Significance
The arbuscular mycorrhizal symbiosis between fungi of the Glomeromycota phylum and plants involves more than two-thirds of all known plant species, including important crop species. This mutualistic symbiosis, involving one of the oldest fungal lineages, is arguably the most ecologically and agriculturally important symbiosis in terrestrial ecosystems. The Glomeromycota are unique in that their spores and coenocytic hyphae contain hundreds of nuclei in a common cytoplasm, which raises important questions about the natural selection, population genetics, and gene expression of these highly unusual organisms. Study of the genome of Rhizophagus irregularis provides insight into genes involved in obligate biotrophy and mycorrhizal symbioses and the evolution of an ancient asexual organism, and thus is of fundamental importance to the field of genome evolution.
Keywords: carbohydrate-active enzymes, effector, fungal evolution, glomales, mutualism
Abstract
The mutualistic symbiosis involving Glomeromycota, a distinctive phylum of early diverging Fungi, is widely hypothesized to have promoted the evolution of land plants during the middle Paleozoic. These arbuscular mycorrhizal fungi (AMF) perform vital functions in the phosphorus cycle that are fundamental to sustainable crop plant productivity. The unusual biological features of AMF have long fascinated evolutionary biologists. The coenocytic hyphae host a community of hundreds of nuclei and reproduce clonally through large multinucleated spores. It has been suggested that the AMF maintain a stable assemblage of several different genomes during the life cycle, but this genomic organization has been questioned. Here we introduce the 153-Mb haploid genome of Rhizophagus irregularis and its repertoire of 28,232 genes. The observed low level of genome polymorphism (0.43 SNP per kb) is not consistent with the occurrence of multiple, highly diverged genomes. The expansion of mating-related genes suggests the existence of cryptic sex-related processes. A comparison of gene categories confirms that R. irregularis is close to the Mucoromycotina. The AMF obligate biotrophy is not explained by genome erosion or any related loss of metabolic complexity in central metabolism, but is marked by a lack of genes encoding plant cell wall-degrading enzymes and of genes involved in toxin and thiamine synthesis. A battery of mycorrhiza-induced secreted proteins is expressed in symbiotic tissues. The present comprehensive repertoire of R. irregularis genes provides a basis for future research on symbiosis-related mechanisms in Glomeromycota.
The arbuscular mycorrhizal symbiosis between fungi in the Glomeromycota, a distinctive phylum of the early diverging Fungi, and plants involves more than two-thirds of all known plant species, including important crop species such as wheat and rice. This mutualistic symbiosis is widely hypothesized to have promoted the evolution of land plants from rootless gametophytes to rooted sporophytes during the mid-Paleozoic (1, 2). These arbuscular mycorrhizal fungi (AMF) perform vital functions in the phosphorus cycle (3) that are fundamental to sustainable crop plant productivity (4). They also drive plant diversity (5). The extraradical mycelium of the symbiont acts as an extension of the root system and increases the uptake of key nutrients, particularly phosphorus (3). Furthermore, because the colonization of plants by AMF also can result in a 20% net increase in photosynthesis, these fungi make a very large, poorly understood contribution to the global carbon cycling budget of terrestrial ecosystems.
The Glomeromycota are unique in that their spores and coenocytic hyphae contain multiple nuclei in a common cytoplasm. With no known sexual cycle (6), AMF reproduce clonally through large and multinucleated spores (Fig. 1), although a conserved meiotic machinery (7, 8) might allow individuals to shuffle their genetic material (9, 10) and reduce their mutational load. Genetic variation has been observed within AMF in ribosomal DNA and in other regions of the genome (11). It has been hypothesized that AMF maintain an assemblage of genetically different nuclei (heterokaryosis) and transmit them from generation to generation (12, 13). Another study failed to find evidence for heterokaryosis, however, and the authors suggested that the genetic variation that they observed potentially could be due to polyploidy, although no studies of ploidy were conducted (14). A recent study of Rhizophagus irregularis (but not isolate DAOM-197198) provided further evidence in favor of heterokaryosis (15), so this remains an open question. Another hypothesis, specific for multicopy ribosomal DNA, is that variation among copies could exist within nuclei of a homokaryotic AM fungus and this is supported by some studies (14, 16).
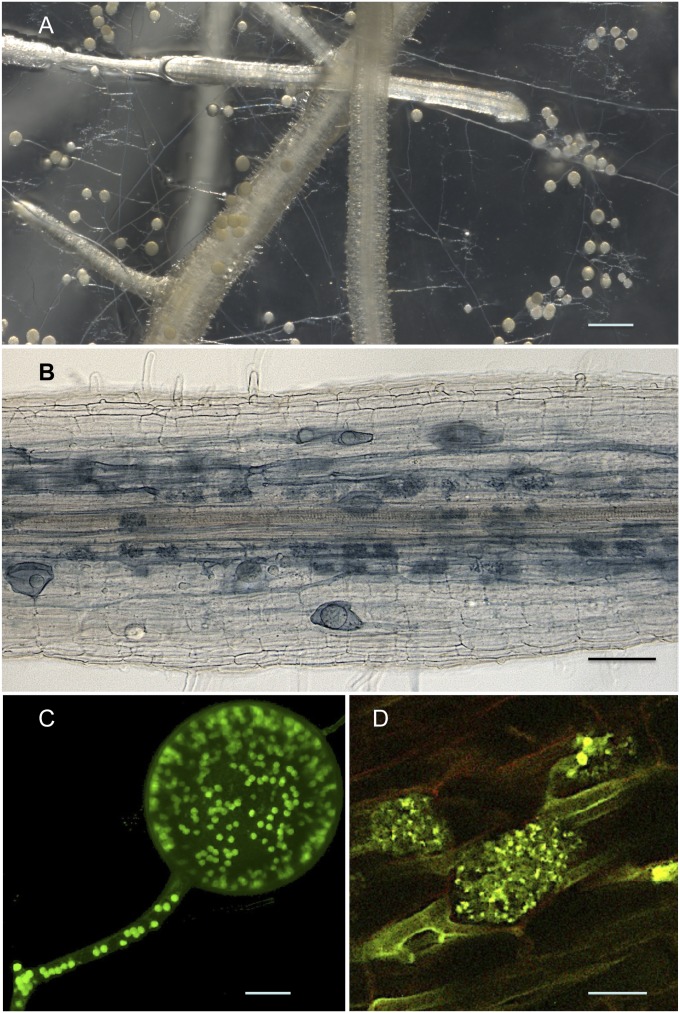
Fig. 1.
(A) In vitro coculture of R. irregularis with carrot roots showing extraradical hyphae and spores. (Scale bar: 750 µm.) (B) Colonized carrot root showing fungal colonization that is restricted to the root cortex where the fungus produces vesicles and/or intraradical spores, and arbuscules. (Scale bar: 100 µm.) (C) Typical multinucleated asexual spore of R. irregularis and its attached coenocytic hyphae observed by confocal laser scanning microscopy. Nuclei were stained using SYTO Green fluorescent dye and are shown as green spots. (Scale bar: 10 µm.) (D) Arbuscules, highly branched structures formed by the fungus inside cortical cells, are considered the main site for nutrient exchanges between the mycorrhizal partners. The yellow-green fungal structures are detected by wheat germ agglutinin-FITC labeling on root sections, whereas PCWs are shown in red. (Scale bar: 10 µm.)
Here we introduce the assembly and annotation of the genome of R. irregularis DAOM-197198 (formerly Glomus intraradices) in association with transcriptome data, and show that it is remarkably different from other sequenced fungal genomes in content and organization. We focus on the genome polymorphism, annotation, and transcript profiling of gene families likely to be involved in symbiosis, to reveal adaptation processes for growth in planta.
Results and Discussion
Genome Assembly.
To investigate the gene repertoire and genomic polymorphism of R. irregularis, we sequenced the genomic DNA from multinucleated hyphae of the strain DAOM-197198 grown in root culture of carrot (Daucus carota) (Fig. 1). The size of the genome assembly is 101 megabases (Mb), and the coding space is 98% complete on the basis of conserved core eukaryotic single-copy genes (17) (SI Appendix, sections 1.1–1.3, Figs. S1–S5). As expected, this genome is rich in A and T bases (A + T content, 72%) (Fig. 2). Based on flow cytometry assays and Feulgen densitometry measurements, the size of the DAOM-197198 genome has been estimated as 154.8 ± 6.2 Mb (18). Using the frequency distribution of 17-base oligomers in the usable sequencing reads to determine sequencing depth (19), we obtained an estimated genome size of 153 Mb (SI Appendix, Fig. S6). Thus, the genome size of R. irregularis is among the largest fungal genome sequenced to date, along with the obligate biotrophic powdery mildews (20) and the ectomycorrhizal symbiont Tuber melanosporum (21). Interestingly, no contigs corresponding to multiple haplotypes were identified. Transposable elements (TEs) compose 11% of the assembly (Fig. 2 and SI Appendix, section 1.5, Table S1 and Figs. S3–S5), but fosmid Sanger sequencing showed a much higher TE abundance (36%), including several retrotransposons. The long (9–25 kb), highly repetitive, and nested nature of the transposons (SI Appendix, Fig. S4) is the main explanation for the observed fragmentation of this assembly. Assuming that the TE abundance of fosmids (36%) holds for the whole genome, this would contain ∼55 Mb of repeated sequences, possibly reflecting a lack of efficient control mechanisms to prevent the expansion of repetitive elements and their subsequent elimination.
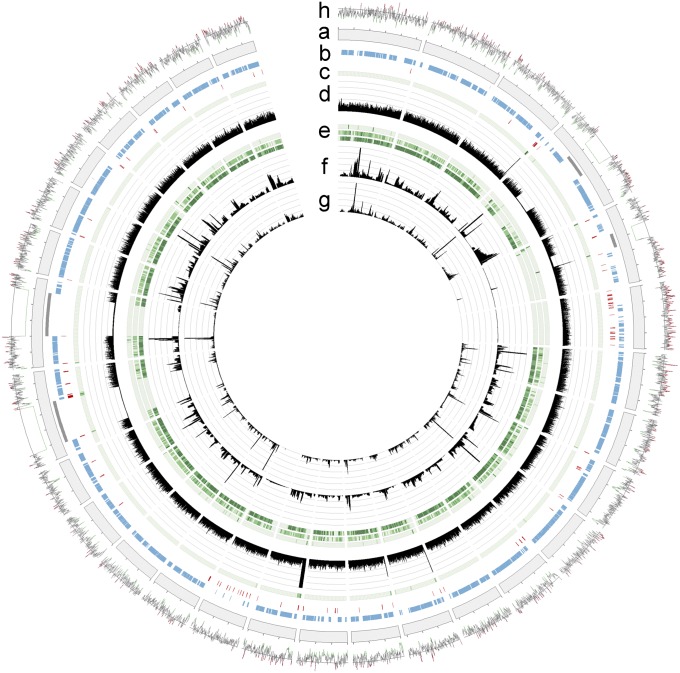
Fig. 2.
Circos circular visualization of the genome assembly. (A) The 30 largest scaffolds of the genome assembly. (B) Locations of gene models (blue), repeated elements (red) and sequence gaps (gray). (C) Genomic SNP density. (D) Read coverage on genome (scale, 0–100). (E) Expressed gene SNP density (outer to inner: RNA-Seq tags from DAOM-197198 spores, R. irregularis C2 spores, and R. diaphanum spores). (F) Transcript coverage, RNA-Seq reads from spores (scale, 0–10,000). (G) Transcript coverage, RNA-Seq reads from symbiotic roots (scale, 0–200). (H) Guanosine and cytosine (GC) content based on a sliding window of 100 bp (red, >40%; green, <20%; midline, 33%).
Genome Polymorphism.
Evidence exists from this and other Glomeromycotan species that these fungi are heterokaryotic, that is, harbor genetically different nuclei (11, 12). To investigate whether large differences could exist among nuclei, we analyzed the assembled contigs for local similarities to other contigs within the assembly. A BLAST search of all contigs against all contigs was carried out. The proportion of contigs sharing significant similarity (>90% identity) with at least one other contig of >1,000 bp was low (5.6%) and involved mostly repetitive sequences. Neither segmental duplication nor distinct haplotypic contigs were detected, suggesting that the assembled data are not composed of multiple genomes.
We also investigated the possibility that alleles had been collapsed during the assembly through genomic and RNA-Seq read mapping and single nucleotide polymorphism (SNP) calling (SI Appendix, section 1.4). The polymorphism in the assembled genome was estimated as 0.43 SNP per kb (Table 1). The SNP rate for expressed genes was identical (0.40 SNP per kb) as measured by mapping DAOM-197198 RNA-Seq reads to the assembly (Table 1), and only a few genes were found to have more than three SNPs (SI Appendix, Table S2). Using identical coverage and quality thresholds, we estimated the SNP rate of the assembled genomes as 0.06 SNP per kb in the homokaryotic ascomycete T. melanosporum and 0.78 SNP per kb in the dikaryotic basidiomycete Laccaria bicolor. These findings allow us to reject the hypothesis that large sequence polymorphism occurs among coexisting nuclei in the DAOM-197198 isolate; however, the density of SNPs suggests some allelic variation among nuclei, likely reflecting the limited opportunities for recombination and related sequence homogenization during the life cycle.
Table 1.
SNP and substitution density in the R. irregularis genome and transcriptome
| SNP or substitution per kb | ||||
|---|---|---|---|---|
| DAOM-197198 genome | DAOM-197198 exons | C2 transcriptome | R. diaphanum transcriptome | |
| Genomic reads | ||||
| Intrastrain SNPs per kb | ||||
| Genomic, raw | 1.26 | |||
| Genomic, filtered* | 0.43 | |||
| Genomic, exons, filtered* | 0.43 | |||
| RNA-Seq reads | ||||
| No. of intrastrain SNPs per kb | ||||
| DAOM-197198 | 0.40 | |||
| C2 | 0.25 | |||
| R. diaphanum | 0.22 | |||
| No. of interstrain SNPs/ substitutions per kb |
||||
| DAOM-197198–C2 | 2.6 | 2.3 | ||
| DAOM-197198–R. diaphanum | 8.0 | 9.3 | ||
| C2–R. diaphanum | 7.7 | 9.2 | ||
Illumina genomic reads from R. irregularis DAOM-197198 were mapped to the R. irregularis Gloin1 assembly. RNA-Seq reads from R. irregularis DAOM-197198, R. irregularis C2, and R. diaphanum were mapped to the R. irregularis DAOM-197198 exons, R. irregularis C2 transcriptome, and R. diaphanum transcriptome, respectively. Fixed differences between species are termed substitutions, and variable positions within species are SNPs.
Filtered values correspond to values without positions with coverage <5 and coverage in the top 5%, to remove potential artifactual SNPs caused by repetitive and paralogous sequences.
In contrast, the SNP/substitution density was 6.5- and 20-fold higher when RNA-Seq reads from the strains R. irregularis C2 (2.6 SNPs per kb) and R. diaphanum (8.0 substitutions per kb), respectively, were mapped to the DAOM-197198 genome (Table 1). We also assessed the intrastrain and interstrain/species SNPs/substitutions by mapping RNA-Seq reads to assembled transcripts. The intrastrain SNP density of the transcripts was 0.40, 0.25, and 0.22 per kb for the DAOM-197198, C2, and MUCL43196 isolates, respectively. The interstrain SNP/substitution density of the transcripts ranged from 2.3 for DAOM-197198 vs. C2 to 9.3 for DAOM-197198 vs. R. diaphanum MUCL43196 (Table 1), confirming significant intraspecific and interspecific genetic variability. Only 291 SNPs/substitutions were shared among all strains analyzed (SI Appendix, Fig. S7).
We calculated the number of synonymous substitutions per synonymous site of paralogs, but found no evidence for any recent whole genome duplication. The distribution of paralog duplication age (SI Appendix, section 1.8, Fig. S8) did not follow the typical steep exponential decay pattern (22) that is the hallmark of gene birth and death by constantly occurring small-scale duplication events. Gene loss in DAOM-197198 may occur at a much lower rate than typically observed; however, eight hidden components contributing to the distribution were detected (SI Appendix, Fig. S8), potentially representing remnants of large-scale duplication events under an alternative scenario. Strikingly, all but the model component representing the youngest paralogs were enriched for genes annotated to contribute to biological processes related to phosphorus metabolism and signaling via phosphorylation (SI Appendix, Fig. S9), the vast majority of which were annotated as kinases. We suggest that the expansion of the kinase-based signaling network observed in the R. irregularis genome (see below) may be derived from frequent retention of duplicated kinase genes over a long period.
The Rhizophagus Gene Repertoire.
Of the 28,232 protein-coding genes predicted (Fig. 2 and SI Appendix, Figs. S1 and S10), 23,561 high-confidence genes had transcriptomic support (RNA-Seq) and/or showed sequence similarity to documented proteins and/or domains (SI Appendix, sections 1.6 and 1.8 and Tables S3 and S4). Only 62% of the high-confidence genes showed sequence similarity to documented proteins and/or domains (SI Appendix, Figs. S11 and S12). Compared with representative sequenced fungi, including the taxonomically related Mucoromycotina, the percentage of proteins encoded by species-specific genes (SI Appendix, Fig. S12) and multigene families (SI Appendix, Fig. S13) in R. irregularis was among the highest observed. Expansion of protein family sizes was prominent in the lineage-specific multigene families (SI Appendix, Fig. S13 and Table S5), but marked gene family expansions were also seen in genes predicted to have roles in signal transduction mechanisms [e.g., tyrosine kinase-like genes (TKLs)], in protein–protein interactions (e.g., Sel1-domain-containing proteins), and RNA interference-related mechanisms (e.g., the Argonaute proteins) (SI Appendix, Tables S6 and S7). Notably, several TKL-containing proteins are associated with Sel1 repeats and were highly expressed in germinating spores and intraradical mycelium (SI Appendix, Fig. S14). Together with RNA-dependent RNA polymerases, DICER (IPR011545), and C-5 cytosine-specific DNA methylases, the unusually high number of Argonaute genes (SI Appendix, Table S7) are likely involved in silencing the abundant TEs.
We identified gene families with a smaller size in R. irregularis compared with other fungi (SI Appendix, Table S8). The inability of R. irregularis to grow in vitro suggests that the obligate biotroph genome may lack genes typically present in autotrophic fungi. Thus, we systematically searched for genes absent in R. irregularis, the obligate biotrophic pathogen Blumeria graminis (20), and early diverging Mucoromycotina and Chytridiomycota genomes (23) but present in the well-annotated yeast (Saccharomyces cerevisiae) genome (SI Appendix, Table S9). Genes encoding enzymes of primary metabolism are retained in R. irregularis [(Kyoto Encyclopedia of Genes and Genomes; KEGG) Metabolic Pathways database; jgi.doe.gov/Rhizophagus], but several key genes are missing in the genome assemblies of both the obligate biotrophs R. irregularis and B. graminis (see below), suggesting that the lack of these genes is an evolutionary adaptation to the obligate biotrophy.
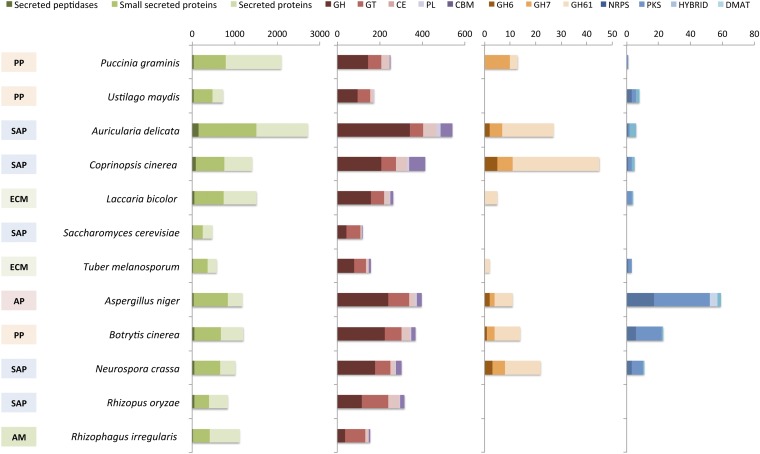
Like obligate biotrophic pathogens (20) and ectomycorrhizal symbionts (21, 24), R. irregularis has a decreased repertoire of genes involved in the degradation of plant cell wall (PCW) polysaccharides and in the biosynthesis of secondary metabolite toxins. None of the glycoside hydrolases (GHs) identified in R. irregularis are involved in degrading PCW polysaccharides. No gene encoding cellobiohydrolases (GH6 and GH7), polysaccharide lyases (PL1, PL3, PL4, and PL9), or proteins with cellulose-binding motif 1 (CBM1) were identified (Fig. 3 and SI Appendix, Table S10 and Fig. S15). Lytic polysaccharide mono-oxygenases (AA9, formerly GH61) that are abundant in the ectomycorrhizal L. bicolor and T. melanosporum are missing from R. irregularis. Similarly, no genes involved in lignin decomposition, such as class II peroxidases (SI Appendix, Fig. S15), were found. No orthologs of bacterial genes coding for enzymes involved in symbiotic lipochitooligosaccharide factors (25) were identified.
Fig. 3.
Numbers of genes devoted to secondary metabolism and genes encoding secreted proteins and cellulose- or hemicellulose-degrading enzymes, identified in the R. irregularis genome and 11 fungal species included in this study. The boxes on the left represent the lifestyle of the selected organisms. SAP, soil saprotrophs; PP, plant pathogens; AP, animal pathogens; ECM, ectomycorrhizal symbionts; AM, arbuscular mycorrhizal symbiont. The colored bars represent the secondary metabolic and PCW degrading enzymes, identified by the key at the top. PKS, polyketide synthases; NRPS, nonribosomal peptide synthases; DMATs, dimethyl allyl tryptophan synthases; HYBRID, PKS-NRPS hybrids; GH, glycoside hydrolases; GT, glycosyltransferases; CE, carbohydrate esterases; PL, polysaccharide lyases; CBM, carbohydrate-binding modules.
Key enzymes that catalyze the biosynthesis of fungal toxins, such as polyketide synthases, modular nonribosomal peptide synthetases, terpene cyclases, and dimethylallyl diphosphate tryptophan synthases, are also lacking in R. irregularis (Fig. 3). Thus, it appears that biotrophy is associated with a convergent loss of secondary metabolic enzymes and PCW-degrading enzymes (20). No previously sequenced plant-interacting biotrophic fungus has such a minimal set of degrading enzymes, however. This finding strongly suggests an evolutionary adaptation to minimize the release of effector molecules that could trigger the plant immune system. The lack of PCW-degrading enzymes in the mycobiont also implies that penetration of the PCW, a prerequisite to the development of the intracellular symbiotic arbuscules, relies on plant enzymes. In addition, R. irregularis has no gene coding for the thiamine biosynthetic pathway (SI Appendix, Table S9). Interestingly, haustorial oomycetes (Albugo laibachii, Hyaloperonospora arabidopsis, and Phytophthora spp.) also have lost the gene for thiamine biosynthesis (26).
No secreted invertase or sucrose transporter was identified, implying that this fungus likely relies on the host plant to provide monosaccharides as a carbon source (27). It is unlikely that the aforementioned gene sets were missed because of incomplete genome coverage, given that the R. irregularis assembly encompasses 98% of conserved core eukaryotic single-copy genes (17). In contrast, genes coding for nitrate and nitrite reductases, nitrate transporter, and sulfite reductase that are missing from B. graminis (20) were found in R. irregularis (SI Appendix, Table S9). Thus, the obligate mycobiont R. irregularis retains the ability to take up and assimilate nutrients from its soil environment, a key issue for a soil-borne fungus providing its host plant with mineral nutrients (28).
Additional genes are missing from R. irregularis and are also lacking in the genomes of Mucoromycotina and Chytridiomycota (SI Appendix, Table S9), suggesting that this is a genomic feature of early diverging fungi. A comparison of the metabolic and cellular (KEGG) gene categories confirmed that R. irregularis is closer to the early diverging fungi, such as the Mucoromycotina, than to Dikarya (SI Appendix, Figs. S16 and S17).
We identified hundreds of fungal symbiosis-related genes by mapping RNA-Seq reads from germinating spores and Medicago-colonized roots (SI Appendix, section. 1.7, Table S4). Of the 22,647 expressed genes, 1,068 (4.7%) were up-regulated in planta by at least twofold (false discovery rate-adjusted P value < 0.05). Most of the highly up-regulated genes code for proteins with no known function which are specific to R. irregularis (SI Appendix, Table S11), but several enzymes and membrane transporters are also induced during the interaction, as shown by eukaryotic orthologous groups (KOG) and InterPro (IPR) analyses of induced transcripts (SI Appendix, Table S12 and Fig. S18). Twenty-nine of the 50 most highly up-regulated fungal transcripts in mycorrhizal roots code for small secreted proteins with a predicted size of <150 aa (SI Appendix, Tables S11, S13, and S14), several of which were detected in laser microdissected arbusculed cells (SI Appendix, Table S11). These mycorrhiza-induced small secreted proteins (MiSSPs) belong to R. irregularis-specific orphan gene families (SI Appendix, Table S15) and may act as effector proteins to manipulate host cell signaling or to suppress defense pathways during infection, as has been shown for the R. irregularis SP7 effector, which interacts with a plant nuclear ethylene-responsive transcriptional factor (29). Although the repertoire of carbohydrate-active enzymes (CAZyme) of R. irregularis is limited, a few genes coding for CBM-containing proteins (LysM), glycosyl transferases (e.g., glycogen synthase, chitin synthase, UDP-glucosyltransferases), and GHs (e.g., α-amylase, lysozyme, glucosaminidase) were dramatically up-regulated (SI Appendix, Table S16), suggesting a role in mycorrhiza metabolism.
Presence of Sex-Related Genes in R. irregularis.
AMF have long been considered ancient clonal organisms, but recent studies have revealed the presence of many AMF homologs of genes known to be involved in sexual reproduction in other fungi, including meiosis-specific genes (7, 30) and an expanded gene family harboring a mating-type-related high mobility group domain (MATA-HMGs) (31). Here we confirm the previous findings of sex-related genes, and also show that the number of MATA-HMGs present in the genome of R. irregularis that harbor a MATA domain, or that share similarities with mating-type (MAT)-like HMGs from other fungi, is much larger than previously identified by mining transcriptome sequences (SI Appendix, section 1.6, Fig. S19 and Table S17). Specifically, 146 AMF genes were found to share similarities with homologs present in the mating type locus of various fungal lineages, including SexM/P from Phycomyces blakesleeanus (Zygomycota) (32). We confirm that none of the MATA-HMG genes are located in close proximity to orthologs of genes known to compose the MAT locus of other fungi, and that six scaffolds harbor MATA-HMGs that are repeated in tandem (31). We investigated the presence of potential idiomorphs in these MATA-HMGs by isolating their respective alleles from three genetically different strains of R. irregularis (strains A4, B3, and C2). Only 3 of the 146 homologs failed to amplify from one of three isolates using nonstringent procedures, raising the intriguing possibility that these represent idiomorphs of a heterothallic AMF mating-type locus.
In conclusion, the occurrence of multiple highly diverged genomes in the multinucleated R. irregularis is not supported by the present study. Ancient whole-genome duplication and TE proliferation likely have promoted massive gene duplications. The R. irregularis genome shares many features with fungi belonging to Mucoromycotina (e.g., homokaryotic organization in coenocytic hyphae, similar core metabolic pathways), suggesting that Glomeromycota have strong phylogenetic relationships with these early diverging fungi. On the other hand, the expression of effector-like MiSSPs and the lack of PCW-degrading and toxin-synthesizing enzymes are hallmarks of R. irregularis. These features suggest a functional converging evolution with phylogenetically unrelated biotrophic pathogens (20, 26) and ectomycorrhizal symbionts (21, 24). In contrast, the obligate biotrophic lifestyle of R. irregularis is not associated with a significant reduction in genes involved in nitrogen and sulfur assimilation, as observed in many obligate biotrophic leaf pathogens (20, 26), but is associated with the high expression of genes involved in nutrient uptake (8). Thus, R. irregularis has the dual ability to interact with the soil environment with respect to mineral nutrient uptake and to integrate the complex cues imposed by its in planta life. The present comprehensive repertoire of R. irregularis genes provides a basis for future research on symbiosis-related mechanisms and the ecological genomics of Glomeromycota.
Methods
Detailed descriptions of materials and methods are provided in SI Appendix, Methods. In brief, the genome of the multinucleated mycelium of R. irregularis DAOM-197198 was sequenced using Sanger, 454, Illumina, and PacBio platforms and assembled using the CLC Genomic Workbench assembler. Gene models were predicted and validated using computational tools, and RNA-Seq transcriptomics were annotated using the Joint Genome Institute annotation pipeline. Gene expression of R. irregularis and Medicago truncatula was assessed using RNA-Seq.
Supplementary Material
Acknowledgments
We acknowledge Y.C. Li, H. Niculita-Herzel, and A. Brachman (from the former Joint Genome Institute Glomus consortium) for their genome analyses that were not included in this study. We also thank the Lausanne University Genomic Technologies Facility, especially K. Harshman and E. Beaudoing, for PacBio sequencing support, and the Génome et Transcriptome-Plateforme Génomique (GeT-PlaGE) Facility of Toulouse, especially N. Marsaud and N. Ladouce, for Illumina sequencing support. The computations were performed at the Institut National de la Recherche Agronomique Nancy Ecogenomics facilities and in part at the Vital-IT Center for high-performance computing of the Swiss Institute of Bioinformatics. E.T. is supported by a postdoctoral fellowship from the European Commission (project EcoFINDERS FP7-264465). This work was supported by the French National Research Agency through the Clusters of Excellence ARBRE (Advanced Research on the Biology of Tree and Forest Ecosystems) (ANR-11-LABX-0002-01) and TULIP (Toward a Unified Theory of Biotic Interactions: Role of Environmental Perturbations) (ANR-10-LABX-41). This work was also funded by grants from the US Department of Energy’s Oak Ridge National Laboratory Scientific Focus Area for Genomics Foundational Sciences (to F.M. and G.A.T.); the Conseil Régional Midi-Pyrénées (to C.R.); the Natural Sciences and Engineering Research Council of Canada (to N.C.); the German Federal Ministry of Education and Research (to S.A.R.); the Swiss National Science Foundation (to I.R.S.); the Italian Regional Project Converging Technologies-BIOBIT (to P.B.); the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.K.); and the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (to K.S.). The work conducted by the US Department of Energy’s Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences have been deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank (accession no. AUPC00000000). RNA-Seq reads have been submitted at the National Center for Biotechnology Information’s Sequence Read Archive (accession nos. SRR1027885, SRX312982 and SRX312214).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313452110/-/DCSupplemental.
References
- 1.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91(25):11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289(5486):1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 3.Bucher M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 2007;173(1):11–26. doi: 10.1111/j.1469-8137.2006.01935.x. [DOI] [PubMed] [Google Scholar]
- 4.Ceballos I, et al. The in vitro mass-produced model mycorrhizal fungus, Rhizophagus irregularis, significantly increases yields of the globally important food security crop cassava. PLoS ONE. 2013;8(8):e70633. doi: 10.1371/journal.pone.0070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosendahl S. Communities, populations and individuals of arbuscular mycorrhizal fungi. New Phytol. 2008;178(2):253–266. doi: 10.1111/j.1469-8137.2008.02378.x. [DOI] [PubMed] [Google Scholar]
- 6.Sanders IR. Evolutionary genetics. No sex please, we’re fungi. Nature. 1999;399(6738):737–739. doi: 10.1038/21544. [DOI] [PubMed] [Google Scholar]
- 7.Halary S, et al. Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biol Evol. 2011;3:950–958. doi: 10.1093/gbe/evr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tisserant E, et al. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 2012;193(3):755–769. doi: 10.1111/j.1469-8137.2011.03948.x. [DOI] [PubMed] [Google Scholar]
- 9.den Bakker HC, Vankuren NW, Morton JB, Pawlowska TE. Clonality and recombination in the life history of an asexual arbuscular mycorrhizal fungus. Mol Biol Evol. 2010;27(11):2474–2486. doi: 10.1093/molbev/msq155. [DOI] [PubMed] [Google Scholar]
- 10.Croll D, Sanders IR. Recombination in Glomus intraradices, a supposed ancient asexual arbuscular mycorrhizal fungus. BMC Evol Biol. 2009;9:13. doi: 10.1186/1471-2148-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders IR, Croll D. Arbuscular mycorrhiza: The challenge to understand the genetics of the fungal partner. Annu Rev Genet. 2010;44:271–292. doi: 10.1146/annurev-genet-102108-134239. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414(6865):745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- 13.Hijri M, Sanders IR. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature. 2005;433(7022):160–163. doi: 10.1038/nature03069. [DOI] [PubMed] [Google Scholar]
- 14.Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427(6976):733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- 15.Ehinger MO, Croll D, Koch AM, Sanders IR. Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol. 2012;196(3):853–861. doi: 10.1111/j.1469-8137.2012.04278.x. [DOI] [PubMed] [Google Scholar]
- 16.Hosny M, Hijri M, Passerieux E, Dulieu H. rDNA units are highly polymorphic in Scutellospora castanea (glomales, zygomycetes) Gene. 1999;226(1):61–71. doi: 10.1016/s0378-1119(98)00562-9. [DOI] [PubMed] [Google Scholar]
- 17.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 18.Sędzielewska KA, et al. Estimation of the Glomus intraradices nuclear DNA content. New Phytol. 2011;192(4):794–797. doi: 10.1111/j.1469-8137.2011.03937.x. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3(2):e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 21.Martin F, et al. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature. 2010;464(7291):1033–1038. doi: 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 22.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 23.Joneson S, Stajich JE, Shiu SH, Rosenblum EB. Genomic transition to pathogenicity in chytrid fungi. PLoS Pathog. 2011;7(11):e1002338. doi: 10.1371/journal.ppat.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452(7183):88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 25.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469(7328):58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 26.Judelson HS. Dynamics and innovations within oomycete genomes: Insights into biology, pathology, and evolution. Eukaryot Cell. 2012;11(11):1304–1312. doi: 10.1128/EC.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helber N, et al. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 2011;23(10):3812–3823. doi: 10.1105/tpc.111.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leigh J, Hodge A, Fitter AH. Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol. 2009;181(1):199–207. doi: 10.1111/j.1469-8137.2008.02630.x. [DOI] [PubMed] [Google Scholar]
- 29.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 30.Riley R, Corradi N. Searching for clues of sexual reproduction in the genomes of arbuscular mycorrhizal fungi. Fungal Ecol. 2013;6:44–49. [Google Scholar]
- 31.Riley R, et al. Extreme diversification of the mating type high-mobility group (MATA-HMG) gene family in a plant-associated arbuscular mycorrhizal fungus. New Phytol. 2013 doi: 10.1111/nph.12462. [DOI] [PubMed] [Google Scholar]
- 32.Idnurm A, Walton FJ, Floyd A, Heitman J. Identification of the sex genes in an early diverged fungus. Nature. 2008;451(7175):193–196. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.