Significance
This study demonstrated that cross-neutralizing anti-poliovirus antibodies bind the site on poliovirus capsid surface that significantly overlaps the binding site of the cellular receptor. A second antibody with similar specificity was isolated by sequential phage display panning, suggesting that cross-reactive anti-poliovirus antibodies may be more prevalent in primates than previously recognized. Binding to the receptor recognition site explains unusually broad specificity of the antibodies. The antibodies bind type 1 and type 2 polioviruses at a slightly different angle, indicating that molecular details of virus–antibody interaction are different and suggesting that further screening or engineering may produce an antibody neutralizing all three serotypes of poliovirus. These results may be used for developing new antiviral strategies for the polio eradication campaign.
Keywords: phage display, passive immunization, antiviral therapy, broadly reactive antibodies
Abstract
Most structural information about poliovirus interaction with neutralizing antibodies was obtained in the 1980s in studies of mouse monoclonal antibodies. Recently we have isolated a number of human/chimpanzee anti-poliovirus antibodies and demonstrated that one of them, MAb A12, could neutralize polioviruses of both serotypes 1 and 2. This communication presents data on isolation of an additional cross-neutralizing antibody (F12) and identification of a previously unknown epitope on the surface of poliovirus virions. Epitope mapping was performed by sequencing of antibody-resistant mutants and by cryo-EM of complexes of virions with Fab fragments. The results have demonstrated that both cross-neutralizing antibodies bind the site located at the bottom of the canyon surrounding the fivefold axis of symmetry that was previously shown to interact with cellular poliovirus receptor CD155. However, the same antibody binds to serotypes 1 and 2 through different specific interactions. It was also shown to interact with type 3 poliovirus, albeit with about 10-fold lower affinity, insufficient for effective neutralization. Antibody interaction with the binding site of the cellular receptor may explain its broad reactivity and suggest that further screening or antibody engineering could lead to a universal antibody capable of neutralizing all three serotypes of poliovirus.
The worldwide campaign to eradicate poliomyelitis seeks to stop transmission of wild strains and to eliminate vaccine-derived polioviruses (VDPVs). The latter continuously emerge from oral polio vaccine (OPV) and threaten sustainability of polio eradication, which will not be complete until OPV use is stopped (1). Besides circulating VDPVs that cause outbreaks of paralytic disease (2), there are also immunodeficiency-associated VDPVs continuously excreted by chronically infected patients with certain types of immune disorders (3), and VDPVs of unknown origin isolated from environmental samples, possibly also excreted by chronic shedders. Treatment of immunodeficient carriers is a critically important challenge that requires development of new therapeutic tools, which could also be used for posteradication risk management and rapid response to potential reemergence of poliovirus.
New treatments could include antiviral drugs and biologicals, for instance antibodies. Passive immunotherapy against polio was shown to be highly effective in the early 1950s (4) but was not used after introduction of vaccines. Recently we isolated neutralizing chimpanzee–human monoclonal antibodies against poliovirus and proposed that they may be used for this purpose (5). Among antibodies isolated by phage-display technology, the antibody designated A12 neutralized two serotypes of poliovirus (types 1 and 2) and could also weakly bind type 3 poliovirus. Here we describe isolation of another cross-neutralizing antibody and demonstrate that they both bind to an epitope that is located in the region on the poliovirus surface that interacts with cellular poliovirus receptor CD155.
Results
Cryo-EM Localization of A12 Epitope.
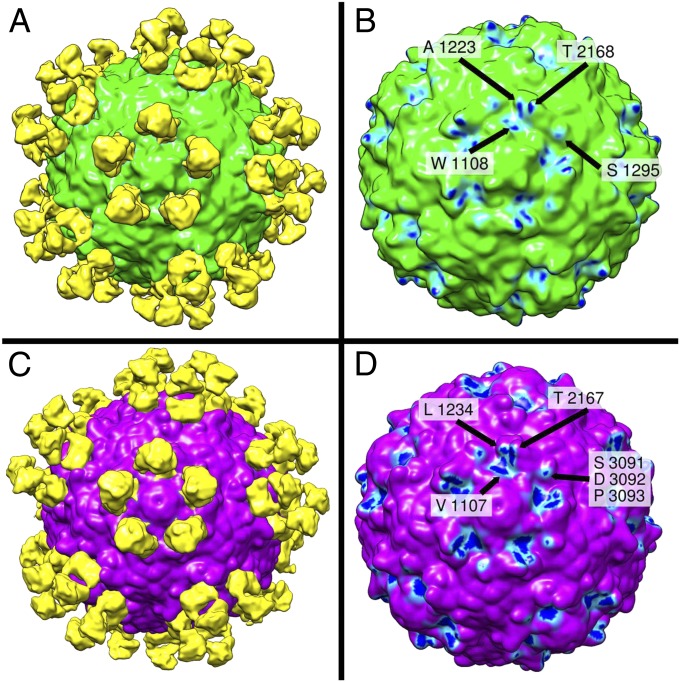
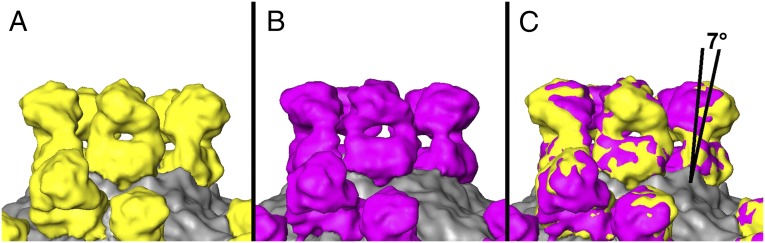
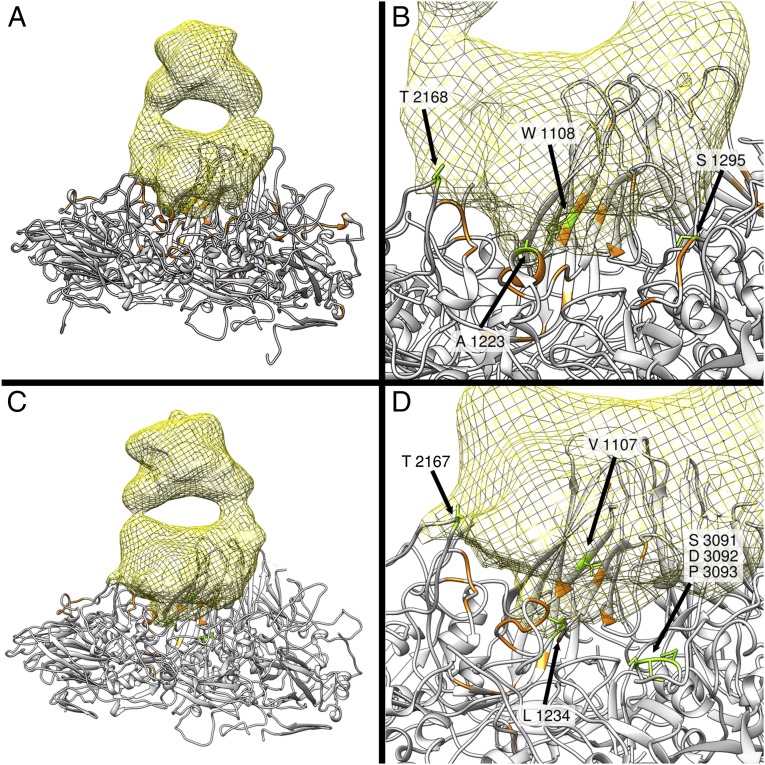
Analysis of neutralization-resistant (escape) mutants generated by treatment of type 1 and type 2 poliovirus with A12 antibody presented in our previous publication (5) indicated that mutations rendering the virus resistant to neutralization are located on the opposite sides of the circular depression surrounding the fivefold axes of symmetry of the virion called the canyon. Cellular receptor (CD155) interacts with amino acid residues at the bottom of the canyon (6–8), and the region has been suggested not to be accessible by antibodies. To confirm epitope localization revealed by escape mutant analysis, we determined the structure of complexes of types 1 and 2 poliovirus with A12 Fab using cryo-EM single-particle reconstruction. These structures showed that A12 Fabs formed circular arrays of five molecules around the fivefold axis of symmetry in the canyon region (Fig. 1 A and C). They were indeed binding to both sides of the canyon and were located directly over its bottom and overlapped with the binding site for the poliovirus receptor CD155. Some mutations that rendered poliovirus resistant to these antibodies or were a part of their footprint on the virion surface were previously found to participate in the interaction between CD155 and poliovirus (9). Fig. 1 B and D show the footprints of Fab on the surface of type 1 and type 2 poliovirus. Cryo-EM analysis of the complexes revealed that the set of five Fab fragments bound to type 2 poliovirus was more compact compared with those bound to type 1 poliovirus, because the angle at which the fragments attach to the virion surface of these two viruses differed by 7° (Fig. 2). This suggests that whereas the general location of the epitope was very similar the exact molecular footprint of the binding was different. Comparison of our cryo-EM reconstructions with 3D structures determined by X-ray analysis (10) revealed that there were some differences between serotypes in the exact amino acid residues interacting with the antibody, and that amino acids previously implicated in CD155 receptor binding (7) were also in close proximity with the A12 Fab fragment (Fig. 3).
Fig. 1.
Surface rendering of the reconstruction of the A12 Fab-decorated capsid of serotype 1 (A) and serotype 2 (C) of poliovirus based on cryo-EM images. Serotype 1 and 2 of poliovirus are colored in green and magenta, respectively. The Fab-related densities around a fivefold axis are shaded yellow. Antibody footprints (B and D) consist of virion regions representing high-density antibody-bound difference map (close interaction with Fab) shown in dark blue, regions representing intermediate density (weaker interaction) shown in light blue, and the low-density regions (no interaction) are shown in green and magenta for types 1 and 2, respectively. They are W1108, A1223, S1295, and T2168 in serotype 1 (B) and V1107, L1234, and T2167 and in serotype 2 (D). In addition, there was another contact between Fab and the outer rim of the canyon of type 2 poliovirus that could not be unambiguously assigned at this resolution and may be formed by any of the three amino acids S3091, D3092, or P3093.
Fig. 2.
A12 binds to serotypes 1 and 2 with a different binding angle. Cryo-EM reconstructions of A12 Fab complexes with type 1 (yellow) (A) and type 2 (magenta) (B) polioviruses and superimposition of the two (C) showing that the binding angles differ by 7°.
Fig. 3.
Ribbon rendering of the reconstruction of complexes of A12 Fab with type 1 (A and B) and type 2 (C and D) poliovirus. Yellow mesh-surface objects in A and C represent Fab fragments binding to one poliovirus capsomer (shown by gray ribbons) consisting of one molecule of each VP1, VP2, VP3, and VP4. B and D show the close-up view of the contact regions. Amino acids marked green are those that interact with A12 Fab; those marked orange interact with poliovirus receptor CD155 (7).
Affinity Measurements.
ELISA and neutralization tests performed with previously described A12 Fab or A12 IgG showed that they strongly bound to types 1 and 2 polioviruses and neutralized them and also interacted with type 3 poliovirus but did not neutralize it. To quantitatively evaluate the binding affinity of A12 to each of the three types of polioviruses, surface plasmon resonance (SPR) studies on a Biacore machine were performed. The initial measurement of the affinity of Fab fragment showed that A12 had a low intrinsic affinity to type 1 poliovirus with a Kd of 2–3 × 10−7 M. In a later experiment, we measured the functional affinities or avidities with A12 IgG. It was found that the functional affinities were increased by 100-fold owing to the bivalent binding, with Kd values in a nanomolar range (Table 1). The A12 IgG bound to type 1 and type 2 viruses with essentially the same functional affinities, whereas more than 10-fold lower functional affinity was observed for interaction between A12 IgG and type 3 viruses. It may be possible that the 10-fold functional affinity difference is the reason why A12 is unable to neutralize the type 3 virus. Most neutralization tests in this study were performed at the antibody concentration of 10 µg/mL. To test whether there is a very low activity against type 3 poliovirus, we have performed a test at 100-fold higher concentration of A12 antibodies (1 mg/mL). No neutralizing activity could be detected, suggesting that activity against Sabin 3 virus was less than 1 unit/mg, or at least 105 lower than against types 1 and 2.
Table 1.
Functional affinities of anti-poliovirus A12 IgG
| Poliovirus serotype | koff, 1/s | kon, 1/Ms | Kd, M |
| 1 | 1.87 × 10−4 | 6.68 × 104 | 2.8 × 10−9 |
| 2 | 1.62 × 10−4 | 5.54 × 104 | 2.92 × 10−9 |
| 3 | 2.91 × 10−4 | 9.28 × 103 | 3.61 × 10−8 |
Binding kinetics of A12 IgG to poliovirus particle was assessed by SPR using a Biacore 1000. The kinetic rate constants of association (kon) and dissociation (koff) were measured from surface binding kinetics, and the equilibrium dissociation constant (Kd) was calculated as the ratio koff/kon.
Isolation of Additional Broadly Reactive Antibodies.
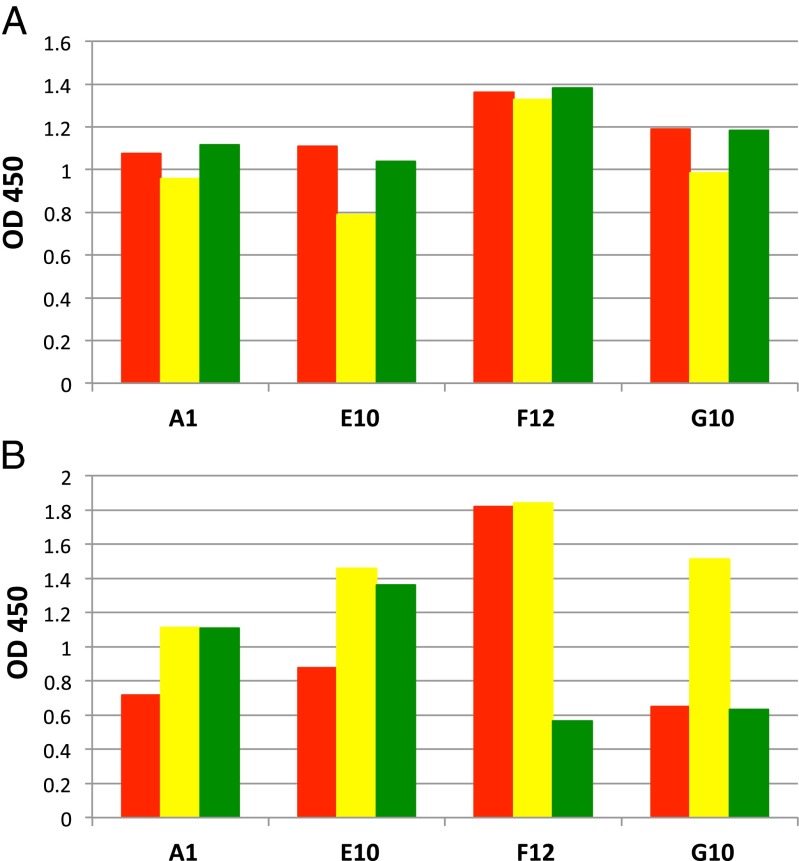
Weak but specific binding of the type 1–type 2 cross-neutralizing antibody to serotype 3 suggested that broadly reactive antibodies that neutralize all three serotypes may exist, prompting us to perform additional screening of Fab fragment libraries produced from B cells of chimpanzees immunized with poliovirus vaccines. The phage display method is well suited for isolation of cross-reactive antibodies by sequentially panning Fab-displaying phage libraries against polioviruses of types 1, 2, and 3. After two cycles of panning, positive clones were screened for binding to all three serotypes by ELISA with phages expressing poliovirus-binding Fab sequences. Nineteen out of 96 clones showed relatively strong binding to all three types with an OD450 value of ≥0.8 in phage ELISA. DNA sequence analysis identified four unique clones: A1, E10, F12, and G10. As shown in Fig. 4A, the four clones bound to all three serotypes in a phage ELISA. These four clones were subsequently processed to produce soluble Fabs and their binding specificities were examined again in ELISA using live poliovirus. It was confirmed that the Fabs were able to react with all three serotypes, but each Fab preferentially interacted with certain serotypes (Fig. 4B). For example, Fabs A1 and E10 bound mostly to types 2 and 3, whereas Fab F12 bound to types 1 and 2 and Fab G10 bound to type 2 much more strongly than the other two serotypes. Based on the strength and the specificity of binding, Fab E10 and F12 were selected for IgG production and neutralization assay. As a result E10 was unable to neutralize any serotype, but F12 was similar to A12 in its ability to neutralize both type1 and type 2 virus, but not type 3 (Table 2). Nucleotide sequencing of the gene coding for F12 Fab showed that it was distinct from the A12 gene (GenBank accession nos. KF410673–KF410676).
Fig. 4.
The binding profile of newly isolated anti-poliovirus antibodies A1, E10, F12, and G10. Each of the three serotypes of OPV (types 1, 2, and 3 represented by red, yellow, and green, respectively) at a concentration of 5 × 106 pfu/mL was captured on a 96-well ELISA plate coated with purified rabbit polyclonal serotype-specific IgG (2 µg/mL) and was reacted with phage-displayed Fabs (A) or with soluble Fab (B) at a concentration of 3 µg/mL. The binding was measured by ELISA. OD, optical density measured at 450 nm.
Table 2.
Neutralization of polioviruses with F12 and A12 antibodies
| Serotype 1 | Serotype 2 | Serotype 3 | ||||
| Sample | Sabin 1 | Mahoney | Sabin 2 | MEF1 | Sabin 3 | Saukett |
| F12 IgG* | 2,896 | 1,825 | 1,448 | 4,598 | <1 | <1 |
| A12 IgG* | 1,024 | 1,218 | 1,218 | 1,722 | <1 | <1 |
| Human serum† | 456 | 1,448 | 287 | 575 | 912 | 2,896 |
Reciprocal titer of antibody solutions at 10 µg/mL.
Reciprocal titer of a pool of human sera from IPV-immunized donors.
Generation of F12 Escape Mutants and Cross-Neutralization.
To localize epitopes recognized by F12 antibody, we have attempted to generate escape mutants by growing Sabin 1 and Sabin 2 viruses in its presence. We were unable to identify any F12-resistant Sabin 1 viruses but could produce Sabin 2 virus stock that was resistant to neutralization with F12. Multiple antibody-resistant clones prepared from this virus by plaque purification were sequenced and shown to contain Lys–Glu mutations at amino acids 109 and 65 of VP1. Amino acid 65 is located at the internal surface of the capsid proteins, whereas amino acid 109 is located on the virion surface at the lower end of the so-called B–C loop. Its position in the 3D structure is close to amino acid 166 in VP1, which was previously found to be involved in A12 antibody binding (5), suggesting that both antibodies interact with the common epitope. The role of mutation at amino acid 65 is unclear; it can either participate in long-range conformational change resulting in abrogating binding of F12 antibodies, or it may provide a compensatory effect if the mutation at amino acid 109 reduces viral fitness. To confirm functional relatedness of epitopes recognized by A12 and F12 antibodies, cross-neutralization of escape mutants by both antibodies was performed. Escape mutants to other monoclonal antibodies were used as a control. Tables 3 and 4 shows that escape mutants of serotypes 1 and 2 raised against A12 were also not neutralized by F12, and vice versa. Escape mutants of type 2 poliovirus could not be neutralized by either A12 or F12, and we were unable to isolate type 1 escape mutants to F12. Thus, it seems that both antibodies bind the same site on the virion surface, despite having different amino acid sequences. Escape mutants generated against chimpanzee/human antibody H2 (5) and mouse antibody 1o (11), which are specific only to type 1, or type 2-specific chimpanzee/human antibodies A6 and B2 (5) could be readily neutralized by A12 and F12 antibodies, showing that these antibodies bind different epitopes.
Table 3.
Cross-neutralization of escape mutants by human monoclonal antibodies (reciprocal neutralization titers): Type 1 poliovirus
| Escape clone | A12 escape mutants | H2 escape mutants | 1o escape mutants | Sabin 1 | |||||
| Mutations | ES15-1/5 | ES15-1/6 | ES13-3 | ES13-5 | ES13-10 | EX4-1 | EX4-2 | EX4-5 | Control |
| V 1166 E | V 1166 E | K 1144 R | K 1144 E | V 1107 A | D 1298 N | D 1298 G | D 1298 G | ||
| I 1090 M | D 1288 N | D 1219 G | S 3058 C | D 3181 Y | L 1234 V | ||||
| Antibody | |||||||||
| A12 | <1 | <1 | 144 | >181 | 45 | 114 | 144 | 144 | 114 |
| F12 | <1 | <1 | >181 | >181 | >181 | >181 | >181 | >181 | >181 |
| H2 | >181 | >181 | <1 | <1 | <1 | 18 | <1 | <1 | 144 |
| 1o | 36 | 11 | 18 | 23 | 3 | <1 | <1 | <1 | 45 |
Table 4.
Cross-neutralization of escape mutants by human monoclonal antibodies (reciprocal neutralization titers): Type 2 poliovirus
| Escape clone | A12 escape mutant | F12 escape mutants | A6 escape mutants | B2 escape mutants | Sabin 2 | |||
| Mutations | ES15-6/3 | ES17-4/3 | ES17-4/10 | ES15-9/1 | ES15-10/3 | ES15-13/2 | ES15/13/5 | Control |
| G 1225 D | K 1109 E | K 1109 E | R 1100 C | R 1100 L | A 1101 D | A 1101 D | ||
| H 1065 R | H 1065 R | N 2165 S | I 4062 T | I 4062 T | ||||
| Antibody | V 2212 A | |||||||
| A12 | <1 | <1 | <1 | 57 | 114 | 91 | 57 | 72 |
| F12 | <1 | <1 | <1 | 114 | >181 | >181 | >181 | >181 |
| A6 | >181 | 144 | 144 | <1 | <1 | 11 | 9 | 45 |
| B2 | 144 | >181 | >181 | <1 | <1 | <1 | <1 | >181 |
Discussion
The antigenic structure of polioviruses was extensively studied in the mid-1980s using mouse monoclonal antibodies (12–14), whereas information about human antibodies remains scarce. One attempt to address this was based on EBV-immortalized human B cells that were shown to produce neutralizing antibodies (15). Some of them could neutralize type 1 and type 2 polioviruses (16), but no attempt to map their epitope was reported at the time. Results presented in this communication suggest that such cross-neutralizing antibodies may be more common than previously recognized. It was therefore important to identify epitopes on the surface of polio virions that are involved in this broadly specific neutralization. Sequencing of escape mutants suggested that the antibodies bind to opposite rims of the circular depression surrounding the fivefold axis of symmetry, called the canyon. Cryo-EM, which was not previously used to study binding of antibodies to poliovirus, confirmed this localization and demonstrated that A12 binds the region that previously was identified as the site interacting with cellular poliovirus receptor CD155. Earlier studies with mouse monoclonal antibodies identified several neutralization antigenic sites in poliovirus, some of them located near the rims of the canyon, but none of them was mapped directly to the bottom of the canyon. It is noteworthy that A12 antibodies interact with amino acids located on the opposite walls of the canyon, which were previously identified as belonging to neutralizing antigenic sites 1 and 2. Although poliovirus antigenic sites represent overlapping continuums of epitopes of individual monoclonal antibodies, they were believed to be functionally distinct regions on the poliovirus surface. Our observations suggest that classification of antigenic sites previously developed based on mouse antibody data may not fully apply to primate antibodies.
In contrast to previous data on location of epitopes of mouse anti-poliovirus antibodies, mapping by cryo-EM of epitopes of several neutralizing antibodies to human rhinovirus 14 showed that Fab fragments also bind both walls of the canyon, albeit not as deeply as A12 antibody. The rhinovirus 14 antibodies directly overlap much of the receptor-binding site, and two arms of the IgG molecule may reach across the twofold axis of symmetry to bind two adjacent pentamers (17). The pattern of A12 binding described in this communication seems to be similar to the binding of rhinovirus 14 antibodies. This may have implications for the mechanism of action of these antibodies, because it is believed that during poliovirus uncoating the RNA molecule is released from the capsid through the temporary umbilical connections that are formed between cellular membrane and the virion region located near the twofold axis of symmetry (18–20). This region would be shielded by a bivalent IgG molecule if its two Fab fragments are bound to canyons surrounding adjacent pentamers. However, our results do not provide clear evidence of binding of bivalent IgG molecules to two epitopes in adjacent pentamers across twofold axis of symmetry, as opposed to binding neighboring epitopes within the same pentamer. A more definite conclusion could be obtained by modeling of IgG binding similar to a recently published study of another poliovirus antibody (21) that requires higher-resolution images, or performing direct cryo-EM study of virus–IgG complexes.
Significant overlap between receptor-binding site and neutralizing antigenic epitopes observed in this work was previously postulated by Wimmer et al. (22). Functional relatedness of receptor binding site and neutralizing epitopes creates a constraint on antigenic variability and may be the reason why polioviruses have only three serotypes. Because all three serotypes of poliovirus bind to CD155, their receptor recognition site must be similar, explaining why antibodies binding this region have a broad neutralizing activity. Although normal cellular receptor binds all three serotypes equally well, some mutations of CD155 reduce its binding to poliovirus in a serotype-specific manner (23), suggesting that the interaction of CD155 with the receptor-recognition site of different serotypes may involve different molecular interactions. The same may be true for A12 antibodies, because they strongly bind to and neutralize types 1 and 2 but only weakly bind type 3 without a measurable neutralizing activity. SPR study of the interaction of A12 antibody with the virus showed that the affinity with which it binds type 1 and 2 is about 10-fold higher than for type 3, possibly explaining why it does not neutralize it. Despite that, A12 antibody binds to the same region on the surface of both type 1 and type 2 poliovirus; the detailed mechanism of binding may slightly differ, as suggested by a small difference in the angle (7°) at which Fab fragment seems to bind the virion (Fig. 3). Therefore, there may be multiple ways antibodies bind related structures of receptor-recognition sites, and modification of complementarity-determining regions of the antibody may alter specificity of this interaction and lead to increased affinity. It will be interesting to test whether antibody engineering could create an A12 variant with increased affinity and the ability to neutralize all three serotypes. Such improved antibody would not only be useful therapeutically, but also can shed light on the mechanisms of neutralization. Further screening for cross-neutralizing antibodies that is currently underway may also yield triple-neutralizing antibodies.
It is noteworthy that despite their strong neutralizing activity, the affinity of the cross-neutralizing antibodies is relatively low for bivalent anti-viral IgG, which often have binding constants that are 10- to 100-fold higher. This may be a trade-off for their broad reactivity and could partially explain why cross-neutralizing antibodies were not discovered before. Phage-display screening that was used in this paper could isolate Fab fragments with relatively weak binding that did not reach the level needed for neutralizing activity. There are also noticeable differences in binding patterns between phage-displayed Fabs and soluble Fabs in ELISA (Fig. 4 A and B). This discrepancy could be explained by the difference between monovalent binding of soluble Fabs versus multivalent binding of phage-displayed Fabs. For low-affinity antibodies, this difference could be more pronounced, resulting in measurable binding with phage-displayed Fabs that cannot be detected with soluble Fabs.
Binding of antibodies to a receptor-recognition site may have an additional significance. Some mutations making virus resistant to the antibodies were also shown to affect its binding to the cellular receptor (9), thereby reducing viral fitness and minimizing generation of escape mutants in vivo. It seems that antibodies targeting virion structures involved in receptor binding tend to be broadly reactive not only for poliovirus, but for other viruses as well. For instance, antibodies binding the receptor-recognition site of influenza hemagglutinin were also shown to neutralize a broad spectrum of influenza strains (24, 25). Similarly, broadly reactive anti-HIV monoclonals also bind structures interacting with cellular receptor CD4 (26, 27). Therefore, the ability to bind critically important and conserved receptor-recognition sites could be a hallmark of “silver bullet” antibodies that could become the basis for effective therapies.
Results presented in this paper reveal the mechanism of action of cross-neutralizing antibodies and suggest that further search may yield antibodies that could neutralize all three serotypes of poliovirus. Our attempts to use sequential phage-display panning to isolate such antibodies resulted in isolation of another antibody that neutralized type 1 and type 2 poliovirus by interacting with the same epitope but failed to identify a universal anti-poliovirus antibody. Future screening and/or engineering of A12 or F12 antibodies to expand their specificity may eventually result in creation of such antibodies. Alternatively, a mixture of antibodies with different mechanisms of action could be used against polioviruses of all three serotypes while preventing emergence of antibody-resistant variants. Future studies should also include clinical evaluation of the ability of these antibodies to cure chronic poliovirus excretors and prevent paralytic disease in individuals exposed to poliovirus. Our results strongly argue in favor of adding broadly specific human monoclonal antibodies to the arsenal of tools used in the final phases of the campaign to eradicate polio and beyond.
Materials and Methods
Antibodies and Viruses.
Poliovirus-neutralizing Fabs A12 and F12 were isolated from a Fab-displaying phage library constructed from bone marrow cells of chimpanzees immunized with polioviruses (ref. 5 and this study). Sequences of heavy and light chains of the two Fabs were deposited to GenBank (accession nos. KF410673–KF410676). The full-length IgG (known as antibody) consisting of chimpanzee variable domain of heavy chain and whole light chain and human IgG1 Fc fragment (GenBank accession no. Y17957) were produced by transiently transfecting 293-T cells. Antibodies were purified by affinity chromatography on protein A columns. Stocks of purified formalin-inactivated Mahoney and MEF-1 polioviruses represented monovalent bulks of inactivated polio vaccine and were kindly provided by a vaccine manufacturer. They were concentrated to 1 mg/mL using Centricon spin columns (Millipore).
Cryo-EM.
Immune complexes were prepared by mixing at a ratio of 2:1 of poliovirus (1 mg/mL) and A12 Fab (0.67 mg/mL) and incubating at room temperature for 1 h. Four-microliter aliquots of sample were applied to freshly glow-discharged 200-mesh r2/2 Quantifoil copper grids suspended by forceps in the FEI Mark IV Vitrobot. Specimens were vitrified, after blotting for 2 s with a blot force of 5 at 80% relative humidity, by plunge-freezing into liquid ethane. Specimens were examined under low dose cryo conditions at 300 kV with a, FEG Titan Krios transmission electron microscope and images were recorded at a nominal magnification of 47 kx corresponding to a pixel size of 1.8 pixels/Å2 and an electron dose of ∼15 e−/Å2. Particles with a defocus range of 1–5 µm were boxed with EMAN2, and then processed using the standard single-particle reconstruction procedure with full contrast transfer function correction and icosahedral symmetry imposed (28). The resolution was estimated as ∼12 Å for both the type 1 and type 2 maps using the “gold standard” resolution criterion (29) as implemented in EMAN2 using the 0.143 Fourier shell correlation threshold. To test for completeness of antibody binding, a reference map was created with one antibody-unoccupied vertex, and refinement was run with no symmetry imposed. Any particles with at least one missing antibody would have preferentially aligned with the artificially cleaned vertex, producing a reduced or missing antibody density in the final refined map. Instead, the density for the missing vertex immediately returned during refinement, indicating 100% antibody occupancy to within the limits of measurement.
Although the resolution of the reconstructions was relatively low, the available atomic model for the virus particle could readily be docked into the cryo-EM reconstruction, and the density indicating the connection point between capsid and antibody was sufficiently localized to identify the interacting residues within a range of a few amino acids. Visualization and analysis of the binding regions were performed in UCSF Chimera (30) using 1asj and 1eah from the Protein Data Bank as the unbound reference models for type 1 and type 2, respectively. The binding angle measurement was done in Amira using the 2D angle measurement tool. The 3D reconstructions were deposited into the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb/) under codes 5670 and 5671 for type 1 and type 2 poliovirus, respectively.
Affinity Measurement by SPR.
Binding kinetics of antibody A12 IgG to poliovirus was assessed by SPR using a Biacore 1000 (GE Healthcare). Running buffer was 10 mM Hepes (pH 7.4), 150 mM NaCl, and 0.005% Tween-20 (HBS-EP buffer). Formaldehyde-inactivated poliovirus (IPV) was coupled to the surface of a CM5 chip using the manufacturer-recommended protocol for amine coupling. Briefly, a 7-min injection at 10 µL/min of a mixture of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 0.1 M NHS was used to activate the surface, then 1–2 µg/mL IPV in 100 mM acetate buffer (pH 3.5) was injected across the surface to achieve 150–200 RU, followed by deactivation of the surface by a 7-min injection of 1 M ethanolamine at 10 µL/min. IgGs at a concentration series from 100 nM to 1,000 nM were passed over the IPV surface for 2 min at 30 µL/min. Following dissociation for 6 min at 30 µL/min, the surface was regenerated by injecting a solution of Glycine Buffer (pH 1.5) for 1 min at 50 µL/min. Data were analyzed with Biacore software (version 2.1.2) using the Langmuir model.
Isolation of Broadly Reactive Antibodies Through a Panning Procedure.
To select additional broadly reactive antibodies, a sequential panning against poliovirus serotypes 1, 2, and 3 was used, reasoning that only the clones that are cross-reactive with three serotypes are enriched after the successive panning. A Fab-displaying phage library constructed previously (5) was first panned against Sabin 1 strain. The phages were eluted, propagated, and panned against Sabin 2 strain. The phages were eluted and used to infect bacteria to produce more phages for the further panning against Sabin 3 strain. This sequential panning was repeated one more time. Next, 96 randomly picked colonies were grown in a 96-well plate and cultured for phage production. Positive cross-reactive clones were identified by phage ELISA with all three serotypes of poliovirus. Subsequently, the sequences of the variable domain of heavy (VH) and light (VL) chains of the identified Fab clones were determined. Clones with unique VH and VL sequences were selected for soluble Fab production. The soluble Fabs were examined for their binding specificities by ELISA. Clones with higher ELISA reactivity were further converted into complete IgGs and were used for in vitro neutralization assays.
Production and Purification of Fab and IgG.
As described previously (5), plasmid DNA was modified to express soluble Fab tagged with six histidines at the C terminus. The Fab was expressed in Escherichia coli in a 250-mL culture for small scale or in a 4-L culture for large scale. The Fab was purified on a nickel column and then on a sulfopropyl (SP) cation-exchange column. Sometimes, the Fab was further purified by gel filtration chromatography on a Superdex column.
For IgG production, light chain and heavy chain genes were cloned into light and heavy expression vectors, separately. The resulting plasmids were used to cotransfect 293-T mammalian cells for transient expression in a serum-free medium. The culture media were collected and IgG was purified by passing the media through a HiTrap column of Mab Select SuRe (GE Healthcare). The IgG was further purified using a cation-exchange SP column. The purity of the Fab and IgG was evaluated by SDS/PAGE, and the protein concentrations were determined by optical density measurements at 280 nm (OD280) assuming that 1.35 A280 corresponds to 1.0 mg/mL.
Microneutralization Test.
Poliovirus-neutralizing antibody titers were determined in a microneutralization test according to the World Health Organizatin procedure (31) with slight modifications. The mAb samples were diluted to 10 μg/mL in DMEM supplemented with 2% FBS and 1% (vol/vol) of antibiotic/antimycotic (all from Invitrogen) and sterilized by filtration through a Spin-X column (Corning). Four parallel series of twofold serial dilutions of the antibodies starting at 10 μg/mL were incubated with 100 TCID50 of respective poliovirus strains in equal volumes for 3 h at 36 °C with 5% CO2. At the end of the incubation 1 × 104 HEp-2C cells were added to each well. The plates were incubated for 10 d at 36 °C, the cytopathic effect (CPE) was assessed microscopically, and neutralizing antibody titers were calculated using the Kärber formula.
Generation of Escape Mutants.
Four fivefold dilutions of monoclonal antibodies starting at 200 µg/mL were incubated with ∼108 CCID50 of virus for 1 h at room temperature, followed by 2 h at 36 °C. Virus samples were inoculated onto Hep-2C cell monolayers and incubated at 36 °C until CPE developed. Supernatants after three freeze–thaw cycles were clarified by centrifugation. For cloning by plaque purification, serial dilutions of mAb-resistant viruses (101–104 CCID50/mL) were inoculated onto Hep-2C monolayers in six-well plates, incubated for 1 h at room temperature, then replaced with 4 mL MEM/0.5% agarose overlay with 6% FBS (all reagents from Invitrogen). Plaques were picked after 48 h of incubation at 36 °C, 5% CO2, transferred into 12-well plates with confluent monolayers, and incubated overnight until CPE developed. Supernatants after three freeze–thaw cycles were clarified by centrifugation and nucleotide sequences were determined by the Sanger method.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, the Food and Drug Administration Center for Biologics Evaluation and Research, and NIH Grant R01-GM080139 (to S.L.).
Footnotes
References
- 1.Dowdle WR, De Gourville E, Kew OM, Pallansch MA, Wood DJ. Polio eradication: The OPV paradox. Rev Med Virol. 2003;13(5):277–291. doi: 10.1002/rmv.401. [DOI] [PubMed] [Google Scholar]
- 2.Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Update on vaccine-derived polioviruses—worldwide, January 2008-June 2009. MMWR Morb Mortal Wkly Rep. 2009;58(36):1002–1006. [PubMed] [Google Scholar]
- 4.Hammon WM. Passive immunization against poliomyelitis. Monogr Ser World Health Organ. 1955;26:357–370. [PubMed] [Google Scholar]
- 5.Chen Z, et al. Chimpanzee-human monoclonal antibodies for treatment of chronic poliovirus excretors and emergency postexposure prophylaxis. J Virol. 2011;85(9):4354–4362. doi: 10.1128/JVI.02553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97(1):79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y, et al. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol. 2003;77(8):4827–4835. doi: 10.1128/JVI.77.8.4827-4835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, et al. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci USA. 2008;105(47):18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan G, Peters D, Racaniello VR. Poliovirus mutants resistant to neutralization with soluble cell receptors. Science. 1990;250(4987):1596–1599. doi: 10.1126/science.2177226. [DOI] [PubMed] [Google Scholar]
- 10.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 11.Minor PD. Antigenic structure of poliovirus. Microbiol Sci. 1986;3(5):141–144. [PubMed] [Google Scholar]
- 12.Emini EA, Jameson BA, Lewis AJ, Larsen GR, Wimmer E. Poliovirus neutralization epitopes: Analysis and localization with neutralizing monoclonal antibodies. J Virol. 1982;43(3):997–1005. doi: 10.1128/jvi.43.3.997-1005.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogle JM, Filman DJ. The antigenic structure of poliovirus. Philos Trans R Soc Lond B Biol Sci. 1989;323(1217):467–478. doi: 10.1098/rstb.1989.0024. [DOI] [PubMed] [Google Scholar]
- 14.Minor PD, Ferguson M, Evans DM, Almond JW, Icenogle JP. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J Gen Virol. 1986;67(Pt 7):1283–1291. doi: 10.1099/0022-1317-67-7-1283. [DOI] [PubMed] [Google Scholar]
- 15.Uhlig H, Rutter G, Dernick R. Self-reactive B lymphocytes detected in young adults, children and newborns after in vitro infection with Epstein-Barr virus. Clin Exp Immunol. 1985;62(1):75–84. [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlig H, Dernick R. Intertypic cross-neutralization of polioviruses by human monoclonal antibodies. Virology. 1988;163(1):214–217. doi: 10.1016/0042-6822(88)90251-6. [DOI] [PubMed] [Google Scholar]
- 17.Che Z, et al. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J Virol. 1998;72(6):4610–4622. doi: 10.1128/jvi.72.6.4610-4622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostina M, Levy H, Filman DJ, Hogle JM. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol. 2011;85(2):776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy HC, Bostina M, Filman DJ, Hogle JM. Catching a virus in the act of RNA release: A novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol. 2010;84(9):4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol. 2013;87(7):3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Cheng N, Hogle JM, Steven AC, Belnap DM. Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy. J Immunol. 2013;191(2):884–891. doi: 10.4049/jimmunol.1202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 23.Harber J, Bernhardt G, Lu H-H, Sgro JY, Wimmer E. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology. 1995;214(2):559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 24.Tsibane T, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8(12):e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20(3):363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20(2):205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157(1):281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Anonymous . Manual for the Virological Investigation of Polio. Geneva: World Health Organization; 1997. [Google Scholar]