Significance
Isoform identification and discovery are an important goal for transcriptome analysis because the majority of human genes express multiple isoforms with context- and tissue-specific functions. Better annotation of isoforms will also benefit downstream analysis such as expression quantification. Current RNA-Seq methods based on short-read sequencing are not reliable for isoform discovery. In this study we developed a new method based on the combined analysis of short reads and long reads generated, respectively, by second- and third-generation sequencing and applied this method to obtain a comprehensive characterization of the transcriptome of the human embryonic stem cell. The results showed that large gain in sensitivity and specificity can be achieved with this strategy.
Keywords: isoform discovery, PacBio, hESC transcriptome, alternative splicing, lncNRA
Abstract
Although transcriptional and posttranscriptional events are detected in RNA-Seq data from second-generation sequencing, full-length mRNA isoforms are not captured. On the other hand, third-generation sequencing, which yields much longer reads, has current limitations of lower raw accuracy and throughput. Here, we combine second-generation sequencing and third-generation sequencing with a custom-designed method for isoform identification and quantification to generate a high-confidence isoform dataset for human embryonic stem cells (hESCs). We report 8,084 RefSeq-annotated isoforms detected as full-length and an additional 5,459 isoforms predicted through statistical inference. Over one-third of these are novel isoforms, including 273 RNAs from gene loci that have not previously been identified. Further characterization of the novel loci indicates that a subset is expressed in pluripotent cells but not in diverse fetal and adult tissues; moreover, their reduced expression perturbs the network of pluripotency-associated genes. Results suggest that gene identification, even in well-characterized human cell lines and tissues, is likely far from complete.
In the 5 y since the introduction of RNA-Seq (1, 2), there have been remarkable advances in our ability to analyze the transcriptome. During this period, additional methods based on next-generation sequencing (NGS) have been developed for the study of many different aspects of RNA biology. These include methods to study RNA species that are ribosome bound (3), nuclear (4), implicated in RNA editing (5), functional noncoding RNAs (6), in protein–RNA binding sites (7), and interacting in microRNA–mRNA complexes (8). Concurrently, the increase in NGS throughput and the development of multiplex sequencing protocols have made RNA-Seq analysis as cost-effective as gene expression microarrays.
Despite these advances, we are still far from achieving the original goals of RNA-Seq analysis, namely the de novo discovery of genes, the assembly of gene isoforms, and the accurate estimation of transcript abundance at the gene or the isoform level. Current RNA-Seq experiments are based on second-generation sequencing (SGS) instruments capable of generating a large number of short reads. From these reads, one obtains two types of information: (i) frequency of reads aligned to a contiguous genomic segment (exonic reads) and (ii) frequency of reads aligned to two contiguous segments of the genome with a single gap of from 60 bp to 400 kbp in size (junction reads) (9–11). If the set of possible isoforms is assumed known (i.e., the gene is well annotated), then it is possible to infer isoform-specific expression from exonic reads and junction reads based on simple statistical models such as the Poisson deconvolution model of Jiang and Wong (12). On the other hand, if the set of isoforms is not known or only partially known, then currently there is great difficulty in isoform quantification based on SGS data. The main reason is due to insufficient length of the SGS reads. The median length of human gene transcripts is about 2,500 bp, which is much longer than the length of a contiguous read (about 250 bp) currently attainable by SGS. In previous work we showed that generally, isoform deconvolution from short-read RNA-Seq data is not an identifiable problem (13), in the sense that isoform expression cannot always be uniquely determined from the set of exons and splice junctions. Thus, strong assumptions are made on the set of candidate isoforms in all current methods for isoform assembly from short reads, including Cufflinks (14), SLIDE (15), and Montebello (16); as a result, the assembled isoforms are of uncertain accuracy. Although hundreds of RNA-Seq datasets are being generated in any given day by diverse groups in academia and industry, their interpretations all depend critically on the completeness and reliability of gene and isoform annotations on the species and cell types being analyzed. Although results from de novo transcript reconstruction algorithms can provide useful hints, they are not accurate enough to stand on their own as definitive evidence for new transcripts.
One may hope that the completeness and reliability of gene annotation are improving commensurably with the exponential increase in the amount and diversity of sequence data from RNA-Seq. However, from release 43 (September 2010) to 49 (September 2011), the number of RefSeq records for mammals increased by 6.4% (17). Does this slow increase imply that the annotations of mammalian transcripts are close to complete? We believe the answer is clearly negative. The slow increase is likely due to underinvestment in new tools and methods to generate full-length transcript sequences. Even in well-studied species such as human and mouse, isoforms with low to moderate abundance are only partially sampled in full-length cDNA sequencing, especially when their expression is specific to a minority of cell types. Ironically, although gene annotations are necessary for the interpretation of short-read RNA-Seq data, the emergence of RNA-Seq itself as the preferred method to examine the transcriptome may be contributing to a slowdown of large-scale cDNA and EST sequencing projects that were once major sources of experimental evidence for gene annotation. This is a fundamental roadblock for further progress in transcriptomic analysis.
The research reported here offers a way out of this impasse. We address the hypothesis that full-length and large partial fragments of transcripts can be sequenced in a massively parallel fashion by combining SGS and third-generation sequencing (TGS). One characteristic of TGS technologies is that they can generate long reads. Pacific Biosciences (PacBio) RS, the first TGS platform to offer very long reads, has an average read length of 2–3 kb and reads of >7 kb are not uncommon (18). Thus, the use of TGS for transcript assembly has always been viewed as an attractive approach. In our preliminary work on this problem, we found that the major barriers in using PacBio for transcriptomic analysis are the relatively high error rate (up to 15%) and moderate throughput [50,000 reads per single molecule real time (SMRT) cell]. To bypass the first barrier, we developed a method to correct errors in PacBio long reads by SGS short reads from the same transcriptome (19). Here, we address the second barrier by proposing new statistical methods for transcript discovery and reconstruction based on the combined information from error-corrected long reads and short-read counts.
We applied our “hybrid sequencing” RNA-Seq method to human embryonic stem cells (hESCs), using the well-characterized H1 hESC line; this line is a Tier 1 ENCODE line whose transcriptome has been extensively studied by EST sequencing (20) and RNA-Seq (21–23). However, even for such a well-characterized cell line, our analysis revealed that its current annotation is very incomplete and there are hundreds of novel genes/long noncoding RNAs (lncRNAs) and thousands of novel isoforms of known genes expressed. In this paper we use the term “novel gene/isoform” to describe a gene loci or gene isoform that is directly detected by PacBio long reads or predicted by our proposed method in the H1 transcriptome, and not found in existing gene annotation databases [RefSeq, Ensembl (24), UCSC Known Genes (25), EST (26) and GENCODE(27)]. The lncRNAs identified by our study are on average much longer and have more exons than those annotated in existing databases, suggesting significant downward bias in the current strategy for genome-wide discovery of lncRNAs. Finally, our experimental validation demonstrated that at least a subset of novel transcripts represents bona fide gene loci.
Results
Gene Isoform Detection and Prediction.
We applied the splice junction detection program SpliceMap (9) to 116,476,819 mappable 100-bp Illumina sequence reads and detected 183,825 junctions with an estimated false positive rate (FPR) of 5% (9). Of the detected junctions, 46,898 (∼25.51%) were not reported in RefSeq. Furthermore, many of them were found in the intergenic regions and formed 2,568 clusters (SI Appendix: Methods, Short Reads Alignment and Exon Junction Detections by SpliceMap). This suggested that existing gene annotations are incomplete and there may exist many novel genes as well as novel isoforms of annotated genes.
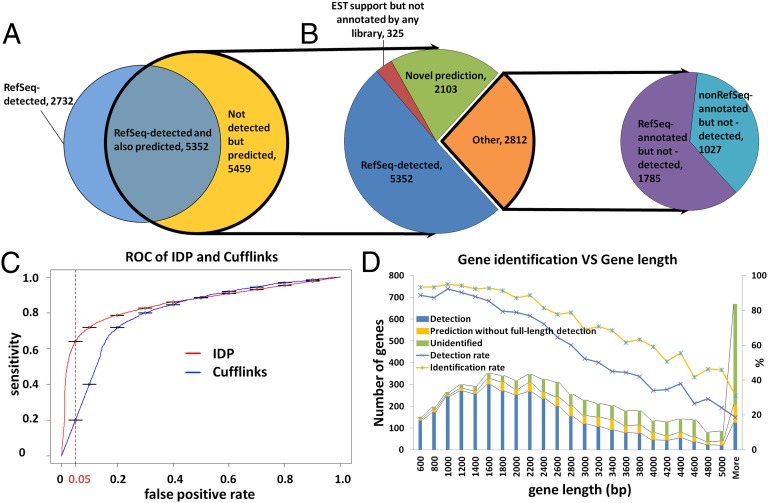
To capture multiexon transcripts, we used the PacBio RS sequencing platform to capture the full-length RNA in hESCs (H1 cell line). We generated full-length cDNA libraries by two different established methods, using polyA RNA as starting material. The prepared double-stranded cDNA was then converted to libraries for PacBio single-molecule real-time sequencing. In total, 7,816,704 raw PacBio RS reads were generated. Error correction of the raw PacBio RS reads was performed by LSC (19). A total of 1,998,716 error-corrected long reads (GSE51861) were mappable to the reference human genome hg19 by BLAT. A total of 781,128 long reads covered at least one junction supported by SpliceMap junction detection or RefSeq annotation. In total, 8,084 gene transcript isoforms from RefSeq were detected directly with a full-length read. We compared these isoforms to the 5,851 multiexon RefSeq genes that are significantly expressed [i.e., having reads per kilobase per million mapped reads (RPKM) >10 based on Illumina short reads] in hESCs and found that although 61.37% of these genes are covered by the directly detected isoforms, the detection rate varies depending on the gene length. The detection rate is 78.18% (3,226) in the 3,689 multiexon genes of size <3,000 bp and 32.71% (707) in the 2,162 multiexon genes of size >3,000 bp (Fig. 1D). Although we also detected many single-exon transcripts, isoform identification of multiexon genes was of more interest for this study and we focused on those cases. Unless otherwise specified, the term “isoform” in the remaining text refers to multiexon RNA isoforms.
Fig. 1.
Gene isoform detection and prediction of hESCs (H1 cell line) by IDP. (A) Venn diagram of IDP detections and predictions (see introductory section for definition of detection and prediction). A total of 8,084 RefSeq isoforms are detected and highlighted in blue. A total of 10,811 predictions are highlighted in yellow and outlined with a thick black line. A total of 5,352 detections of RefSeq isoforms are also predicted by IDP. (B). Pie chart of annotated isoforms and novel isoforms in IDP predictions. IDP predictions rescue 1,785 RefSeq-annotated isoforms (in purple) that cannot be detected directly at full length. In addition, there are 1,027 predictions that are not annotated in RefSeq but are found in Ensembl, Known Genes, or GENCODE (cyan). Finally, 2,428 novel isoforms (green and red) are identified, 325 of which have EST support (red). (C) ROC performance analysis of IDP and Cufflinks. IDP predictions have much higher sensitivity in the acceptable FPR range from 5% to 10%. When FPR is controlled to 5%, the IDP prediction sensitivity is as high as ∼62%, whereas the corresponding Cufflinks sensitivity is only about 20%. (D) RefSeq gene identification rate decreases with the gene length. Combining detections and predictions, the overall identification rate by IDP is ∼73% (yellow line with blue star markers). IDP prediction rescues a significant number of isoforms from long genes that are not directly detected.
To identify more isoforms for long genes, we developed a statistical isoform prediction method to construct possible isoform candidates from the union of long reads and short reads with spliced alignment (SI Appendix: Methods, IDP—Isoform Detection and Prediction) (http://www.stanford.edu/people/kinfai/IDP/IDP.html). Using this method we obtained 5,459 isoform predictions (Fig. 1A), which significantly expanded the sensitivity of isoform identification beyond direct detection. We indicated a RefSeq isoform as identified by IDP if either it was directly detected by sequencing or there was a predicted isoform with an identical set of exon–exon junctions. The isoform-based identification rate of significantly expressed genes now improved to 73.70% (compared with 61.4% by direct detection). More specifically, for long genes (>3,000 bp) the identification rate improved from 32.70% to 50.23%, whereas for short genes (<3,000 bp) the identification rate improved from 71% to 90% (Fig. 1D). As we keep optimizing LSC and IDP, we will keep updating the hESC transcriptome at our website (http://www.stanford.edu/people/kinfai/IDP/hESC.html), including novel isoforms, novel genes, novel lncRNA, and gene/isoform profiling.
We compared the performance of IDP isoform prediction against Cufflinks, which is currently the most widely used method for isoform identification based on short reads [Fig. 1C; see SI Appendix for details of receiver operating characteristic (ROC) analysis]. At the same specificity of 5% false positive rate, IDP had a much higher sensitivity (62% true positive) compared with Cufflinks (20% true positive). This result shows that IDP is effective in using the information from the PacBio long reads to significantly improve isoform identification.
The predicted (i.e., not directly detected) isoforms from IDP included a large number of annotated isoforms that are from RefSEq. (1,785 isoforms) or other annotation libraries [1,027 isoforms; combined Ensembl, GENCODE, and University of California, Santa Cruz (UCSC) Known Genes]. There is also a small group (325) of nonannotated but EST-supported isoforms. More importantly, 2,103 predicted isoforms are not reported by any existing annotations. The identification of these isoforms provides a more comprehensive characterization of the hESC transcriptome. As discussed below, the novel isoforms also lead to better estimates of isoform-specific gene expression.
Novel Genes.
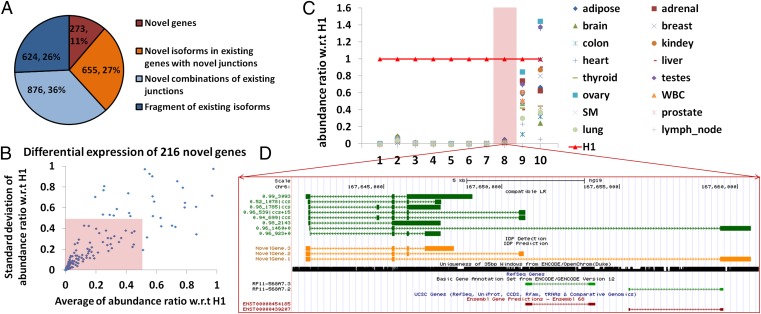
Of the 2,103 identified novel isoforms that are not reported by any existing annotations, 273 isoforms were transcribed from 216 novel nonannotated gene loci, i.e., all of the splicing junctions are not reported in any existing annotations (Fig. 2A). Using the short-read data, we computed their representation in RNA-Seq data from 16 adult human tissues (50 bp; Illumina Human Body Map) and their abundance ratio with respect to H1. We began by plotting the abundance ratios of the 10 novel genes with the highest expression; 8 of these 10 genes had <5% relative expression levels in all 16 adult tissues compared with hESCs, which indicated that their expression was highly specific to hESCs and thus they may be valuable candidates for novel pluripotency markers (Fig. 2C). For example, several long reads with up to four junctions were mapped to the locus chr6:167,641,267–167,660,912 (hg19, the same below), where no annotated genes in RefSeq, Ensembl, UCSC Known Genes, or GENCODE are reported. The long reads indicated complex expression from this locus with at least three different isoforms transcribed (Fig. 2D). The RPKM of this gene was 31.94 in hESCs, a value much higher than the averaged RPKM (0.53) in the other 16 adult tissues. For the other 2 novel genes (of 10), the expression level in 16 tissues was comparable to that of hESCs, and both of them had isoforms containing three or more junctions. Next, to determine the hESC-specific expression of all 216 novel genes, we calculated their abundance ratio (over the expression in the 16 adult tissues) and we plotted it vs. the SD of abundance ratios for each one. Sixty-eight percent of the novel genes (146/216) had an average abundance ratio smaller than 0.5 with SD smaller than 0.5 (inside the pink box in Fig. 2B), which indicated that they had relative specific expression in hESCs. Because their expression is specific to H1 hESCs, they may not have been easily identified in the previous research. We noted that 32% of the novel genes (70/216) were not hESC specific; they had comparable abundance in the other human tissues.
Fig. 2.
Novel gene identifications. (A) A total of 2,428 novel isoforms are categorized according to the use of the annotated junctions. A total of 273 isoforms from 216 novel genes are observed (in brown). A total of 655 novel isoforms use at least one junction from annotated genes (in orange). A total of 876 novel isoforms are novel combinations of annotated junctions. Six hundred twenty-four are fragments of annotated isoforms. (B) Differential expression of 216 novel genes in H1. The abundance ratio of a novel gene in a given tissue is defined as its abundance in this tissue divided by its abundance in H1. One hundred forty-six novel genes (68%, inside the pink box) have an averaged abundance (among 16 human tissues) ratio smaller than 0.5 with SD smaller than 0.5. (C) Relative expression levels of the top 10 novel genes (10 highest expressions in hESCs) in 16 human tissues. The reference expression levels are expressions in hESCs (highlighted in red line with triangles). Eight novel genes have high expression specifically in hESCs (example 8 in D) whereas the other 2 have significant expression across many different tissues. The gene structure of the eighth novel gene is visualized in D. (D) Novel gene at chr6:167,641,267–167,660,912. The dark green track shows the nonredundant long reads, each of which represents an alignment. The arrow refers to the alignment of the read relative to the reference (i.e., aligned to reference or to reverse complement of reference) and is not the direction of transcription. The naming of nonredundant long reads is A_B|ccs ± D, where A is the percentage identity of BLAT alignment, B is the length of alignment, and D is the distance between the mappable part of the long read and the polyA/polyT detection (“+” is the forward strand and downstream whereas “−” is the reverse strand and upstream). PacBio circular consensus sequence (CCS) reads are labeled with “ccs”. The orange track shows IDP predictions. The 35-bp mappability of this locus is in black. The light green track is GENCODE annotation and the brown one is Ensembl. RefSeq (light purple track) and UCSC Genes are also displayed but they have no annotated genes in this locus and thus no IDP detections (referenced to RefSeq, red track) are displayed either. The track display settings of other figures are the same.
hESC-Specific Novel Genes.
To confirm that the novel genes have expression specific to hESCs, we further characterized 23 of those showing the highest expression. For this purpose, we examined expression by quantitative PCR in two independent hESC lines (H1 and H9), one induced pluripotent stem cell (iPSC) line (iPSC line RiPSC.HUF1), and a collection of cDNAs obtained from different adult human tissues. All 23 targets, denoted as HPAT (Human Pluripotent Associated Transcript) genes, were abundantly expressed in the pluripotent cell lines (H1, H9, and RiPSC.HUF1) as opposed to adult tissues where they were either undetectable or expressed at very low levels (Fig. 3A and SI Appendix, Fig. S9A). Controls included genes whose expression is highly enriched in pluripotent stem cells and tissue-specific markers (SI Appendix, Fig. S9 B and C).
Fig. 3.
Gene expression validation of HPAT. (A) Gene expression analysis by qPCR was performed on two hESC samples (H1 and H9), one iPSC line (RIPSC.HUF1), and a collection of cDNAs from fetal and adult tissues. (B) Gene expression profiling of HPAT genes in single blastomeres of eight-cell embryos and blastocysts. E1 and E2 denote the embryos from which blastomeres were isolated. (C) Reactivation of genes during the reprogramming process. Cells were analyzed at different time points of mRNA-mediated iPSCs derivation.
We also examined expression during human development, in single blastomeres of eight-cell embryos and in human blastocysts (Fig. 3B). We observed three different categories of expression: genes (HPAT1, HPAT15, and HPAT19) detected in the blastocysts but not in the eight-cell-stage blastomeres, genes abundantly expressed in both blastocysts and eight-cell embryos (HPAT2 and HPAT3), and genes that were not expressed in either stage of preimplantation development; controls were as shown (SI Appendix, Fig. S9D). To further investigate the role of the genes in pluripotency, we also examined their expression during reprogramming of neonatal fibroblasts toward iPSCs. We observed that several of these genes, which were not expressed in parental fibroblasts, were activated during reprogramming (between day 10 and day 12) with a kinetic very similar to that observed for the pluripotency-associated genes like NANOG, POU5F1, and DNMT3B (Fig. 3C).
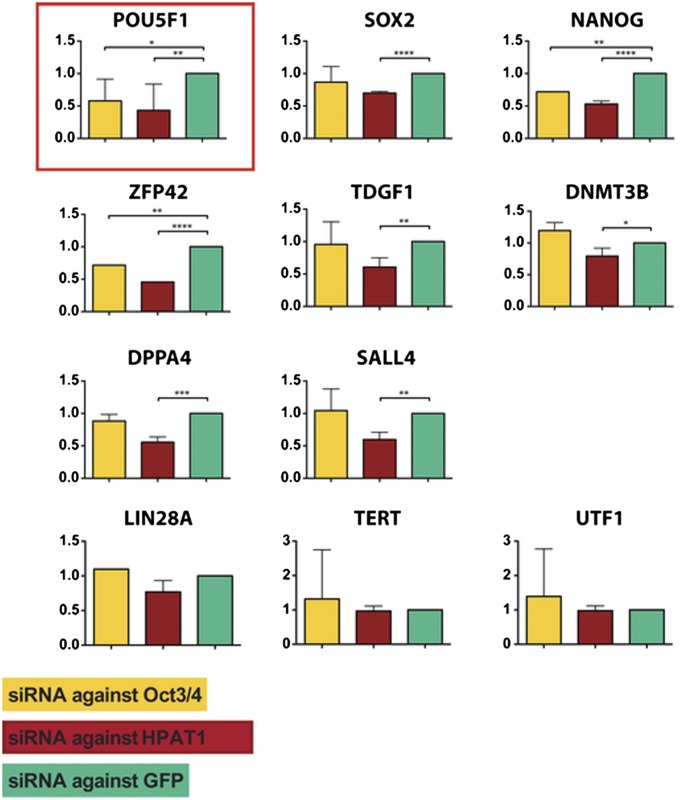
To confirm that the identified HPAT genes encoded functional elements, we performed knockdown experiments. For this purpose, we transfected H1 cells with siRNAs and analyzed gene expression 24 h posttransfection. Results indicated that reduction of the novel hESC-specific gene (HPAT1) resulted in significant down-regulation of expression of several pluripotency-associated factors including POU5F1, SOX2, NANOG, DPPA3, DNMT3B, SALL4, and TDGF1, in a fashion similar to that observed in a control sample where we down-regulated POU5F1 by transfecting a pool of four commercially available siRNAs (Fig. 4). Other genes like LIN28A, TERT, and UTF1 were not affected (Fig. 4), indicating a targeted effect of HPAT1 on the transcriptional network of pluripotency genes. Interestingly, the down-regulation of HPAT1 also resulted in down-regulation of approximately half of the novel genes whereas the other half were unaffected (SI Appendix, Fig. S10 A and B).
Fig. 4.
Functional effect of down-regulation of HPAT genes. Gene expression analysis by qPCR of different pluripotency-associated genes after down-regulation of GFP (negative control, in green), OCT4 (positive control, in yellow), and HPAT1 (in red). siRNAs for GFP and HPAT1 were derived by in vitro dicer-mediated digestion of the corresponding double-stranded mRNAs; siRNAs for OCT4 were designed in silico. Analysis was performed 24 h after the transfection.
Novel Gene Isoforms.
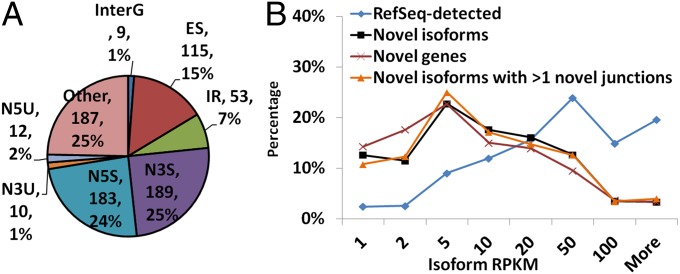
In addition to the novel genes, 655 identified isoforms contained both annotated junctions and novel junctions and thus represent novel isoforms of known genes. The novel junctions increase the diversity of the existing known isoforms. A total of 758 novel junctions from this subgroup of isoforms were categorized by type (Fig. 5A). Twenty-four percent and 25% of the novel junctions resulted from novel splice sites at the 5′ end and the 3′ end, respectively. Seven percent of the junctions accounted for intron retention events. Fifteen percent resulted from exon-skipping events. Nine novel junctions were defined “intergenic proximal” because they fell upstream or downstream of the annotated genes, thus extending the gene locus with novel exons and novel isoforms (Fig. 6). None of these novel splices have been reported in previous annotations.
Fig. 5.
Novel isoform identifications. (A) Pie chart of different novel junction types in 655 novel isoforms of existing genes. N5U, novel 5′-UTR; N3U, novel 3′-UTR; N5S, novel 5′-splice site; N3S, novel 3′-splice site; IR, intron retention; ES, exon skipping; InterG, intergenic proximal. Examples are in Fig. 6. (B) Abundance distributions of novel isoform predictions and annotated detections. Approximately 35% novel isoforms have RPKM >10.
Fig. 6.
Novel isoforms of existing genes with six different types of novel junctions. The genome browser setting is the same as in Fig. 2D. The GENCODE annotation is in the dark blue track. The novel junction uses are highlighted by a pink dashed box and are not reported by existing annotations but are supported by both long reads and short reads. Other: a novel exon in novel isoform TMEM142.2 is detected with two novel flanking junctions that are categorized in the “Other” group in Fig. 2C. InterG: A novel junction in novel isoform INO80C.1 is categorized as “intergenic proximal” junctions from annotated genes, but is a 3′-end junction of a novel isoform of INO80C (note that this gene is in the reverse strand). IR: A novel junction in novel isoform YTHDF1.1 indicates a splice within an annotated exon; i.e., the RefSeq annotation has a retained intron relative to YTHDF1.1. N3S: A novel junction in novel isoform MBD1.1 has a novel 3′-splice site (note that this gene is in the reverse strand). N5S: A novel junction in novel isoform TTC32.1 has a novel 5′-splice site (note that this gene is in the reverse strand). ES: Novel isoform HSD17B14.1 contains a novel junction that skips an annotated exon.
Without any novel junctions, novel combinations of annotated junctions can generate novel isoforms as well. A total of 876 novel isoforms are identified in this category. This finding indicates that the completeness of junction libraries does not guarantee the completeness of gene isoform identification. Although SGS data have shown impressive performance in junction detection at local regions, new techniques such as IDP from hybrid-sequencing data are needed in isoform identifications.
Compared with annotated isoforms, the expression levels of the novel genes and novel isoforms identified by IDP are lower overall (Fig. 5B). However, a significant proportion (35.58%) have reasonably high expression levels (RPKM >10). Thus, previous approaches have yielded incomplete annotations even for highly expressed genes. IDP will be useful in filling this gap.
Quantification of Isoform Abundance.
Estimates of isoform-specific gene expression from high-throughput SGS data rely on statistical models in which a short read that is consistent with two or more of the annotated isoforms is regarded as being more likely to be generated from the more abundant isoforms (12). Isoform abundance estimates based on these models are highly sensitive to the annotation. Two types of isoform annotation libraries are often used: (i) reference annotation libraries such as RefSeq or Ensembl or (ii) candidate isoform sets inferred computationally by SGS analysis tools such as Cufflinks and SLIDE. Because a reference annotation library is constructed based on all data from the same species, not all annotated isoforms in the library are truly expressed in a sample. The inclusion of annotated but not truly expressed isoforms can greatly increase the variability of the abundance estimates of the expressed isoforms—as discussed in section 4.2 of ref. 12, this shows up as a widening of the 95% interval for an isoform’s abundance when additional isoforms are included in the model. On the other hand, if the reference library is incomplete, then any nonannotated but truly expressed isoforms are completely missed and their contribution to short-read coverage may be incorrectly assigned to the annotated isoforms. These two effects can lead to serious errors in the abundance estimates based on a reference library. Finally, for multiexon genes there are usually many isoforms consistent with the exon and junction reads from SGS data. As a result, candidate isoform sets inferred directly from the SGS data may include many false isoforms that cause incorrect abundance estimation. In contrast, in our approach the error-corrected long reads are ideal for narrowing down the isoforms expressed in a sample, thus enabling much more reliable abundance quantification from SGS reads.
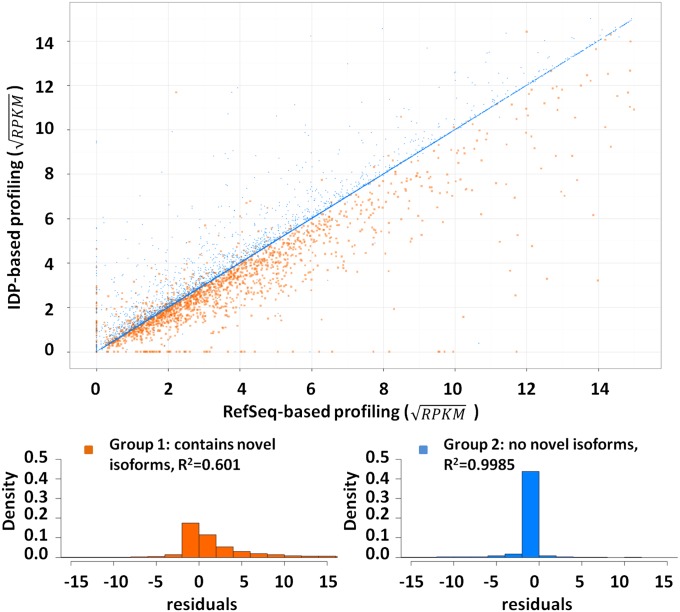
To evaluate the bias in quantification due to incomplete annotation, we computed the isoform expression levels from SGS short reads based on two different isoform libraries: RefSeq annotation and IDP isoform identification from hESC data (Fig. 7). There are 9,775 isoforms appearing in both libraries (common isoforms). Of these, 1,763 are from genes with at least one novel isoform (group 1, with novel isoforms) and 8,012 are from genes without any novel isoform discoveries (group 2, without novel isoforms). For genes in group 2, the expression of a common isoform, computed via the RefSeq library, is well correlated with that computed based on the IDP library (R2 = 0.9985). In contrast, for group 1 genes this correlation is much lower (R2 = 0.6008). Therefore, novel gene isoform discoveries have important effects in the accuracy of abundance estimations.
Fig. 7.
Isoform abundance estimation by IDP-identified hESC transcriptome and RefSeq annotation. The gene abundances of “common isoforms” (main text) are rescaled by a square-root transformation. The genes without novel isoforms (group 2) have stable abundance estimation and have high R2 of 0.9985 in linear regression (blue dots). In contrast, novel isoforms found in existing genes lead to a large range of abundance corrections. The residuals of linear regression show different distributions between the two groups. Group 1 (genes with novel isoforms) is highlighted in orange and group 2 in blue. The residuals of group 2 concentrate around 0, which indicates a small difference between two computations. However, group 1 has a heavy tail at the positive range. That is, most abundances of group 1 are corrected to lower values because the SGS reads must be shared with novel isoforms.
The linear regression of all 9,775 common isoforms shows the difference of abundance correction between group 1 and group 2 (Fig. 7). IDP-sample-specific estimations in group 1 genes tend to have lower RPKM because some SGS short reads must now be shared with the novel isoforms. This confirms that novel isoform identifications have a significant impact on the isoform abundance estimation within a gene. Therefore, our sample-specific IDP pipeline also leads to a more reliable isoform-specific expression estimation that will benefit downstream analyses.
Use of Gene Isoforms and Exon Junctions.
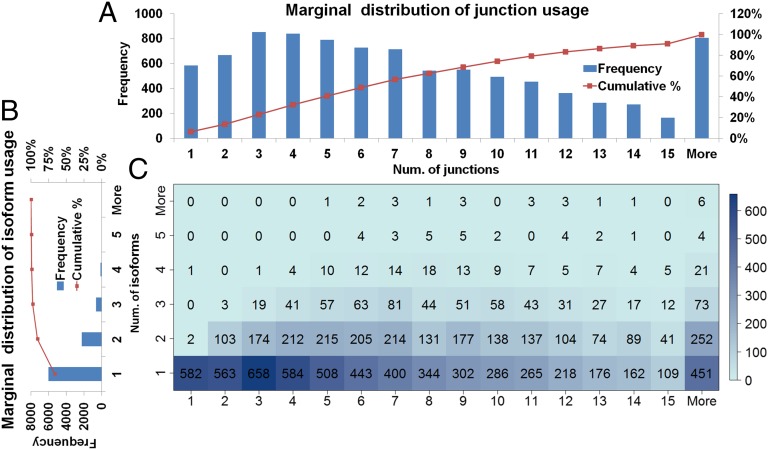
Although there are ∼20,000 multiexon genes with 35,000 ∼ 40,000 isoforms in RefSeq, the total number of isoforms identified in hESCs is only 13,292. A total of 12,976 isoforms (RefSeq annotated or unannotated) were identified from 4,312 genes of 5,851 RefSeq multiexon genes with RPKM >10. The identification rate is 73.70% among these RefSeq genes (Gene Isoform Detection and Prediction). A total of 91.18% express only one or two dominantly expressed isoforms in hESCs (Fig. 8). Gene functions within a cell may depend only on a subset of possible isoforms. Thus, there must exist sophisticated transcriptional and posttranscriptional regulatory mechanisms to select and express a correct subset of isoforms in certain cell conditions. On the other hand, some genes express many isoforms in hESCs, such as 14 isoforms from the ncRNA gene NCRNA00188 and 13 from mortality factor MORF4L2.
Fig. 8.
(A) Marginal distributions of numbers of junction use. (B) Marginal distributions of numbers of isoform use. (C) Joint distribution displayed as heat map of number of genes by number of junctions and number of isoforms. The numbers of genes are given in each bin. Most genes express only one to two isoforms. Note that the number of junctions and the number of expressed isoforms within a gene have no significant correlation.
In contrast to limited use of isoforms, the number of junctions used within a gene has a much larger range from 1 to 50. Thus, the IDP pipeline with hybrid sequencing data has identification power not only in simple genes but also in the complex genes with many alternative splices, such as the 49-junction gene RBM5 (RNA-binding protein 5) and the 50-junction gene DPP4 (adenosine deaminase complexing protein). This advantage of obtaining global gene structure with so many junctions is due to the TGS long reads and is not achievable by SGS short reads. In addition, the median number of junctions within a gene identified by IDP is 7, which is the same as in RefSeq annotation.
It is interesting to note, however, that the number of junctions within a gene has no significant correlation with the number of expressed isoforms (Fig. 8). This suggests that the number of expressed isoforms is largely determined by the need of the cell and does not depend on the complexity of the gene junctions.
Isoforms of Pluripotent Stem Cell Markers.
Many genes are expressed with great specificity in hESCs. Reliable and sensitive gene isoform identification enables improved observation of the use of isoforms in hESCs. Here we report isoforms from 15 pluripotent stem cell markers (Table 1). As with most genes, these markers have significant expression of only one to two annotated isoforms, most of which can be detected directly by error-corrected full-length cDNA reads. Two annotated isoforms KLF4 and E-cadherin are the exceptions and are predicted by IDP.
Table 1.
Isoform identification of pluripotency markers
| Pluripotency marker | RefSeq isoform identified | Novel isoforms identified |
| NODAL | NM_018055 | |
| TDGF1 | NM_001174136, NM_003212 | |
| PRMT5 | NM_001039619, NM_006109 | |
| KLF4 | NM_004235 | |
| TEAD4 | NM_003213, NM_201441 | |
| E-CADHERIN | NM_004360 | |
| LIN28A | NM_024674 | |
| ALPL/TNAP | NM_001127501, NM_000478 | |
| POU5F1 | NM_203289, NM_002701 | |
| NANOG | NM_024865 | |
| DNMT3B | Novel isoforms are identified with intron retention events | |
| ZFP42 | NM_174900 | A novel isoform has an alternative splicing site at the last exon |
| SALL4 | NM_020436 | A novel isoform (with 20% gene abundance) contains a novel exon |
| DPPA4 | NM_018189 | A novel isoform skips three exons |
| TERT | A novel isoform skips exon 7 and exon 8 and the exon-skipping junction is of high expression |
The transcriptional regulator of pluripotency, POU5F1, expresses two isoforms, which are distinct from the RefSeq-annotated isoform NM_001173531. In addition, a few long reads are observed that exhibit intron retentions within two detected isoforms (NM_203289 and NM_002701). However, the corresponding intronic coverage is not high, so these intron retention events may be from preprocessed transcripts. Similar intron retention events with low expression also exist in NANOG, TERT, and DNMT3B.
Compared with intron retentions that occur at low frequency, novel isoforms with highly expressed exon-skipping events are more interesting. For example, in the DPPA4 locus, a RefSeq-annotated isoform is expressed but a novel isoform skipping three exons contributes ∼17% of the total gene abundance (SI Appendix, Fig. S11A). In TERT, a novel isoform displaying cassette exon skipping junctions contributes as much as 54% of the gene abundance. Alternative splicing may be one mechanism of regulation of the telomerase activity of TERT.
As a third example, we note that a novel exon (not in RefSeq) is observed in SALL4. Although this exon was reported in an isoform of SALL4 in Ensembl, the Ensembl-annotated isoform is a partial fragment of the IDP-predicted isoform (SI Appendix, Fig. S11B).
ncRNA Identification.
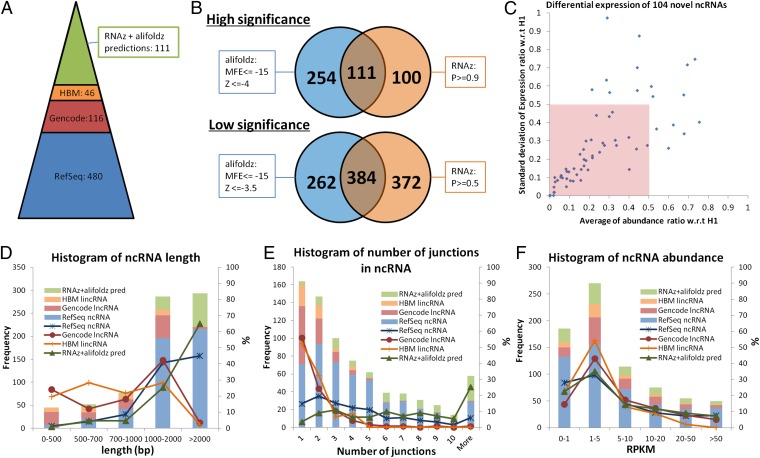
In addition to genes coding for proteins, 480 annotated (RefSeq) multiexon ncRNAs were identified from IDP, 85% (406/480) of which are direct detections. The remaining 3,455 non–RefSeq-annotated IDP multiexon isoform predictions (green plus red plus light blue fractions in Fig. 1B: 2,103 + 325 + 1,027 = 3,455) are compared with the GENCODE lncRNA library (V12), which was created through manual curation of available cDNA and EST data. A total of 116 are found within this lncRNA library (Fig. 9A). In addition to RefSeq and GENCODE ncRNAs, the remaining IDP isoforms were compared with the Human Body Map (HBM) long intergenic noncoding RNA (lincRNA) library, which unifies existing annotation sources with transcripts assembled from SGS RNA-Seq data collected across 24 tissues and cell types (28). Forty-six are found from this lincRNA library. To further study the remaining predictions, two functional RNA structure prediction tools, RNAz (29, 30) and alifoldz (31), were applied to predict the secondary structure potential of the novel ncRNAs remaining in the IDP output. Previous research had suggested the use of two stringency levels for predictions from two methods (32) (Fig. 9B and SI Appendix, Methods). To have a more confident ncRNA library for hESC study, we used a high-stringency mode for each method [P ≥ 0.9 for RNAz; minimum free energy (MFE) ≤ −15 and Z score ≤ −4 for alifoldz]. We then took the intersection of the two sets of predictions. In total, 111 IDP novel isoforms (P value for overlap <0.0001) are predicted as structured ncRNAs by both methods. We also computed their abundance ratio in 16 human tissues with respect to H1. The abundance ratios of 104 novel ncRNAs from all 16 human tissues with respect to (w.r.t.) H1 can be computed, whereas 7 targets have insufficient short-read coverage in H1. A total of 48.08% (50/104) of novel ncRNAs have an averaged abundance ratio smaller than 0.5 with SD smaller than 0.5 (pink box in Fig. 9C), which indicates that these genes tend to have relatively higher expression in H1.
Fig. 9.
Noncoding RNA identification: the distributions of length, number of junctions, and abundance. (A) Annotated ncRNA identifications and novel ncRNA predictions. A total of 480 multiexon RefSeq-annotated ncRNAs are identified from H1. After filtering out RefSeq isoforms, the remaining IDP output contains 116 GENCODE-annotated lncRNAs. After filtering out RefSeq and GENCODE isoforms, 46 HBM lincRNAs are identified. The intersection of high-significance RNAz and alifoldz predictions of the remaining novel isoforms contains 111 putative ncRNAs. (B) RNAz and alifoldz are used to identify the ncRNA from 2,428 isoform predictions. Two stringency levels are suggested by the developers. For all subsequent analyses, we use the intersection of the high-stringency outputs from the two methods as our predicted ncRNAs. (C) Differential expressions of 104 novel ncRNAs w.r.t. H1. Seven of 111 novel ncRNA predictions are not included, because of insufficient short-read coverage in H1. Fifty novel ncRNAs (inside the pink box) have an averaged abundance ratio smaller than 0.5 with SD smaller than 0.5. (D) Length distribution of IDP-identified isoforms of RefSeq ncRNA, GENCODE lncRNA, HBM lincRNA, and RNAz/alifoldz predictions. (E) Distribution of number of junctions of IDP-identified isoforms of RefSeq ncRNA, GENCODE lncRNA, HBM lincRNA, and RNAz/alifoldz predictions. (F) Abundance distribution of IDP-identified isoforms of RefSeq ncRNA, GENCODE lncRNA, HBM lincRNA, and RNAz/alifoldz predictions.
GENCODE lncRNAs and HBM lincRNAs found in hESCs have very different length distributions compared with ncRNAs predicted by our method or found in RefSeq annotations (Fig. 9D). Only 4 of the GENCODE lncRNAs (3.4%) and 1 HBM lincRNA (2.2%) are longer than 2,000 bp, whereas 216 RefSeq-annotated (45%) IDP isoforms and 72 predictions (65%) from novel IDP isoforms are longer than 2,000 bp with the averaged lengths around 2,300 bp (Table 2). Furthermore, the isoforms from the latter two IDP groups have more junctions: 183 (38%) RefSeq-annotated IDP hits and 187 predictions (70%) from novel IDP hits have more than 4 junctions (that is, at least 6 exons), whereas only 6 of GENCODE lncRNAs and none of the HBM lincRNAs have more than 4 junctions (Fig. 9E). The average number of junctions is 1.72 in the HBM lincRNA group and 1.91 in GENCODE lncRNAs, which is far smaller than 4.52 and 7.67 in the RefSeq group and IDP novel ncRNAs.
Table 2.
The mean and median values of lengths, number of junctions, and abundances of four categories of ncRNAs
| ncRNA Database | Length, bp | No. junctions | Abundance, RPKM |
| RefSeq | 2,248, 1,832 | 4.52, 4 | 55.45, 3.03 |
| GENCODE | 1,012, 919 | 1.91, 1 | 10.83, 4.40 |
| HBM | 838, 832 | 1.72, 1 | 4.63, 2.27 |
| IDP, novel ncRNA | 2,402, 2,454 | 7.67, 7 | 17.97, 3.56 |
Isoforms in HBM lincRNA and GENCODE lncRNA libraries, regardless of tissue type, are generally much shorter and contain fewer junctions compared with those identified by IDP in hESCs (Fig. 9 D and E). However, such striking differences are not seen in the abundance distribution (Fig. 9F). These results suggest that SGS data and manual curation of cDNA and EST data are likely to underestimate the size and complexity of ncRNA isoforms. By combining SGS and TGS data, our approach can provide a much more accurate characterization of ncRNAs. Finally, a significant percentage (27.93%, 31 of 111) of the novel IDP lncRNA isoforms predicted by our method are highly expressed (RPKM >10, Fig. 9F). Further investigation of this set of novel, long, and highly expressed ncRNAs will be worthwhile.
Discussion
We have identified de novo isoform discovery and assembly as a fundamental obstacle to progress in RNA-Seq analysis. Innovations on both the conceptual level and the technical level are presented to overcome this obstacle. On the conceptual level, we distinguish two different types of RNA-Seq analysis and argue that it is desirable to develop different experimental and computational approaches for them separately. In discovery, analysis aims to identify all genes and gene isoforms that are being transcribed in a particular cell sample. Once it is done well for a particular species and cell type, it will contribute to the improvement of gene annotation for that species and will benefit all subsequent RNA analysis on that species. For quantification, on the other hand, analysis aims to estimate expression (preferably also allele-specific) levels of the gene isoforms in an annotation that specifies the set of isoforms actually present. It is anticipated that quantification analysis will be performed on a very large number of samples in translational research and in clinical applications. In these contexts it is very important to have high throughput and low cost.
A large amount of statistical and computational research has been performed over the past 5 y on how to analyze short-read RNA-Seq data for both types of analyses. It appears that many effective methods for quantification are now readily available. Most of these methods have incorporated variations of the concepts of RPKM from the Wold laboratory (1) as an expression index and the Poisson deconvolution model (12) as the statistical framework for isoform-level inference. As for discovery, several algorithms, such as Tophat, SpliceMap, and MapSplice, have been developed to map splice junctions with high sensitivity and specificity. Based on the detected junctions and short-read counts, several methods such as Cufflinks and SLIDE have also been proposed to assemble the set of expressed isoforms. Our belief is that although these methods can provide useful information, the assembly problem cannot be solved effectively based on short-read RNA-Seq data alone. Instead, we pursued a hybrid TGS-SGS sequencing approach that at this moment is more costly than SGS alone. However, the decoupling of the task of discovery from quantification allowed us to conclude that even with its higher cost, hybrid sequencing is an attractive approach for discovery, as the cost will be offset by the gained accuracy in all subsequent quantification analysis due to the resulting improvement of annotation.
On a technical level, we have developed an analysis methodology to combine the specific strengths of TGS and SGS, represented by PacBio and Illumina sequencing, respectively. Our methodology involves new statistical modeling and inference. Although the error-corrected long reads are capable of capturing many transcripts in full length, there will always be a substantial fraction of long reads that capture only partial transcript fragments. This is particularly likely if the transcript is very long. It is important to be able to combine the information from the full-length long reads and partial transcript fragment reads. Furthermore, there is also statistical information useful for isoform reconstruction, and this information can be substantial because the number of short reads will be an order of magnitude larger than the number of long reads. To effectively use all three types of reads for isoform reconstruction, we developed statistical models for how such reads are sampled from the underlying isoforms and derive appropriate algorithms to infer the isoforms based on such models. To our knowledge, such models are not available currently, and the development of these represents significant innovation in statistical methodology.
Finally, to test our method, we generated 2 million mappable long reads on H1 hESCs and analyzed them in conjunction with existing short-read RNA-Seq data. Our analysis not only illustrates the effectiveness of our method, but also provides a rich resource for stem cell biologists interested in a deeper characterization of the ESC transcriptome. Indeed, further characterization of the novel loci identified here indicates that a subset is specifically and highly expressed in pluripotent human stem cells and embryos. Moreover, silencing of these genes modulates pluripotency gene expression. Thus, at least a subset of these gene loci may have functional roles in both human development and pluripotent stem cells. Taken together, these results demonstrated the effectiveness of our methodology and suggest that gene identification, even in well-characterized cell lines and tissues, is far from complete.
Experimental Procedures
Cell Culture.
Male human embryonic stem cells (H1) were used for our study. The cells were routinely cultured in feeder-free conditions on Matrigel (BD) and in mTeSR1 (Stem Cell Technologies). RNA-Seq was performed on cells between passages 50 and 55. The undifferentiated state of the hESCs was assayed by immunofluorescence for transcription factors OCT4 and NANOG and for the surface markers SSEA4, TRA-1-60, and TRA-1-81 (SI Appendix, Fig. S12). Pluripotency, assessed by teratoma assay, revealed the capacity of hESCs to form in vivo derivatives of the three germ layers (SI Appendix, Fig. S13).
RNA and cDNA Preparation.
Total RNA was prepared by TRIzol extraction (Ambion; www.ncbi.nlm.nih.gov/pubmed/2440339) and treated with RNase-free DNase I to degrade contaminating genomic DNA. This was followed by a second extraction with acid-phenol-chloroform and the RNA was precipitated in 0.5 M ammonium acetate and 2.5 vol ethanol. The integrity of the RNA was tested by a Bioanalyzer RNA Nano assay (Agilent). PolyA RNA was purified from the total RNA (>100 µg), using magnetic oligo-dT beads (Dynal) according to the manufacturer’s recommended protocol.
Two different manufacturer’s kits and accompanying protocols for generating full-length cDNAs were used: Clontech SMARTer PCR and the Invitrogen SuperScript Full-Length cDNA kit. Each uses a retroviral reverse transcriptase to initiate cDNA synthesis at the 3′ polyA tail, but the two kits use different mechanisms to capture the 5′-end information. The Clontech method relies upon template switching to an exogenous tag oligonucleotide when the reverse transcriptase reaches the 5′ end of the molecule. The Invitrogen method employs an RNase I digestion and subsequent 7-methyl-guanosine affinity step to purify the RNA-DNA hybrids that represent a full copy of the transcript. This is followed by a DNA ligase step to add a defined sequence to the 3′ ends of the full-length cDNA, before reverse transcription. For each method, 1 µg of polyA RNA served as the input material into the recommended protocol and the manufacturers’ methods were followed up through second-strand cDNA synthesis. At this point, PCR (12–15 cycles; Phusion DNA polymerase; New England Biolabs) was used to amplify the double-stranded cDNA by using specific primers directed to the defined sequences present at the 5′ and 3′ ends of the cDNAs. Long extension times of 3.5 min ensure that longer molecules can be captured as well. The cDNA libraries were assayed by fluorimetry and Bioanalyzer assays. Both Clontech and Invitrogen kits are reproducible. For example, housekeeping genes at different sizes (ACTB, 1,808 bp; GADPH, 1,310 bp; SARS, 2 291 bp; and IARS, NM_002161, 4,406 bp and NM_013417, 4,510 bp) are detected by full-length long reads from the two preparations (SI Appendix, Figs. S14–S17).
Size selection was used with each of the cDNA libraries to enrich for larger cDNA molecules. Briefly, 500 ng of double-stranded cDNA was separated on a 1× TAE agarose gel and stained with SybrSafe. The gel was visualized on blue light to avoid DNA damage and, using a size standard in a parallel lane, size fractions were cut from the gel from 1 kb to 2 kb, from 2 kb to 3 kb, and from 3 kb to 10 kb. The DNA was recovered from each fraction, using a silica column kit (Zymo), and the DNA was then subjected to additional PCR to enrich for molecules in each size fraction. For both non–size-selected and size-selected PCR reactions, the double-stranded cDNA was purified via AMPure PB beads (Pacific Biosciences), using a 0.5× ratio of bead slurry to PCR.
Single-Molecule Sequencing.
Sequencing templates were prepared using the manufacturer’s SMRTbell template protocol (Pacific Biosciences Large Template Prep kit), with a minimum of 1 µg of dsDNA input into each library procedure. After exonuclease digestion of incomplete SMRTbell templates, the libraries were purified by three sequential 0.5× AMPure PB purifications (Pacific Biosciences.) Libraries were quantified by fluorimetry and assayed for quantity and size distribution by Bioanalyzer. Sequencing was performed on the Pacific Biosciences RS, using standard protocols for C2 chemistry with the C2 or XL enzyme.
Supplementary Material
Acknowledgments
We thank Dr. Arnold Ludwig Hayer, Dr. Tobias Meyer, Dr. Laughing Bear Torrez, and Dr. Diana Cepeda for help in designing siRNA experiments and for technical help in performing the knockdown experiments. K.F.A. and W.H.W. were supported by the National Human Genome Research Institute (R01HG005717). B.A.W. was partially supported by the California Institute of Technology Beckman Center for Functional Genomics. The Illumina short-read data were generated as part of the Encode project (National Human Genome Research Institute Grants U54 HG004576 and U54 HG006998 to Barbara Wold). V.S., J.D.D., and R.A.R.-P. were supported by the National Heart, Lung, and Blood Institute (U01HL100397) and the National Institute of Child Health and Human Development (U54 HD068158).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51861).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320101110/-/DCSupplemental.
References
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell JA, et al. Nuclear RNA sequencing of the mouse erythroid cell transcriptome. PLoS ONE. 2012;7(11):e49274. doi: 10.1371/journal.pone.0049274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333(6038):53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rederstorff M, et al. RNPomics: Defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38(10):e113. doi: 10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38(14):4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25(8):1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiller D, Jiang H, Xu W, Wong WH. Identifiability of isoform deconvolution from junction arrays and RNA-Seq. Bioinformatics. 2009;25(23):3056–3059. doi: 10.1093/bioinformatics/btp544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JJ, Jiang C-R, Brown BJ, Huang H, Bickel PJ. Sparse linear modeling of RNA-seq data for isoform discovery and abundance estimation. Proc Natl Acad Sci USA. 2011;108:19867–19872. doi: 10.1073/pnas.1113972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller D, Wong WH. Simultaneous isoform discovery and quantification from RNA-seq. Stat Biosci. 2013;5(1):100–118. doi: 10.1007/s12561-012-9069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40(Database issue):D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English AC, et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7(11):e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Au KF, Underwood JG, Lee L, Wong WH. Improving PacBio long read accuracy by short read alignment. PLoS ONE. 2012;7(10):e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura T, et al. Monitoring early differentiation events in human embryonic stem cells by massively parallel signature sequencing and expressed sequence tag scan. Stem Cells Dev. 2004;13(6):694–715. doi: 10.1089/scd.2004.13.694. [DOI] [PubMed] [Google Scholar]
- 21. Harrow J, et al. (2006) GENCODE: Producing a reference annotation for ENCODE. Genome Biol 7(Suppl 1):S4.1–9. [DOI] [PMC free article] [PubMed]
- 22.Bernstein BE, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu F, et al. The UCSC known genes. Bioinformatics. 2006;22(9):1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 26.Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC. Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat Genet. 1993;4(4):373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- 27.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci USA. 2005;102(7):2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruber AR, Findeiß S, Washietl S, Hofacker IL, Stadler PF. RNAz 2.0: Improved noncoding RNA detection. Pac Symp Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]
- 31.Washietl S, Hofacker IL. Consensus folding of aligned sequences as a new measure for the detection of functional RNAs by comparative genomics. J Mol Biol. 2004;342(1):19–30. doi: 10.1016/j.jmb.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Washietl S, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17(6):852–864. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.