Significance
Sunlight is an essential environmental factor for photosynthetic plants and ultimately for life on Earth, which is sustained through plants as fundamental source of food. However, plants have a love/hate relationship with sunlight and must be protected from potentially harmful UV-B radiation. The UV-B photoreceptor UVR8 is of great importance in mounting UV-protective responses and thus for survival in sunlight. Based on our understanding of UVR8 signaling, we have engineered a UVR8 variant that is constitutively active in transgenic plants. The generation of a constitutively active photoreceptor variant is an important step in understanding the molecular signaling mechanism and may hold opportunities for crop improvement.
Keywords: signal transduction, abiotic stress, ultraviolet-B
Abstract
Arabidopsis thaliana UV RESISTANCE LOCUS 8 (UVR8) is a UV-B photoreceptor that initiates photomorphogenic responses underlying acclimation and UV-B tolerance in plants. UVR8 is a homodimer in its ground state, and UV-B exposure results in its instantaneous monomerization followed by interaction with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), a major factor in UV-B signaling. UV-B photoreception by UVR8 is based on intrinsic tryptophan aromatic amino acid residues, with tryptophan-285 as the main chromophore. We generated transgenic plants expressing UVR8 with a single amino acid change of tryptophan-285 to alanine. UVR8W285A appears monomeric and shows UV-B–independent interaction with COP1. Phenotypically, the plants expressing UVR8W285A exhibit constitutive photomorphogenesis associated with constitutive activation of target genes, elevated levels of anthocyanins, and enhanced, acclimation-independent UV-B tolerance. Moreover, we have identified COP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 and 2 (RUP1 and RUP2), and the SUPPRESSOR OF PHYA-105 (SPA) family as proteins copurifying with UVR8W285A. Whereas COP1, RUP1, and RUP2 are known to directly interact with UVR8, we show that SPA1 interacts with UVR8 indirectly through COP1. We conclude that UVR8W285A is a constitutively active UVR8 photoreceptor variant in Arabidopsis, as is consistent with the crucial importance of monomer formation and COP1 binding for UVR8 activity.
Plants react to UV-B radiation (UV-B; 280–315 nm) with a photomorphogenic response that generates acclimation to this environmental stress factor (1–3). The associated specific signaling pathway is characterized molecularly by the involvement of the UV RESISTANCE LOCUS 8 (UVR8) photoreceptor (4, 5). Loss of UVR8 in Arabidopsis results in the loss of a broad range of molecular and physiological UV-B responses, including reduced UV-B acclimation and tolerance (6–11). Perception of UV-B photons by UVR8 homodimers results in UVR8 monomerization (4). The crystal structure of UVR8 shows that the homodimer is maintained by salt-bridge interactions between charged amino acids at the dimeric interaction surface (12, 13). Destabilization of the salt bridges upon absorption of UV-B photons by tryptophan-285, and to a lesser extent tryptophan-233, underlies UVR8 monomerization and signal initiation (12, 13). The UVR8 photoreceptor can revert to the ground state in vivo by redimerization (14, 15). This process is facilitated by REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 and 2 (RUP1 and RUP2), consequently disrupting the key interaction of UVR8 with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (14, 16). The reversibility of UVR8 between inactive homodimer and active monomer conformations results in continuous sensitivity to the ambient UV-B environment (14, 15). It can be assumed that UVR8 cycles between the dimeric and monomeric forms in vivo, and thus the resulting UVR8 dimer/monomer photoequilibrium is a measure of the ambient UV-B levels experienced by the plant.
Activated UVR8 interacts with COP1 (8), which is an E3 ubiquitin ligase with important activity as a repressor of photomorphogenesis in the dark (17) and a key role in promoting UV-B signaling (18). Mutations in COP1 or UVR8 affect the interaction and impair UV-B signaling (8, 19). The COP1 interaction domain recently was found to be a region of 27 amino acids in the C terminus of UVR8 (19). UVR8–COP1 interaction is associated with stabilization of the bZIP transcription factor ELONGATED HYPOCOTYL 5 (HY5), which also plays an important role in UV-B signaling (7, 18, 20, 21). Together with FHY3, HY5 also positively regulates COP1 expression in response to UV-B (22), but its transcriptional activity is feedback-inhibited by the B-box protein BBX24 (23).
Mutation of the tryptophan-285 chromophore to phenyalanine renders UVR8W285F constitutively homodimeric and inactive (4, 12, 13, 24). In marked contrast, the mutation of tryptophan-285 to alanine (UVR8W285A) was found to be monomeric in vivo and to interact constitutively with COP1 in yeast (4), in mammalian cells (25), and in plants (24). A recent report has addressed the physiological effect of expressing GFP-UVR8W285A in transgenic plants (uvr8-1/Pro35S::GFP-UVR8W285A) (24). Despite the apparent monomeric form of GFP-UVR8W285A and associated constitutive interaction with COP1 in planta, lines expressing GFP-UVR8W285A were not altered in growth phenotype and lack only UV-B responsiveness (24). Therefore, it was concluded that monomer formation and COP1 binding are not sufficient for UVR8 function (24).
In a search for a constitutively active UVR8 variant, we generated uvr8-7/Pro35S::UVR8W285A transgenic lines. We show here that expression of UVR8W285A results in a constitutive photomorphogenic phenotype, including hypocotyl growth inhibition, target gene expression, and elevated levels of anthocyanins. As a consequence, lines overexpressing UVR8W285A are constitutively acclimated and thus display an enhanced basal UV-B tolerance.
Results
UVR8W285 Appears Partially Monomeric and Interacts Constitutively with COP1 in Vivo.
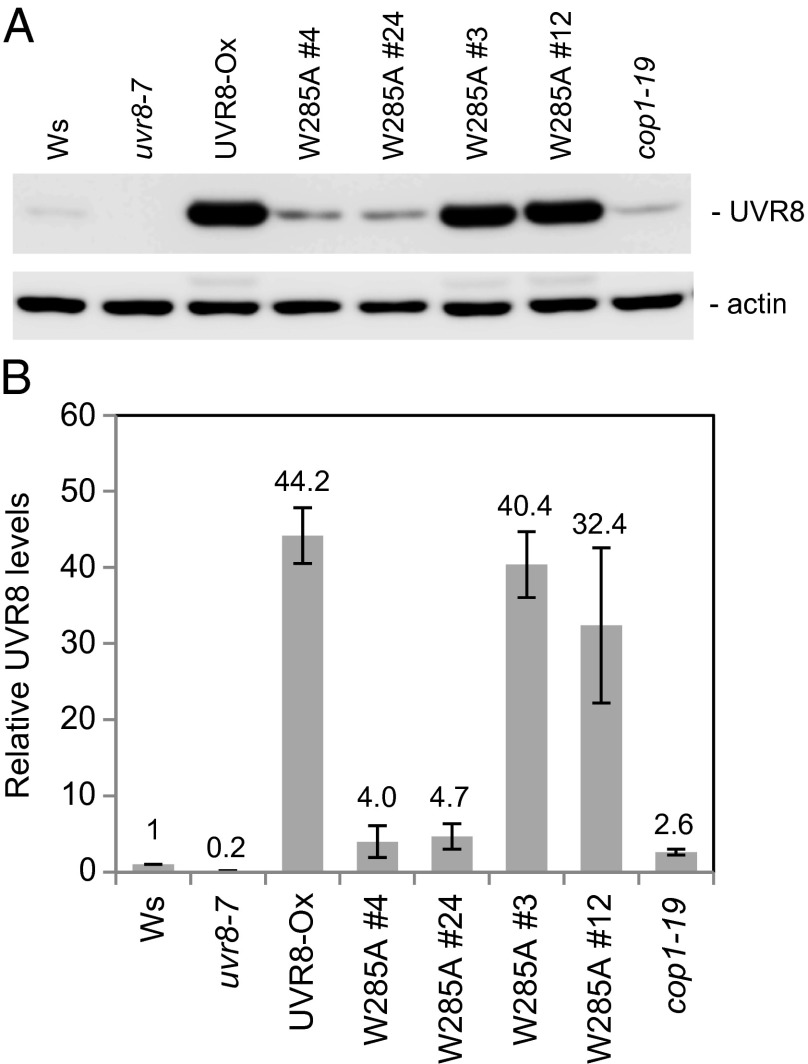
To analyze the in vivo function of UVR8W285A, we transformed the uvr8-7 null mutant with the UVR8W285A coding sequence driven by the constitutive CaMV 35S-promoter (uvr8-7/Pro35S::UVR8W285A). Among the transgenic lines, we carefully selected lines having either low levels of UVR8W285A overexpression relative to endogenous UVR8 expression in wild-type plants (e.g., lines #4 and #24) or high levels of UVR8W285A overexpression similar to those in a control line overexpressing UVR8 (UVR8-Ox; uvr8-7/Pro35S::UVR8) (e.g., lines #3, #6, and #12) (Figs. 1 and 2A).
Fig. 1.
Expression of UVR8W285A in transgenic plants. (A) UVR8 and UVR8W285A protein levels in two transgenic UVR8W285A lines with low levels of overexpression (W285A #4 and #24), in two transgenic UVR8W285A lines with high levels of overexpression (W285A #3 and #12), in the wild-type line (Ws), in a control line overexpressing UVR8 (UVR8-Ox), and in the cop1-19 mutant line. (B) Quantification of UVR8 and UVR8W285A protein levels in biological triplicates, relative to the wild type (Ws) UVR8 level; error bars represent SD.
Fig. 2.
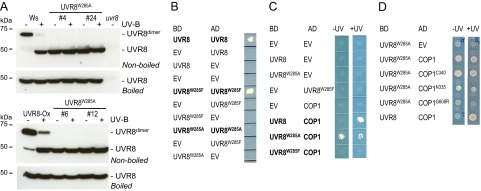
UVR8W285A appears as a constitutive monomer and constitutively interacts with COP1 in transgenic plants. (A) Four-day-old seedlings were irradiated for 15 min with (+) or without (−) supplementary broadband UV-B. The dimer was observed on Western blots from SDS-PAGE without sample boiling (Upper), and the total amount of UVR8 protein was detected in samples denatured by boiling (Lower). UVR8 protein levels of wild-type (Ws) plants are compared with those in Pro35S::UVR8W285A lines #4 and #24 (W285A #4 and #24) (Upper), and UVR8 protein levels of a line overexpressing Pro35S::UVR8 (UVR8-Ox) are compared with levels in lines overexpressing Pro35S::UVR8W285A (#6 and #12, Lower). uvr8-7 is shown as negative control demonstrating the specificity of the detected bands. (B) UVR8W285A does not form homodimers in vivo in yeast two-hybrid growth assays, in contrast to UVR8 and UVR8W285F. (C) UVR8W285A interacts with COP1 in a UV-B–independent manner, in contrast to wild-type UVR8, whereas UVR8W285F shows no interaction with COP1. (D) UVR8W285A interacts with the 340 C-terminal amino acids of COP1 in a UV-B–independent manner in a yeast two-hybrid growth assay. EV, empty vector control.
UVR8W285A in the transgenic lines appears as a constitutive monomer after SDS-PAGE without sample boiling (Fig. 2A), as previously described for UVR8W285A expressed in yeast (4) and GFP-UVR8W285A in transgenic plants (24). This configuration is in marked contrast to that of UVR8W285F, which appears as a constitutive homodimer in plant transgenic lines (Fig. S1 A and B) (24), as it does in yeast (4). Notably, although SDS-PAGE is a very convenient assay for cell extracts, there is some discrepancy when the results are compared with size-exclusion chromatography of purified UVR8W285A in vitro: SDS-PAGE identified UVR8W285A as monomeric, whereas size-exclusion chromatography identified it as homodimeric (12, 13). This discrepancy indicates that UVR8W285A may not be monomeric per se but that the dimer is strongly destabilized. However, UVR8 dimerization assays in an yeast two-hybrid system indicated that UVR8W285A does not homodimerize detectably in vivo, in contrast to UVR8 and UVR8W285F (Fig. 2B). Although we cannot exclude the possibility that UVR8W285A forms weak dimers that are undetectable in yeast two-hybrid assays, our data suggest that UVR8W285A is, at least in part, a functional monomer in the cellular context in vivo. In planta, UVR8 and UVR8W285A are likely to be associated with additional proteins. Indeed, size-exclusion chromatography of protein extracts from transgenic seedlings indicated that UVR8 and UVR8W285F were present in native complexes with an apparent molecular mass <158 kDa, whereas fractions containing UVR8W285A were shifted to a lower molecular mass, and UVR8W285A was detected in fractions that correspond to monomeric size (Fig. S1C). Similarly, in 2D Blue Native/SDS-PAGE, both UVR8 and UVR8W285A were detected in native complexes of about 146 kDa, in which UVR8 appears as dimer and UVR8W285A as monomer (second-dimension SDS-PAGE, nonboiled) (Fig. S1D). We conclude that both UVR8 and UVR8W285A are associated with interacting proteins in vivo rather than being present as isolated dimers or monomers, respectively.
UVR8W285A interacts constitutively with COP1 in yeast, whereas the UVR8W285F mutation prevents UV-B–dependent interaction with COP1 (Fig. 2C) (4). UVR8W285A was found interact constitutively with the C-terminal WD40-repeat domain of COP1 (C-terminal 340 amino acids; COP1C340) but not with the N-terminal RING/coiled-coil domains (COP1N335) (Fig. 2D). In further support of a UVR8–COP1C340 interaction (4, 8), a single amino acid mutation corresponding to the cop1-19 allele (COP1G608R) (8) abolished interaction with UVR8W285A in yeast (Fig. 2D). In agreement with the yeast data, COP1 coimmunoprecipitates with GFP- and TAP-tagged UVR8W285A independently of UV-B in plant cells (ref. 24, and see below), in contrast to wild-type UVR8, which coimmunoprecipitates with COP1 only under UV-B (4, 8, 14, 19, 24). Taken together, these results indicate that at least a fraction of UVR8W285A is constitutively monomeric in planta and interacts with the WD40-repeat domain of COP1.
Expression of UVR8W285A Results in Constitutive UV-B Responses in Seedlings in both Light and Dark Conditions.
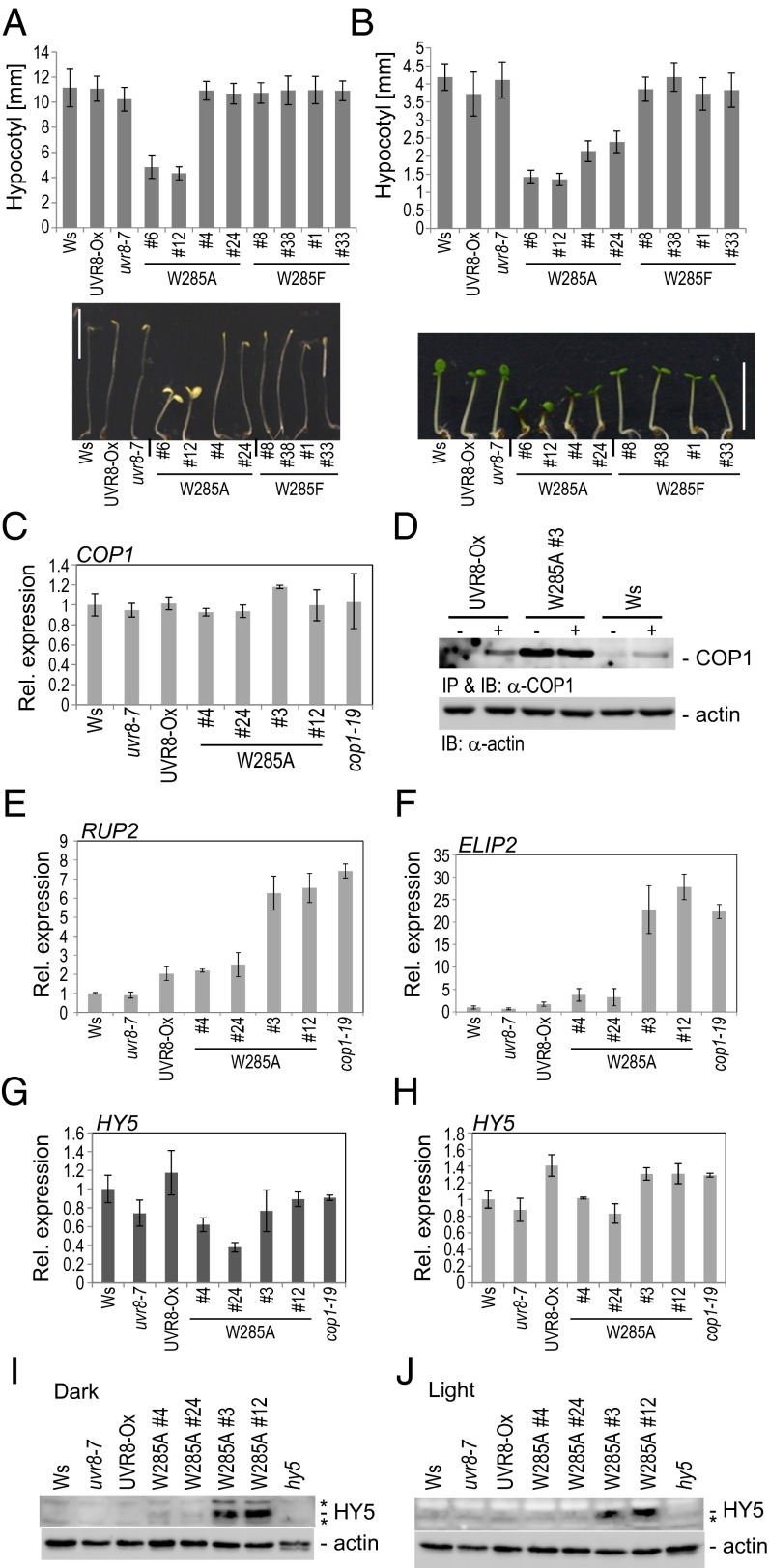
The UVR8-COP1 interaction is an early and crucial step in the UV-B photoreceptor signaling pathway (8). Therefore, we tested whether UVR8W285A is constitutively active in planta. UV-B induces gene-expression changes and a series of photomorphogenic responses, including hypocotyl growth inhibition and the accumulation of anthocyanins (2). Seedlings with low overexpression of UVR8W285A (lines #4 and #24) exhibited significantly shorter hypocotyls than wild-type seedlings in weak white light, but no difference was seen in dark-grown seedlings (Fig. 3 A and B). However, higher overexpression of UVR8W285A (lines #6 and #12) led to a constitutive photomorphogenic (cop) phenotype even in darkness, as displayed by a short hypocotyl and open and expanded cotyledons (Fig. 3A). This phenotype is reminiscent of the cop1-mutant phenotype (26), although COP1 mRNA and COP1 protein levels were not reduced in the line overexpressing UVR8W285A (Fig. 3 C and D). In this line, the cop1-like phenotype is not caused by low levels of COP1. Rather, COP1 was higher in lines overexpressing UVR8W285A grown in white light (Fig. 3D), as is in agreement with the posttranslational COP1 stabilization seen in UV-B–irradiated wild-type seedlings (8). Importantly, overexpression of UVR8 or UVR8W285F did not result in a cop1-like phenotype (Fig. 3 A and B).
Fig. 3.
UVR8W285A expression results in a constitutive photomorphogenic response. (A and B) Analysis of hypocotyl length and quantification of 4-d-old seedlings grown in the dark (A) or in continuous white light (3.6 µmol⋅m−2⋅s−1) (B). Histograms show average hypocotyl length; error bars represent SD (n > 30). (Scale bar, 5 mm.) (C) Quantitative RT-PCR analysis of COP1 mRNA levels in 4-d-old UVR8W285A transgenic seedlings under white light compared with wild-type seedlings (Ws). (D) Immunoprecipitation and Western blot analysis of COP1 protein levels in 7-d-old seedlings grown in white light (−UV-B) or supplemented for 6 h with narrowband UV-B before harvesting (+UV-B). (E and F) Quantitative RT-PCR comparison of RUP2 and ELIP2 mRNA levels in 4-d-old wild-type (Ws) and UVR8W285A transgenic plants grown in white light. (G and H) Quantitative RT-PCR comparison of HY5 mRNA levels in 4-d-old wild-type (Ws) and UVR8W285A transgenic plants grown in the dark (G) or in white light (H). (I and J) Immunoblot analysis of HY5 protein levels in 7-d-old seedlings grown in the dark (I) or in white light (J). Asterisks indicate unspecific bands.
The UV-B–dependent interaction of UVR8 with COP1 is followed by a transcriptional response (2). Therefore we analyzed the expression of UVR8-dependent, UV-B–responsive marker genes in the lines expressing UVR8W285A. The RUP2 and EARLY LIGHT-INDUCIBLE PROTEIN 2 (ELIP2) marker genes tested were both constitutively expressed at elevated levels in lines overexpressing UVR8W285A (Fig. 3 E and F and Fig. S2).
HY5 is a crucial transcriptional regulator in the UV-B signaling pathway (20). Transcriptionally, the HY5 gene is induced early, and the HY5 protein is posttranslationally stabilized upon UV-B exposure, despite the parallel nuclear accumulation of COP1 (8, 18, 20). Indeed, HY5 accumulated substantially in lines overexpressing UVR8W285A, despite wild-type levels of HY5 mRNA (Fig. 3 G–J) and elevated levels of COP1 (Fig. 3D). The wild-type level of HY5 mRNA is consistent with the rapid and transient kinetics of HY5 gene induction by UV-B, corresponding to the basal level after 4 d exposure (8, 16, 18).
Thus, we conclude that UVR8W285A is a constitutively active UV-B photoreceptor in planta, which leads to UV-B–associated phenotypic (hypocotyl growth inhibition) and molecular (COP1 and HY5 stabilization and target gene expression) responses in the absence of UV-B.
UVR8W285A Expression Results in Elevated Constitutive Levels of Chalcone Synthase and Anthocyanins.
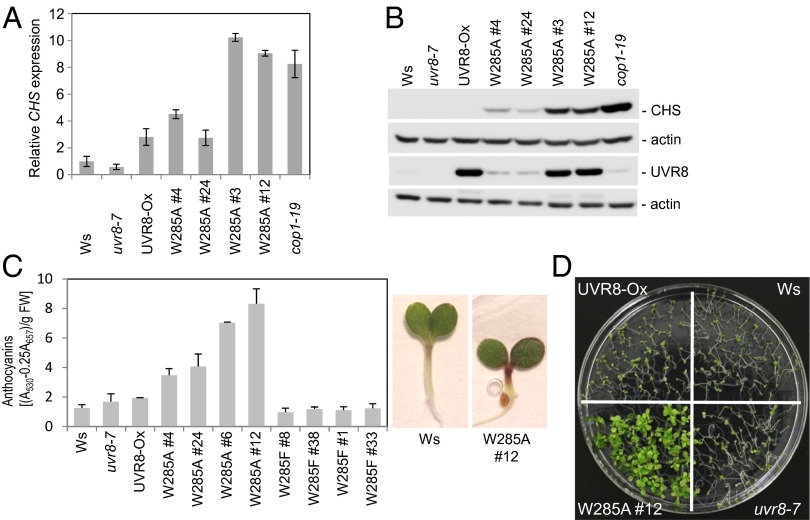
UV-B also is known to induce a series of changes in metabolite levels, including the accumulation of UV-protective pigments such as anthocyanins (2). CHALCONE SYNTHASE (CHS) is a key enzyme in the phenylpropanoid biosynthesis pathway leading to anthocyanins that is regulated largely at the transcriptional level in response to UV-B (27, 28). In agreement with their apparent constitutive UV-B photomorphogenic responses, plants overexpressing UVR8W285A showed higher CHS mRNA levels than the wild-type plants, uvr8-7 mutants, or plants overexpressing UVR8 (Fig. 4A). Elevated levels of CHS in seedlings expressing UVR8W285A also were apparent at the protein level (Fig. 4B). In agreement with the changes in CHS, the overexpression of UVR8W285A resulted in elevated levels of anthocyanins in the absence of UV-B, representing a constitutive UV-B response (Fig. 4C). In contrast, neither UVR8 nor UVR8W285F overexpression resulted in constitutively elevated levels of anthocyanins (Fig. 4C).
Fig. 4.
UVR8W285A expression results in elevated constitutive levels of chalcone synthase and anthocyanins as well as elevated UV-B tolerance. (A and B) Analysis of CHS mRNA levels (A) and CHS protein levels (B) in 4-d-old seedlings grown in white light. (C) Photometric determination of the anthocyanin content of 4-d-old Arabidopsis seedlings. Data shown are the mean values of three independent biological replicates; error bars indicate SD. Representative images showing the elevated anthocyanin content (purple pigmentation) in a 4-d-old transgenic plant overexpressing UVR8W285A (Right). (D) UV-B tolerance of non–UV-B–acclimated 7-d-old seedlings treated for 3 h with broadband UV-B. After UV-B stress treatment, the seedlings were allowed to recover for 1 wk without UV-B before the image was captured.
Constitutive Acclimation Results in Enhanced UV-B Tolerance in Lines Overexpressing UVR8W285A.
The UVR8-dependent UV-B signaling pathway acclimates plants to UV-B and, as a consequence, enhances UV-B stress tolerance (8, 16, 29). Because the phenotype of lines overexpressing UVR8W285A suggested constitutive acclimation to UV-B, we subjected 7-d-old seedlings to UV-B stress without prior acclimation. Indeed, the overexpression of UVR8W285A resulted in constitutively elevated basal UV-B tolerance, in contrast to wild-type seedlings, the uvr8-7 null mutant, and lines overexpressing UVR8 (Fig. 4D). Thus, we conclude that overexpression of UVR8W285A (but not of UVR8) is sufficient to trigger a combination of responses that usually are associated with UV-B–induced photomorphogenesis and acclimation and that render plants UV-B tolerant.
Lines Overexpressing UVR8W285A Show a Dwarfed Growth Phenotype.
UVR8W285A-overexpression phenotypes were not restricted to the seedling stage. When grown on soil for 5 wk, lines overexpressing UVR8W285A had a dwarfed growth phenotype reminiscent of cop1-mutant plants (Fig. 5). The lines expressing low levels of UVR8W285A also were stunted, including short petioles, but to a much lower extent (Fig. 5). We conclude that expression of UVR8W285A results in a dwarf growth phenotype at the adult stage, likely associated with its constitutive interaction with COP1.
Fig. 5.
UVR8W285A overexpression results in dwarf growth of adult plants. Representative 5-wk-old plants grown in standard growth conditions (with a UV-B filter) in a phytochamber under short days (8 h/16 h light/dark).
Tandem Affinity Purification-Tagged UVR8W285A Copurifies with the COP1–SUPPRESSOR OF PHYA-105 Complex.
Our physiological and molecular analyses of lines expressing UVR8W285A demonstrate that UVR8W285A mimics active UVR8. Therefore, we used a tandem affinity purification (TAP)-based screening approach to isolate proteins interacting with UVR8W285A in Arabidopsis cells. We transformed Arabidopsis suspension-cultured cells with GS-TAP–tagged UVR8W285A and identified interacting proteins by copurification and MS (30). As expected, COP1, RUP1, and RUP2 were present in complex(es) with UVR8W285A (Fig. 6A). The only further copurifying proteins were the WD40-repeat protein SPA1 (SUPPRESSOR OF PHYA-105 1) and the related SPA2, SPA3, and SPA4 (Fig. 6A). The SPA1–SPA4 quartet is known to interact with COP1 and to be required for its activities (21, 31, 32). The copurification of SPA1 with UVR8W285A from suspension cultures was supported further by the coimmunoprecipitation of SPA1 with endogenous UVR8 in wild-type seedlings. Coimmunoprecipitation of SPA1 with UVR8 occurred specifically in response to UV-B, similar to coimmunoprecipitation of COP1 with UVR8 (Fig. 6B). Importantly, we did not detect coimmunoprecipitation of SPA1 with UVR8 in a cop1-mutant background (Fig. 6B). We conclude that, although UVR8 interacts directly with COP1, it interacts with SPA1 indirectly through COP1. Moreover, the COP1–SPA complex remains intact upon interaction with UVR8W285A as well as upon interaction with UVR8 after activation by UV-B.
Fig. 6.
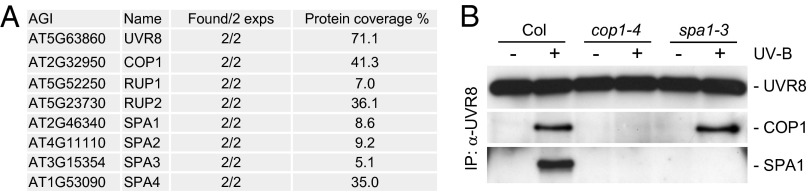
UVR8/UVR8W285A “interactors” include SPA family proteins, but interaction with SPA1 is indirect via COP1. (A) Proteins identified after TAP purification with GS-TAP-UVR8W285A expressed in Arabidopsis cell-suspension cultures [PSB-L/PKSA::GS(rhino)-UVR8W285A]. Known TAP background proteins were filtered from the list. AGI, Arabidopsis Genome Initiative identifier. See Table S1 for details of protein identification. (B) Coimmunoprecipitation of COP1 and SPA1 using UVR8 antibodies in extracts from 7-d-old wild-type (Col), cop1-4, and spa1-3 seedlings. Seedlings were irradiated for 6 h with supplemental narrowband UV-B and for 15 min with (+) or without (−) broadband UV-B.
Discussion
The generation of constitutively active photoreceptor variants has provided useful tools and important information about the signaling and responses of diverse photoreceptors, including plant cryptochrome blue-light receptors (33, 34), plant phytochrome red-/far-red-light receptors (35–37), and mammalian rhodopsin (38). The observation that plants expressing UVR8W285A exhibit constitutive photomorphogenetic development in the dark and exaggerated photomorphogenesis in light devoid of UV-B demonstrates that the functional consequence of this chromophore mutation is the UV-B–independent activation of the UVR8 photoreceptor pathway.
The crystal structures of the core of UVR8, as well as UVR8W285A and UVR8W285F, have been determined recently (12, 13). Interestingly, the overall structures of the UVR8 variants UVR8W285F and UVR8W285A are almost identical to that of the UVR8 core domain (13). Analysis of local structural features surrounding the chromophore residue tryptophan-285 revealed no significant conformational differences between UVR8 and UVR8W285F, but major changes in critical residues have occurred in the variant UVR8W285A (13). Because the solved UVR8 core structure does not include the C-terminal 40 amino acids, it presently is not known how these changes in UVR8W285A have affected the C-terminal C27 region, which is crucial for UVR8–COP1 interaction as well as for UVR8 activity in vivo (19). It has been suggested that the C27 region is hidden from COP1 in homodimeric UVR8 and that UV-B–induced monomerization and associated conformational changes expose the C27 region to interaction with COP1 (19). We thus hypothesize that the C27 region is exposed in UVR8W285A independently of UV-B, resulting in UV-B–independent signaling involving constitutive interaction with COP1. In contrast, UVR8W285F does not monomerize or expose the C27 region to interaction with COP1 in response to UV-B (phenylalanine does not absorb in the UV-B range). In agreement with this notion, UVR8W285F transgenic lines remain unresponsive to UV-B (ref. 24 and this study).
It was shown previously that COP1 forms protein complexes with the SPA quartet (SPA1–SPA4) and that the SPA proteins are required for COP1 activity (31). Whether the SPA proteins also are essential for COP1 activity in the response to UV-B is controversial at present (18, 21). Independent of this question, although cryptochrome photoreceptors impinge on COP1 activity by light-dependent interaction via SPA proteins (39), UVR8 presently is the only photoreceptor showing light-dependent interaction directly with COP1 (2, 4). Moreover, we show here that UVR8 interacts with the COP1–SPA complex and not with COP1 in isolation. This result further indicates that UVR8 must affect COP1 activity in response to UV-B in the presence of interacting SPA proteins. Recent findings suggest that light-dependent reorganization of protein complexes containing the COP1–SPA core underlie the switch to UV-B–specific COP1 function (21). However, the exact molecular mechanism by which the UVR8–COP1 interaction transduces the UV-B signal remains to be determined.
It is noteworthy that a constitutive photomorphogenic phenotype was not found in uvr8-1/Pro35S::GFP-UVR8W285A transgenic lines in an independent study (24), in marked contrast to the clear phenotype of the uvr8-7/Pro35S::UVR8W285A lines reported here. This difference is particularly surprising because GFP-UVR8W285A was reported to appear as a constitutive monomer in the transgenic plants and to interact strongly with COP1 in the absence of UV-B (24). Obvious differences between the two reports are the background accessions [uvr8-7 in Wassilewskija and uvr8-1 in Landsberg erecta (Ler)] and the presence of an N-terminal 27-kDa GFP tag. Given that Ler responds to UV-B in a UVR8-dependent manner and that GFP-UVR8W285A was found to interact with COP1 in the absence of UV-B apparently as strongly as GFP-UVR8 interacts in the presence of UV-B (24), the background accession is the less likely reason for the observed differences. We hypothesize instead that the N terminus of UVR8 contributes to UVR8 activity and that this activity is partially impaired in N-terminal fusions of GFP. The fact that GFP-UVR8 can complement uvr8 mutants (7) suggests that it is still at least partially active and/or that UV-B activation of GFP-UVR8 is stronger than the synthetic activation mimicry in GFP-UVR8W285A, and thus higher levels of GFP-UVR8W285A overexpression may be needed to detect a constitutive photomorphogenic response. Independent of this suggestion, we also conclude that the constitutively photomorphogenic phenotype of the active UVR8W285A allele is not caused simply by COP1 interaction per se, because this interaction also is clearly apparent in GFP-UVR8W285A lines (24). Thus, UVR8W285A must affect COP1 more specifically, perhaps involving the N terminus of the protein (potentially partly impaired in N-terminal GFP fusions) and not only the C27 region. The UVR8 structure indeed suggests that the enigmatic N- and C-terminal regions are in close proximity (12, 13), but any potential interplay remains to be determined.
Interestingly, we initially observed a substantial phenotypic difference between wild-type plants, lines overexpressing UVR8, and uvr8-7 mutants under standard growth conditions using fluorescent lamps (Fig. S3A). We tested whether this difference was associated with the very low levels of UV-B issued by such lamps (Fig. S3B) and found, in fact, that the difference in growth was reduced by inserting a UV-B filter (Fig. S3 A and C). This response demonstrates the high sensitivity of the plant UVR8 photoreceptor system and provides a cautionary note regarding analysis of lines overexpressing UVR8 under “standard conditions.” As expected of a chromophore mutation, a UV-B filter made no difference to transgenic lines expressing UVR8W285A, which were equally dwarfed in both conditions (Fig. S3A).
We conclude that UVR8W285A undergoes spontaneous changes in molecular conformation, mainly from homodimer to monomer, that activate the signaling cascade involving interaction with COP1. This study provides a UVR8 mutant form that can stimulate the UV-B signaling pathway spontaneously at a level high enough to produce a constitutive photomorphogenic phenotype. This result not only underlines the great importance of monomer formation and COP1 binding for UVR8 activity in plants but also provides a promising tool to elucidate further the molecular mechanism of UVR8 signaling. Moreover, the elevated levels of anthocyanins recorded, as well as the enhanced UV-B tolerance, indicate that UVR8W285A may offer considerable potential for crop improvement.
Materials and Methods
Protein immunoprecipitation, protein gel blot analysis, size-exclusion chromatography, Blue Native/SDS-PAGE, yeast two-hybrid assays, real-time PCR, anthocyanin analysis, TAP, and LC-MS/MS analysis are described in SI Materials and Methods.
Plant Material.
The uvr8-7, cop1-19, cop1-4, and spa1-3 mutants were described previously (8, 26, 40). The UVR8W285A and UVR8W285F versions were generated by site-directed mutagenesis (4) and were cloned into Gateway-compatible pB2GW7 (41). The mutated constructs were verified by sequencing and then were introduced into the uvr8-7 mutant to generate uvr8-7/Pro35S::UVR8W285A and uvr8-7/Pro35S::UVR8W285F. The uvr8-7/Pro35S::UVR8 control overexpression line (UVR8-Ox) was described previously (8). The transgenic lines described in this work were determined genetically to have the transgenes integrated at a single locus.
Growth Conditions and UV-B Irradiation.
Arabidopsis seeds were surface-sterilized and sown on half-strength Murashige and Skoog basal salt medium (MS; Duchefa) containing 1% (wt/vol) sucrose and 1% (wt/vol) phytagel (Sigma). Seeds were stratified for 2 d at 4 °C and were germinated at 22 °C in a standard growth chamber under constant white light.
UV-B stress-tolerance experiments were performed essentially as described previously (29): 7-d-old seedlings were irradiated for 3 h using broadband UV-B lamps (Philips TL40W/12RS; 2 mW⋅cm−2, measured with a VLX-3W UV light meter equipped with a CX-312 sensor; Vilber Lourmat) and were transferred to standard white light for 7-d recovery before images were captured.
Hypocotyl lengths were measured on 4-d-old seedlings (n > 30) using ImageJ software (http://rsbweb.nih.gov/ij/), as described previously (8, 18).
For the adult phenotype, Arabidopsis plants were cultured in a growth chamber at 22 °C with 75% humidity in short-day conditions (8 h/16 h light/dark) with a 1:1 ratio of Osram L 58W/840 cool and Osram L 58W/830 warm white light lamps (see Fig. S3B for the spectrum as measured with an Ocean Optics QE65000 spectrometer). To filter out residual UV-B, the fluorescent lamps were covered when indicated with UV-B filter foil no. 226 (Lee Filters).
Supplementary Material
Acknowledgments
We thank Patrick King for editing the manuscript, Richard Chappuis for technical assistance, and Ute Hoecker for providing the SPA1 antibody. This study was supported by the State of Geneva, by European Research Council (ERC) Grant 310539 under the European Union’s Seventh Framework Programme, and by Swiss National Science Foundation (SNF) Grant SNF 31003A-132902 (both to R.U.). The CUSO (Conférence Universitaire de Suisse Occidentale) Doctoral Program in Molecular Plant Sciences supported a short-term research visit by M.B. to the G.D.J. laboratory.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314336110/-/DCSupplemental.
References
- 1.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 2.Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17(4):230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Li J, et al. UV-B-induced photomorphogenesis in Arabidopsis. Protein Cell. 2013;4(7):485–492. doi: 10.1007/s13238-013-3036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 5.Tilbrook K, et al. The UVR8 UV-B photoreceptor: Perception, signaling and response. Arabidopsis Book. 2013;11:e0164. doi: 10.1199/tab.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130(1):234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102(50):18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wargent JJ, Gegas VC, Jenkins GI, Doonan JH, Paul ND. UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 2009;183(2):315–326. doi: 10.1111/j.1469-8137.2009.02855.x. [DOI] [PubMed] [Google Scholar]
- 10.Fehér B, et al. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 2011;67(1):37–48. doi: 10.1111/j.1365-313X.2011.04573.x. [DOI] [PubMed] [Google Scholar]
- 11.Demkura PV, Ballaré CL. UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant. 2012;5(3):642–652. doi: 10.1093/mp/sss025. [DOI] [PubMed] [Google Scholar]
- 12.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484(7393):214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 14.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA. 2013;110(3):1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161(1):547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber H, et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(46):20132–20137. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18(8):1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cloix C, et al. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA. 2012;109(40):16366–16370. doi: 10.1073/pnas.1210898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101(5):1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, et al. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA. 2013;110(41):16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24(11):4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, et al. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012;22(6):1046–1057. doi: 10.1038/cr.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Hara A, Jenkins GI. In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell. 2012;24(9):3755–3766. doi: 10.1105/tpc.112.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 26.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71(5):791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 27.Stracke R, et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010;33(1):88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- 28.Kreuzaler F, Ragg H, Fautz E, Kuhn DN, Hahlbrock K. UV-induction of chalcone synthase mRNA in cell suspension cultures of Petroselinum hortense. Proc Natl Acad Sci USA. 1983;80(9):2591–2593. doi: 10.1073/pnas.80.9.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González Besteiro MA, Bartels S, Albert A, Ulm R. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 2011;68(4):727–737. doi: 10.1111/j.1365-313X.2011.04725.x. [DOI] [PubMed] [Google Scholar]
- 30.Van Leene J, Witters E, Inzé D, De Jaeger G. Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 2008;13(10):517–520. doi: 10.1016/j.tplants.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16(9):2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20(9):2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13(12):2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang HQ, et al. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103(5):815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Su YS, Lagarias JC. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant. 2009;2(1):166–182. doi: 10.1093/mp/ssn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19(7):2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rausenberger J, et al. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146(5):813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Dizhoor AM, et al. Night blindness and the mechanism of constitutive signaling of mutant G90D rhodopsin. J Neurosci. 2008;28(45):11662–11672. doi: 10.1523/JNEUROSCI.4006-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16(12):684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10(1):19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.