure for the RNA protein complex. In alphaviruses, the C-terminal part of the capsid proteins forms the structural framework of the nucleocapsids (9, 11). The RV capsid protein not only packages the RNA genome but also is involved in viral transcription and replication. It regulates viral RNA replication by interacting with the virus-encoded nonstructural proteins (12). Interaction with host proteins is required for its other functions. For example, it inhibits apoptosis through forming complexes with Bax (13) and potentially other host proteins such as p32 and prostate apoptosis response protein (14). The RV capsid protein also blocks the import of precursor proteins into mitochondria through an as-yet-unknown mechanism (15). Finally, interaction with the translation initiation factor, poly(A)-binding protein, may serve to block protein synthesis at discrete intracellular loci or prevent further translation of viral genomes (16).
Fig. 1.
Rubella virus capsid protein structure. (A) RV structural proteins as in the initial polyprotein precursor. (B) Position of the structural proteins across the ER membrane, after posttranslational cleavage, along with the E1 (yellow) and E2 (blue) signal peptides and the E1 (green) and E2 (red) transmembrane domains. (C) Ribbon diagram of the RV capsid monomer. The β-strands are marked A–E, starting from the N terminus. The α-helix is marked as H. (D) Superimposed ribbon diagrams of the monomer as seen in the C2 (cyan) and P22121 (magenta) crystals. (E) Diagram of the RV dimer structure as seen in the P22121 crystal. Boldface letters are names assigned to the secondary structural elements; the letters in the magnified C and D β-strands refer to amino acid abbreviations. (F and G) Ribbon diagram of the RV capsid protein dimer. The two figures are 180° related to each other. The black oval shape (q2) indicates a quasi-twofold axis. The disulfide bonds are shown in blue. The monomer chains are represented in cyan and magenta. C, D, F, and G were prepared using PyMOL (37).
Results and Discussion
Crystal Structure of the RV Capsid Protein.
The RV capsid protein, excluding the E2 signal peptide, is a 277-amino-acid residue protein that forms disulfide-linked dimers (6, 17). The C-terminal part of the protein (amino acids 127–277) was expressed and purified from bacteria (Materials and Methods). The purified RV capsid protein fragment forms a dimer (Fig. S1). The structure of this truncated RV capsid protein was determined in three different crystal forms with space groups C2, P22121, and C2221 that had 3, 1, and 2.5 dimers per asymmetric unit, respectively (Table S1).
Each capsid monomer consists of five antiparallel β-strands—A, B, C, D, and E—as well as a two-turn α-helix H between strands B and C (Fig. 1C). The strands and helix are connected by loops AB, BH, HC, CD, and DE. The BH loop is by far the largest, consisting of residues 176–197. Although the core structure (missing the BH loop) of each of the 13 independent RV capsid monomers from the three different crystal forms are essentially the same (Table S2), the BH loop structure is different in each of the crystal forms (Fig. 1D) and is disordered in the C2221 crystal form. The P22121 crystal structure has a one residue translation in β-strand C, at the edge of the β-sheet, with respect to strand D in one of the monomers (Fig. 1E). This translation requires a 180° flip of the peptide bonds in the C strand. A variety of tests were conducted to confirm these unexpected amino acid assignments. The presence of Trp, Arg, and Lys residues in the C β-strand helped to confirm the correctness of the structure. The impact of this change was not as large as might have been expected because the two basic residues are oriented in the plane of the sheet rather than being perpendicular to the sheet.
All three crystal forms show that the RV capsid protein forms a twofold symmetric dimer, consistent with the previous observation that the RV capsid protein forms dimers when isolated from the virion (17). The capsid monomers are linked through disulfide bonds between Cys-153 and Cys-197. Within a dimer the β-sheet is continuous with the residues 169–175 of β-strand B from one monomer being hydrogen bonded with the same but antiparallel β-strand B in the other monomer (Fig. 1 E–G). The resultant 10-stranded, left-handed twisted sheet forms a partially open β-barrel with the two helices lying in the center of the barrel. The β-strands A and B from one monomer are inserted into the BH loop of the other monomer, forming a tightly bound dimeric structure (Fig. 1F). A DALI search (18) did not find any significant similarity between the RV capsid structure with other known protein structures.
Oligomerization of Capsid Protein Dimers.
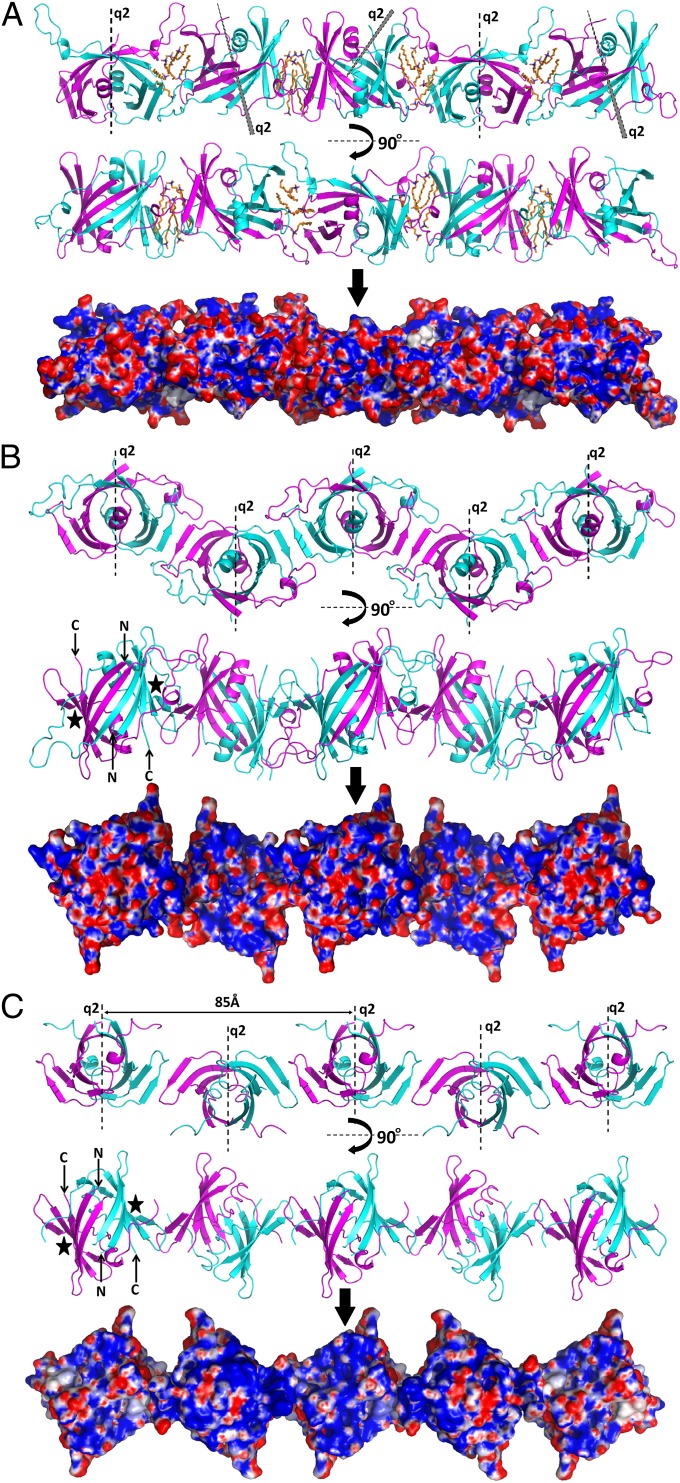
All three crystal forms of the RV capsid protein contain rows of closely packed dimers in which neighboring dimers are held together by hydrogen bonds and hydrophobic contacts (Fig. 2). The interactions between the rows are formed mostly by hydrophilic contacts and are fewer than between the dimers within a row. In the C2 crystal form, adjacent dimers in a row are related by an approximately 120° rotation and make hydrophobic contacts between Trp-221 in one dimer with Trp-189 and Pro-191 in the neighboring dimer’s BH loop. The rows in the C2221 and in the P22121 crystal forms are essentially the same with adjacent dimers in a row related by approximately twofold symmetry perpendicular to the direction of the row. In the C2221 crystal form, the dimers interact through hydrogen bonds between the terminal β-strands C (amino acids 218–222) in neighboring molecules. In the P22121 crystal form, the dimers are in a staggered row-like arrangement similar to that in the C2221 crystal form and also involve hydrogen bond interactions between Val-220 of one monomer with Gly-219 of the neighboring monomer. The similar rows of dimers in the P22121 and C2221 crystal forms have a surface that is rich in positively charged amino acid residues (Fig. 2 B and C).
Fig. 2.
Rows of RV capsid protein dimer. Rows in the crystal forms (A) C2, (B) P22121, and (C) C2221 with their dimer axes parallel and perpendicular to the plane of the paper, respectively. For each crystal form, the molecular surface that presumably faces the viral membrane is shown, after removal of the variable BH loop in B, colored according to its electrostatic potential. The potentials (38) at 300 K are in the range +125 mV (+5 kT/e) (blue) to −125 mV (−5 kT/e) (red) where k = Boltzmann’s constant, T = temperature, e = electronic charge, and mV = millivolts. The detergent molecules in the C2 space group are shown in orange. A black star indicates the site of the detergent-binding region that possibly also binds the cytosolic part of E2. The dashed lines correspond to quasi-twofold axes (q2). Figure panels were prepared using PyMOL (37).
In many enveloped viruses, the lipid membrane faces a positively charged protein surface. Of particular relevance to RV is the nucleocapsid of Sindbis virus (9) and Ross River virus (11), alphaviruses whose nucleocapsid surface is positively charged and surrounded by lipid membranes. A second example is the double-stranded DNA containing Paramecium bursaria Chlorella virus-1 whose capsid protein hugs the exterior of the internal lipid membrane (19). A third example is the matrix protein of the pleomorphic Newcastle disease virus, which is closely associated with the interior of the viral lipid envelope (20). Thus, it seems likely that the RV capsid protein also assembles with a predominantly positive surface close to and facing the viral membrane.
If we assume that the positive surface of the capsid protein rows in P22121 and C2221 crystal forms is positioned parallel to the RV membrane, then the capsid protein dimer axes would be roughly perpendicular to the viral membrane. Furthermore, alternate dimer axes are in opposite directions, implying that every second dimer makes close contacts with the membrane. These contacts are separated by ∼85Å (Fig. 2C).
Orientation of the Capsid Protein in the Virion.
In infected cells, the C terminus of the RV capsid protein remains attached to the E2 signal peptide, which anchors this viral protein to intracellular membranes (8). This implies that the C terminus of the RV capsid structure should be oriented toward the membrane surface in the virions. Hence, the optimum way to orient the dimer structure would be for the dimer axes to be perpendicular to the membrane, thus placing the C termini of the monomers at approximately equal distances from the membrane (Figs. 2C and 3C). The distance to the end of the E2 signal peptide in the membrane can then be easily spanned by the disordered 27 amino acids at the C termini (Fig. S2) of the capsid protein dimer. As the N-terminal region of the RV capsid protein interacts with the viral RNA (1), placing the dimer axis perpendicular to the membrane would also position the exposed amino terminus of both monomers within the dimer at a similar distance from the interior of the virus (Figs. 2C and 3C). This observation is consistent with the dimer orientations in the rows seen in the orthorhombic crystal forms.
Fig. 3.
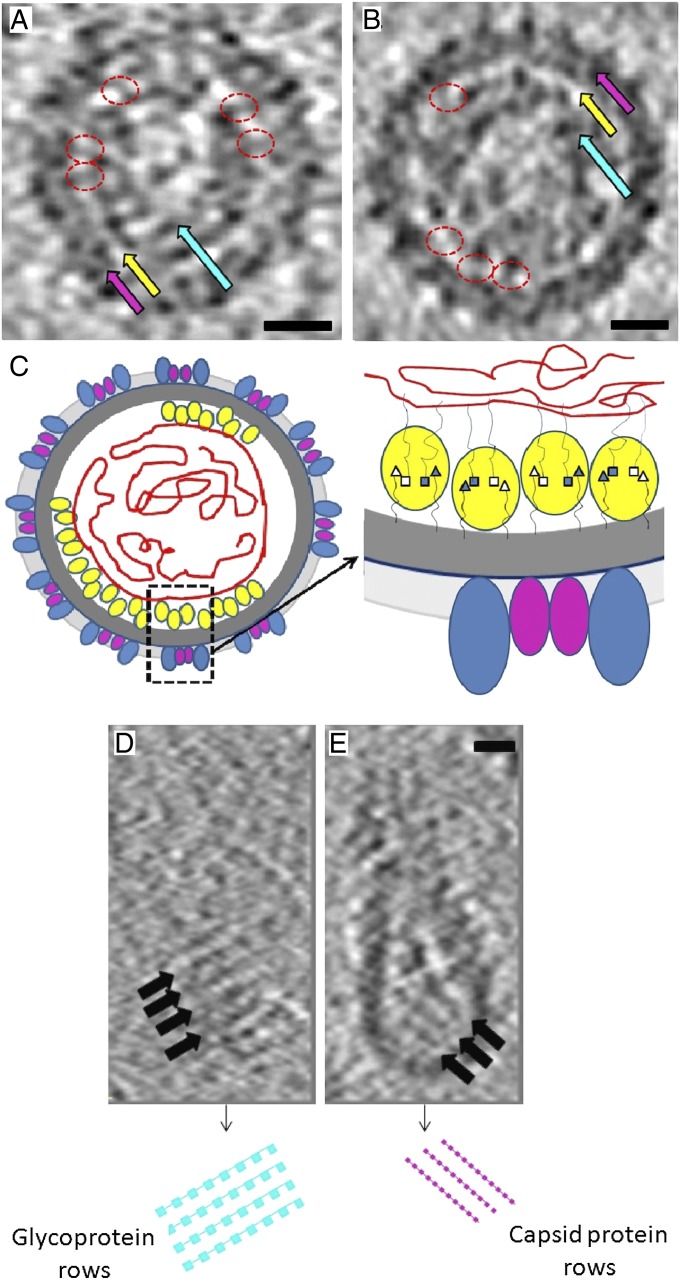
RV tomograms. (A and B) Central section of two RV particle tomograms. Magenta arrows: glycoprotein plus membrane layer (average thickness = 100 Å). Yellow arrows: gap between membrane and internal RNA core. The C-terminal domain of the capsid protein is seen as regions of density crossing the gap (circled in red). Cyan arrows: region occupied by RNA plus N-terminal domain of the capsid protein. (C) Diagram of a RV virion cross-section showing the ectodomains of E1 (blue) and E2 (magenta) glycoproteins, the capsid protein dimer (yellow), and genomic RNA (red). The gray region denotes the membrane layer (dark gray) plus the glycoproteins. In the magnified section, the polypeptide connections (thin lines) of the N termini (□) and C termini (Δ) of the capsid protein to the RNA and membrane, respectively, are shown. The filled and open shapes denote the termini pointing toward and away from the plane of the paper. (D) A section near the surface of an RV virion at a distance of 345 Å from the particle center. (E) A section at a distance of 210 Å from the particle center and below the external surface of the virion shown in D. The separation of pixels in the tomogram is 15 Å. (Scale bar, 170 Å.) Dark regions indicate high density.
Identification of Capsid Protein Rows in Cryoelectron Tomograms of RV.
Cryoelectron microscopic (9, 11) and tomographic (4) reconstructions of alphaviruses show that there is a 40-Å-wide gap at a radius of ∼190Å between the lipid membrane and the nucleocapsid RNA. The alphavirus capsid protein pentamers and hexamers are situated in this gap and produce regions of higher density crossing the gap at intervals of around 80 Å. A similar gap with thickness ranging from 40 to 70 Å, crossed by regions of higher density separated by about 90 Å, exists in RV tomograms (4) (Fig. 3 A and B), except that the mean radius of the gap is at ∼240 Å instead of 190 Å. The similarity of these features in alphaviruses and RV suggests that the positive surface of the capsid protein rows of dimers is close to the lipid membrane and that the alternate capsid protein dimers span the gap in tomograms. This observation overrides our previous conclusions based on volume calculations to determine the site of the capsid protein in the virion (4).
Some RV tomograms show short, parallel ridges separated by ∼90 Å below the viral membrane. These ridges likely correspond to the rows of capsid protein dimers that occur in the crystal structures (Fig. 3 D and E). Although the rows of dimers found in the crystals are linear, it is possible that, in virions, the rows are slightly bent as would be required for the formation of spherical or elongated RV particles. Tomograms of RV particles show ridges, separated by about 90 Å, on the viral surface formed by the glycoproteins (4). These ridges are approximately orthogonal to the rows of capsid protein dimers (Fig. 3 D and E). Thus, the arrangement of the glycoproteins E2 and E1 in RV virions is correlated with the organization of the capsid proteins (Fig. 3C).
Oligomerization of Capsid Protein Dimers Is Essential for Virus Assembly and Infection.
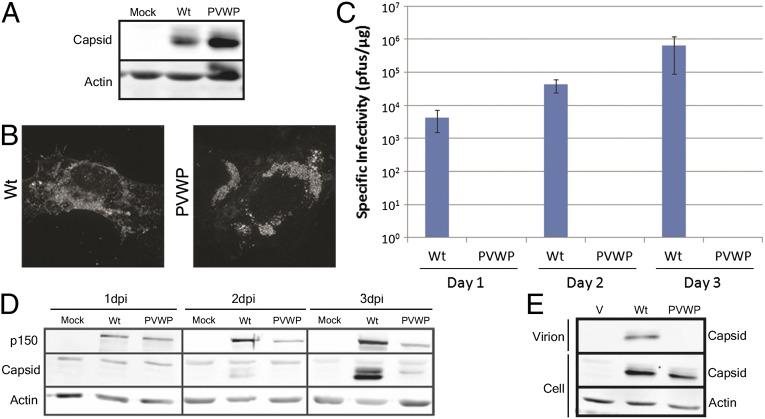
If the organization of capsid protein into rows of dimers as in the two orthorhombic crystal forms is important for the assembly of the RV virion (Fig. 2C), disrupting the hydrogen bonding between adjacent dimers should have a detrimental effect on virus assembly and/or infectivity. The interdimer molecular interface involves amino acid residues 219–222 (GVWG) (Fig. S3). To determine whether the interdimer interface of the capsid is important for infectivity, the RV capsid gene was modified to encode two proline substitutions (PVWP) that should disrupt the hydrogen-bonding interactions. Plasmid-based expression of the PVWP capsid protein in transfected cells verified that this mutant was identical in apparent molecular weight to wild-type (WT) capsid protein (Fig. 4A). The mutant protein also exhibited an identical subcellular localization to the WT capsid protein (Fig. 4B).
Fig. 4.
Interaction between neighboring capsid dimers is essential for infectious virus production. (A) Lysates of Vero cells transfected with plasmids encoding wild-type (Wt) or the PVWP mutant capsid protein were subjected to immunoblotting with anti-RV serum. Actin was used as a loading control. (B) Indirect immunofluorescence was used to detect localization of WT and the mutant capsid protein in transfected cells. (C) Vero cells were transfected with in-vitro–synthesized infectious RNA encoding WT or PVWP mutant RV genome. Viral titers were determined 1–3 d posttransfection, and the specific infectivities (plaque-forming units/microgram of DNA) of the infectious clones were calculated. (D) Immunoblot analyses of cells transfected with viral RNAs showing expression of capsid and the viral replicase protein p150. Actin was used as a loading control. (E) Vero cells were transfected with plasmids encoding WT or PVWP capsid and glycoproteins E2 and E1 or vector alone (V). Forty-eight hours posttransfection, virus-like particles were recovered from media by ultracentrifugation. Cell lysates and virus particle (virion) fractions were subjected to immunoblot analyses.
Cells were transfected with in-vitro–synthesized WT and PVWP genomic RNAs. The resulting viral titers were determined at 1–3 d posttransfection. Whereas the specific infectivity of WT RV genomic RNA approached 106 plaque-forming units per microgram, cells transfected with PVWP genomic RNA did not produce detectable amounts of infectious virus (Fig. 4C). The production of p150, a viral replicase protein that is translated directly from the viral genomic RNA, was assayed to monitor the efficacy of RNA transfection. Expression of p150 was similar for the WT and mutant after 1 d. The p150 levels remained relatively constant over time in cells transfected with the mutant RV genome. In contrast, in cells transfected with WT RV RNA, levels of p150 steadily increased along with capsid levels in a time-dependent manner, a scenario that is consistent with assembly of infectious virus and the infection of neighboring cells (Fig. 4D). Compared with capsid in the WT RV RNA-transfected cells, levels of PVWP capsid were low because no infectious virus was being made, and therefore mutant capsid was made only in the cells that were originally transfected with the “infectious” RNA (Fig. 4D). To determine if the PVWP capsid protein could support assembly and secretion of virus particles, Vero cells were transfected with plasmids encoding the WT or PVWP mutant capsid protein plus glycoproteins E2 and E1. We have previously shown that coexpression of the RV structural proteins results in the assembly of virus-like particles in the Golgi, followed by secretion into the medium (21). The virus-like particles, which are antigenically and immunologically indistinguishable from infectious RV virions (22), can easily be recovered from the cell media by ultracentrifugation (23, 24). Using this assay, it was observed that virus-like particles were secreted only from cells expressing WT capsid, E2, and E1 (Fig. 4E). Together, these data suggest that the interface between capsid dimers observed in the orthorhombic crystal forms is critical for assembly of capsid protein into infectious virions.
The site of the mutations is not in the dimer-to-dimer interface of the capsid protein rows in the monoclinic form. Thus, the impact shown in the mutational analyses further demonstrates that the rows of dimers in the orthorhombic, but not the monoclinic, crystal form are relevant to the virus.
Probable Location of the Glycoprotein-Binding Region on the Capsid Protein.
In alphaviruses, the glycoproteins have a cytoplasmic domain that is anchored in the nucleocapsid (9). Similarly, in RV the short predicted transmembrane and/or cytoplasmic domains of the RV E2 glycoprotein may serve as an anchor on the capsid protein (23). The cytoplasmic domain of E2 has been shown to be essential for budding of RV into the Golgi apparatus (23). In the C2 crystal structure, three detergent molecules derived from the crystallization solution are associated with each capsid monomer. It is possible that this binding site in the RV capsid functions similarly to the pocket in alphavirus capsid proteins that interact with the C-terminal region of E2 (25). The binding site for the detergent molecules in the RV capsid protein consists of hydrophobic residues and a few conserved acidic residues. Appropriately, the C-terminal cytoplasmic region of the RV E2 protein contains mostly positively, highly conserved, charged residues (RRACRRR) that complement this site. However, this site is occupied only in the C2 crystal form, as this is the only form in which there is a suitable ligand in the crystallization solution that might bind into the pocket. In the P22121 and C2221 crystal forms, the binding site would be on the side of the observed capsid protein rows. Therefore, this site would not be facing the membrane in the virus (Fig. 2), but would be accessible to the cytoplasmic tail of E2.
Evolution of the Viral Structural Proteins.
Although the core structure of the RV capsid protein is the same in all of the crystal forms reported here, there is variability in the relative positions of the secondary structure elements in the different crystal structures (Table S2). This variability indicates flexibility in the capsid protein structure that may be required for its numerous cellular functions (26) by facilitating its interaction with a large cohort of host-cell proteins such as Bax, mitochondrial matrix protein p32, prostate apoptosis response protein, and poly(A)-binding protein. However, the RV capsid structure has not yet yielded any detailed insight into the mechanisms of its cellular functions, but its function as a viral capsid protein provides fascinating insight into the evolution of virus structure.
The E1 glycoprotein of alphaviruses and RV (27, 28) and the E glycoprotein of flaviviruses (29) have similar structures. This common surface glycoprotein motif suggests that these structures evolved from an ancestral structure (28) that can attach to potential host cells and initiate infection by fusion with the host cell. In contrast, the capsid protein structures of alpha-, flavi-, and rubella viruses are completely different. The alphavirus capsid protein is a β-sheet structure with a chymotrypsin-like fold (30) that forms an icosahedral shell around the viral genome, whereas the flavivirus capsid protein is an α-helical structure (31) that is closely associated with the genome but does not form a protective shell. The RV capsid protein structure as reported here has a polypeptide fold that is different to both the alpha- and flavivirus capsid proteins. The common function of the capsid proteins in these viruses is to neutralize the charge on the genome, but the task of confining the genome is most obvious for alphaviruses, less clear for RV, and absent for flaviviruses. In addition, the capsid proteins of these viruses have gained other cellular functions that facilitate virus replication and assembly inside host cells. These observations show that the ability to attach and fuse with potential host cells has been conserved in the glycoproteins of these viruses whereas the capsid proteins have evolved independently, resulting in acquisition of different nonstructural functions.
Materials and Methods
Expression and Purification of RV Capsid Protein.
The nucleotide sequence encoding the capsid protein of RV strain M33 was modified by removing the bases corresponding to the basic amino terminal residues 1–126 and inserted into a pGEX plasmid with an N-terminal GST tag. This clone was grown in Escherichia coli BL21(DE3) cells. The fusion protein was isolated from the cell lysate using a GST affinity column. The GST tag was cleaved using thrombin. RV capsid protein was further purified using gel filtration chromatography. The protein was concentrated to 5 mg/mL for crystallization trials. Further details are given in SI Materials and Methods.
Crystallization, Data Collection, and Structure Determination.
Crystallization trials were set up using Hampton crystallization screen kits. Crystals were obtained as clusters of thin plates in 50 mM Tris (pH 7.5), 24% isopropanol, and 25% PEG-4000. The thin plates were broken from the cluster and used for data collection. The selenomethionine-derivatized protein was produced and purified similar to the native protein. It also formed crystal clusters similar to the native protein but, in some cases, formed longer cuboid-shaped crystals. The cuboid-shaped crystals were used for data collection.
After extensive screening, thin-plate crystals of RV capsid protein were also obtained in the presence of 50 mM Tris, 200 mM NaCl, 25% PEG-3350, and 1% lauryl-dimethylamine oxide. These crystals were soaked in 2 mM uranium nitrate hexahydrate overnight at room temperature.
All diffraction data were collected at the Advanced Proton Source, GM/CA, beamline 23 ID-D and ID-B, Argonne National Laboratory, Chicago. Initial phases were determined using single isomorphous replacement with anomalous scattering. The determination of the heavy atom parameters and subsequent phasing was performed using Phenix Autosol (32). Phases were improved by density modification using noncrystallographic symmetry with the same program. The atomic model was built using the program COOT (33). The structure was refined using phenix.refine (34). The structures of RV capsid protein in the P22121 and C2221 space groups were subsequently determined by molecular using the Phaser software (35). Further details are available in SI Materials and Methods.
Cryoelectron Tomography of RV Virions.
Rubella virus purification and collection of images for electron tomography was performed as described by Battisti et al. (4). The 3D reconstructions and analyses of the tomograms were performed using the IMOD software package (36).
Mutational Studies of the Capsid Protein.
Plasmids encoding the mutant PVWP capsid protein in the context of the structural protein cassette (pCMV5-24S PVWP) or the full-length genomic clone (pBRM33-PVWP) were constructed by replacing the ∼400-bp BsrGI-BstEII fragment of the capsid cDNA with a synthetic oligonucleotide. Vero cells were transfected with plasmids or in-vitro–synthesized infectious RNA. Capsid level expression was assayed by immunoblotting and indirect immunofluorescence. Viral titers were determined via plaque assay. Secretion of RV-like particles from transfected Vero cells was assayed as described (24) except that the WT and PVWP mutant capsid proteins were expressed from a single plasmid encoding all three RV structural proteins (C, E2, and E1). Further explanation is in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sheryl Kelly for help in the preparation of the manuscript and Dr. Teryl K. Frey for supplying the initial rubella virus stock. We also thank Dr. Thomas Klose for help in preparation of the figures. This work was supported by National Institutes of Health Grant AI-095366. Technical developments were supported by National Science Foundation Grant MCB-1014547 (to M.G.R.). T.C.H. holds a Canada Research Chair, and research funding was provided by the Canadian Institutes of Health Research (CIHR). S.D.W. holds doctoral scholarship awards from CIHR and Alberta Innovates Health Solutions. Use of the Advanced Photon Source, an Office of Science User Facility operated for the Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the DOE under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported here have been deposited in the Protein Data Bank, www.pdb.org (PDB codes 4HAR, 4HBE, and 4HBO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316681110/-/DCSupplemental.
References
- 1.Hobman TC, Chantler J. Rubella virus. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol I. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1069–1100. [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Progress toward control of rubella and prevention of congenital rubella syndrome: Worldwide, 2009. MMWR Morb Mortal Wkly Rep. 2010;59(40):1307–1310. [PubMed] [Google Scholar]
- 3.Dominguez G, Wang CY, Frey TK. Sequence of the genome RNA of rubella virus: Evidence for genetic rearrangement during togavirus evolution. Virology. 1990;177(1):225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battisti AJ, et al. Cryo-electron tomography of rubella virus. J Virol. 2012;86(20):11078–11085. doi: 10.1128/JVI.01390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strauss JH, Strauss EG. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey TK. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oker-Blom C, Ulmanen I, Kääriäinen L, Pettersson RF. Rubella virus 40S genome RNA specifies a 24S subgenomic mRNA that codes for a precursor to structural proteins. J Virol. 1984;49(2):403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suomalainen M, Garoff H, Baron MD. The E2 signal sequence of rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64(11):5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay S, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14(1):63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 11.Cheng RH, et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80(4):621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MH, Icenogle JP. Rubella virus capsid protein modulates viral genome replication and virus infectivity. J Virol. 2004;78(8):4314–4322. doi: 10.1128/JVI.78.8.4314-4322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilkow CS, Goping IS, Hobman TC. The Rubella virus capsid is an anti-apoptotic protein that attenuates the pore-forming ability of Bax. PLoS Pathog. 2011;7(2):e1001291. doi: 10.1371/journal.ppat.1001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beatch MD, Hobman TC. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J Virol. 2000;74(12):5569–5576. doi: 10.1128/jvi.74.12.5569-5576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilkow CS, et al. The rubella virus capsid protein inhibits mitochondrial import. J Virol. 2010;84(1):119–130. doi: 10.1128/JVI.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilkow CS, Mancinelli V, Beatch MD, Hobman TC. Rubella virus capsid protein interacts with poly(a)-binding protein and inhibits translation. J Virol. 2008;82(9):4284–4294. doi: 10.1128/JVI.02732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron MD, Forsell K. Oligomerization of the structural proteins of rubella virus. Virology. 1991;185(2):811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99(23):14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battisti AJ, et al. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci USA. 2012;109(35):13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobman TC, et al. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202(2):574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 22.Qiu Z, Ou D, Hobman TC, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68(6):4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garbutt M, Law LM, Chan H, Hobman TC. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J Virol. 1999;73(5):3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law LM, Duncan R, Esmaili A, Nakhasi HL, Hobman TC. Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J Virol. 2001;75(4):1978–1983. doi: 10.1128/JVI.75.4.1978-1983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, et al. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4(5):531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 26.Ilkow CS, Willows SD, Hobman TC. Rubella virus capsid protein: A small protein with big functions. Future Microbiol. 2010;5(4):571–584. doi: 10.2217/fmb.10.27. [DOI] [PubMed] [Google Scholar]
- 27.Lescar J, et al. The fusion glycoprotein shell of Semliki Forest virus: An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 28.DuBois RM, et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493(7433):552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- 29.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 30.Choi HK, et al. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354(6348):37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 31.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA. 2004;101(10):3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12, No 1):2126–2132. [DOI] [PubMed]
- 34.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 37. Schrödinger, LLC (2010) The PyMOL Molecular Graphics System (Schrödinger, LLC, San Diego), Version 1.5.
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.