Significance
Cardiac failure is a leading cause of mortality worldwide and a major financial burden for healthcare systems. New tools for understanding cardiovascular disease and developing better therapeutic approaches are therefore needed. To this end, transcriptional regulation has been extensively studied in cardiac hypertrophy and failure, but there is still a lack of understanding of the epigenetic framework in which transcription factors act. Our report adds significant knowledge to the field because we demonstrate, in vivo, that a complex and specific epigenetic signature regulates gene expression by modulating promoters and enhancers, a large number of which have been described here. These findings advance our understanding of the mechanisms underlying this pathology.
Keywords: epigenetic regulation, histone acetylation, histone methylation
Abstract
Cardiac hypertrophy, initially an adaptive response of the myocardium to stress, can progress to heart failure. The epigenetic signature underlying this phenomenon is poorly understood. Here, we report on the genome-wide distribution of seven histone modifications in adult mouse cardiomyocytes subjected to a prohypertrophy stimulus in vivo. We found a set of promoters with an epigenetic pattern that distinguishes specific functional classes of genes regulated in hypertrophy and identified 9,207 candidate active enhancers whose activity was modulated. We also analyzed the transcriptional network within which these genetic elements act to orchestrate hypertrophy gene expression, finding a role for myocyte enhancer factor (MEF)2C and MEF2A in regulating enhancers. We propose that the epigenetic landscape is a key determinant of gene expression reprogramming in cardiac hypertrophy and provide a basis for understanding the role of chromatin in regulating this phenomenon.
Heart failure, a leading cause of mortality worldwide, is frequently accompanied by cardiac hypertrophy, a process characterized by the expression of genes normally present during the fetal stage and the repression of certain genes expressed in adults (1, 2). Although epigenetics is important in regulating transcription, our understanding of its role in cardiac hypertrophy remains scant (3).
Histone marks, such as acetylation (ac) and methylation (me) of lysine (K) residues of histone H3, play an important role in regulating transcription. Combinations of these marks create an “epigenetic code” that defines the transcriptional status of genes: high levels of monoacetylated (ac) Lys-9 and Lys-14 (H3K9ac and H3K14ac) and trimethylated (me3) Lys-4 (H3K4me3) in promoter regions, and trimethylated Lys-36 (H3K36me3) and di-methylated (me2) Lys-79 (H3K79me2) in the body of genes, are detected in transcriptionally active regions. In contrast, an elevated level of deacetylated histones and histone H3 trimethylated on Lys-9 and Lys-27 (H3K9me3 and H3K27me3) are associated with inactive regions (4–7).
Histone modifications also influence the activity of enhancers involved in regulating gene expression during development and cell differentiation. In human embryonic stem cells, specific epigenetic signatures define the activity of enhancer elements involved in the early stages of embryogenesis: H3K27ac is a mark of active (or class 1) enhancers, whereas a high level of H3K27me3 and the absence of H3K27ac are found in “poised” (or class 2) enhancers (8). The acetyltransferase and transcriptional coactivator p300/CBP binds active enhancers in several tissues and organs, including heart, during development (9). Recently, a large set of enhancer elements involved in regulating cardiomyocyte gene expression during differentiation in vitro was identified (10).
Hitherto, most research on the epigenetics of heart failure has focused on the role of histone deacetylases (HDACs; e.g., HDAC5, HDAC9, and HDAC2) and histone acetyl transferases (HATs; e.g., p300). These studies demonstrated a key role for these enzymes in the cardiac hypertrophy program and, thus, suggest a role for histone acetylation in triggering the transcriptional changes occurring in cardiac hypertrophy and heart failure (11–14). However, we still do not know which genes are regulated by histone modifications or how histone modifications influence transcription in cardiac hypertrophy.
Here, we describe epigenetic changes occurring in adult mouse cardiomyocytes subjected to a prohypertrophy stimulus in vivo.
Results
Epigenetic Profile Changes in Hypertrophic Cardiomyocytes.
To gain insight into the role epigenetics plays in heart failure, we generated genome-wide chromatin-state maps for normal and hypertrophic cardiomyocytes and compared them with the gene expression profiles of those cells (Fig. 1A). To this end, we performed chromatin immunoprecipitation (ChIP) coupled with massively parallel DNA sequencing (ChIP-seq) on cardiomyocytes isolated from the left ventricle of mice that had been subjected to transverse aortic constriction (TAC), a surgical procedure that induces cardiac hypertrophy and then heart failure. Cardiomyocytes were isolated 1 wk after TAC, a time corresponding to the maximal activation of compensatory cardiac mechanisms (15, 16). The purity of isolated cardiomyocytes was at least 95%, as evaluated by counting the number of cells with and without the typical rod-shaped appearance (Fig. S1A). ChIP-seq was performed using antibodies against H3K9ac, H3K27ac, and H3K4me3, three marks associated with active regulatory regions; H3K79me2, a mark associated preferentially with transcribed genes (17); and H3K9me2, H3K9me3, and H3K27me3, which are associated with repressed regions (4–7).
Fig. 1.
Genome-wide analysis of histone marks in adult cardiomyocytes isolated from pressure-overloaded and control mouse hearts. (A) Schematic of the experimental workflow. (B) Hierarchical clustering of H3K9ac (9ac), H3K27ac (27ac), H3K4me3 (4me3), H3K79me2 (79me2), H3K9me2 (9me2), H3K9me3 (9me3), and H3K27me3 (27me3) on the basis of their genomic distribution in normal (sham) and hypertrophic (TAC) cardiomyocytes. The heat map was generated with the DiffBind program. Also see Fig. S1B and Dataset S1.
To identify the genomic regions undergoing a change of their histone mark profile on cardiac hypertrophy, we used two algorithms, MACS (model-based analysis of ChIP-seq) (18) and SICER (spatial clustering approach for the identification of ChIP-enriched regions) (19), to minimize the bias inherent in this procedure and compared the obtained peak datasets with DiffBind (differential binding analysis of ChIP-seq peak data) (20). Hierarchical clustering analysis using affinity (read count) data revealed that the histone marks studied were significantly redistributed during cardiac hypertrophy (Fig. 1B). This result was supported by MA [M(log ratios)A(mean average)] plots, which evidenced the high number of differential histone modification sites in sham versus TAC cardiomyocytes (Fig. S1B and Dataset S1). We found that 9.1% of the genome of hypertrophic cardiomyocytes had a change in the distribution of at least a single histone mark (Fig. S1C), with all histone marks analyzed becoming redistributed to transcription start sites (TSSs) (±4 kb) (Fig. S2A).
To obtain information on the biological role of genomic regions found with redistributed histone marks, we performed gene ontology (GO) analysis with the Genomic Regions Enrichment of Annotations Tool (21) (GREAT; default setting, 5 + 1 kb basal, up to 1 Mb extension) (Fig. S2B). The sites with differential distribution of H3K9ac, H3K27ac, or H3K4me3 mapped preferentially to regulatory domains of genes involved in heart function or epigenetic regulation of gene expression, whereas those with altered H3K79me2, H3K9me2, H3K9me3, or H3K27me3 profiles were preferentially associated with genes regulating signal transduction (e.g., regulation of Rab-GTPase activity and regulation of ARF GTPase activity) and organization and regulation of sarcomeric structure. Moreover, GO analysis for mouse phenotype revealed that sites with differential distribution of H3K9ac, H3K4me3, H3K79me2, or H3K27me3 were associated with hypertrophic heart phenotypes, such as thick ventricular wall, altered heart contraction, impaired skeletal muscle contractility, and abnormal cardiac stroke volume. Together, these findings demonstrate that pressure-overload hypertrophy is associated with major chromatin remodeling of a wide array of genes regulating cardiac function.
Histone Modifications Regulate the Promoter Activity of Genes Modulated by Pressure Overload.
Next, we determined whether this chromatin remodeling was responsible for regulating the gene expression changes associated with cardiac hypertrophy. To this end, we first performed RNA sequencing (RNA-seq) on polyadenylated RNA extracted from cardiomyocytes isolated from the same mouse hearts used for epigenetic profiling. We identified 1,109 genes differentially expressed with a log-fold change in expression ≥1.3 in TAC cardiomyocytes (adjusted P value < 0.05); of these, 65.46% (726 genes) were up-regulated and 34.53% (383 genes) were down-regulated (Fig. S3 A and B and Dataset S2).
We then examined the transcriptional level of those genes that had undergone a change at their promoter region (i.e., ±2 kb around the TSS) in only one of the seven histone marks (Fig. S3C). We found that a decrease in H3K9ac or H3K27ac was significantly associated with transcription repression in TAC cardiomyocytes; conversely, a decrease in either H3K9me3 or H3K27me3 was associated with genes that were more actively transcribed. Moreover, either a loss or a gain of H3K79me2 at promoters could be associated with more-expressed genes in TAC cardiomyocytes. Because H3K79me2 distributes to the body of genes as well as promoters, we also analyzed the relationship between gene expression and changes in the distribution of this histone mark at the former region (Fig. S3C). Consistent with previous reports (6, 22), an increase of H3K79me2 at gene bodies correlated significantly with a more-expressed gene set.
Quantitatively, of the 1,109 genes modulated in TAC cardiomyocytes, we found that 596 genes had an alteration of at least one histone mark at their promoter. Three hundred twenty-five (29.30%) of these genes had a transcriptional level and an epigenetic landscape consistent with the epigenetic code theory (23) for the seven histone marks studied. One-way hierarchical clustering of the 325 genes on the basis of fold change in expression revealed the presence of two main gene clusters (Fig. 2A): one comprised 135 genes that were up-regulated in TAC cardiomyocytes and that either gained activating marks or lost repressor marks at their promoters, and the other contained 190 down-regulated genes having an epigenetic profile with an opposite trend to that of the first cluster (i.e., increased repressor marks or decreased activator marks). Moreover, histone pattern analysis of the two clusters revealed that transcriptional activation in TAC cardiomyocytes was influenced to an equal extent by each of the seven histone marks, whereas repression of gene expression was influenced more by a decrease in H3K9ac or an increase in either H3K9me3 or H3K27me3 than by changes in the other epigenetic modifications (Fig. 2B). Notably, in both clusters, promoters presenting with changes in activator marks did not have any alteration of repressive ones, and vice versa, indicating that these classes of histone marks regulate distinct genes in cardiac hypertrophy. GO analysis revealed that a significant number of the genes activated by the epigenetic modifications were implicated in cell adhesion, cytoskeletal formation, and heart development; in contrast, a significant number of repressed genes were involved in oxidative stress and gene transcription regulation (Fig. S3D). Thus, histone H3 marks provide an epigenetic signature that distinguishes genes regulated in cardiac hypertrophy, including important players of this pathology, such as Rgs2, Tcap, Bmp2, and Tgfbr1 (Fig. 2C). For some of these cardiomyocyte-specific genes, we confirmed the epigenetic changes with ChIP performed on total heart from mice after 1 wk of TAC (Fig. S4).
Fig. 2.
Analysis of epigenetic changes occurring at the promoter of genes modulated in cardiac hypertrophy. (A) Heat map of the changes in the histone mark profiles occurring at the promoters of 325 genes found modulated in TAC cardiomyocytes in accordance with the epigenetic code for the seven modifications studied. The genes were ordered on the basis of fold change in expression. (B) Heat maps of hierarchical clustering of the 135 up-regulated genes (Left) and the 190 down-regulated genes (Right) on the basis of the changes in expression of the histone marks. The diagrams below the heat maps give schematic representations of the results. (C) Examples of the distribution of reads obtained by ChIP-seq and RNA-seq in ten loci related to cardiac hypertrophy and heart development in sham and TAC cardiomyocytes. The distributions of the histone marks were normalized to input and library dimension. Uniform scales are used for each histone mark and for mRNA. S, sham; T, TAC; 9ac, H3 monoacetylated at lysine 9; 79me2, H3 di-methylated at lysine 79; 9me3, H3 trimethylated at lysine 9; 27ac, H3 monoacetylated at lysine 27.
Identification of Active Enhancers Specific for Cardiac Hypertrophy.
The enhancers associated with hypertrophy are unknown. Because active enhancers can be distinguished by the presence of H3K27ac (24, 25), we mined our ChIP-seq dataset to identify those enhancers. We found 9,207 putative active enhancers mapping to noncoding intra- and intergenic regions (Fig. S5A and Dataset S2). This set of predicted enhancers overlaps 5.8% with p300/CBP+ enhancers identified during heart development and 53.3% with H3K4me1+/H3K27ac+ enhancers found associated with cardiomyogenic differentiation (9, 10) (Fig. S5B).
Our set of H3K27ac+ enhancers was associated with genes involved in cellular processes underlying cardiac hypertrophy, such as metabolic process, gene expression, and cytoskeleton organization (Fig. S5C). This finding was supported by GO of the human phenotype, which revealed that genes associated with these enhancers were involved in several cardiac disease states (Fig. S5C). Moreover, an analysis of evolutionary conservation of the enhancers revealed that, similar to what has already been observed for cardiac enhancers associated with heart development and cardiomyocyte differentiation (10, 26), these genetic elements are poorly conserved in placental mammals and vertebrates [phylogenetic analysis with space/time models conservation (PhastCons) score, 12% and 9%, respectively]. However, we did find a large set of enhancers (1,560 elements) conserved in humans and mice (Fig. S5D and SI Materials and Methods). Therefore, we have brought to light a large number of hitherto undescribed putative enhancer elements specific to the cardiomyocyte and highly conserved in man.
To determine whether these genetic elements regulate the gene expression program of hypertrophy, we classified the enhancers on the basis of the presence/absence of H3K27ac and H3K27me3. H3K27ac+ enhancers were deemed “active,” or class 1, and H3K27me3+ enhancers were considered “poised,” or class 2, whereas H3K27ac−/H3K27me3− enhancers were classed as “intermediate” (8, 25). Consistent with this classification, the gene set associated with class 1 enhancers was expressed at a level significantly higher than that associated with intermediate enhancers in both sham and TAC cardiomyocytes (Fig. S6A). Of note, there were few enhancers (34 of 9,207) belonging to class 2.
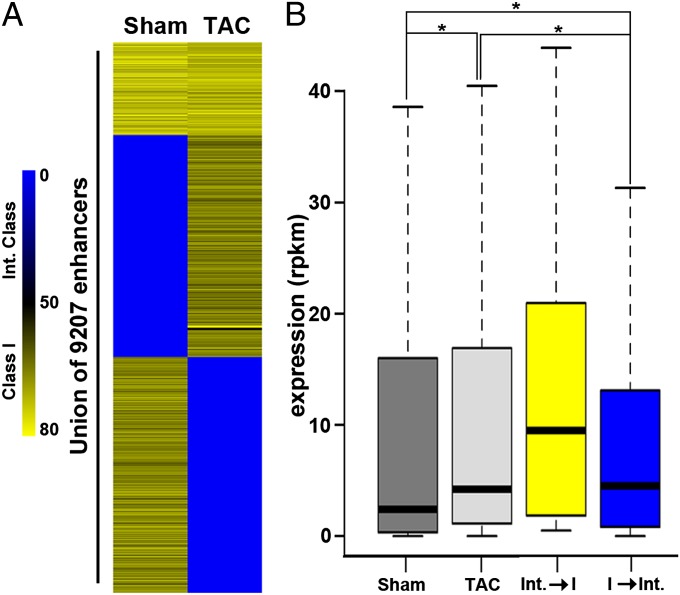
With the induction of cardiac hypertrophy, a large fraction of enhancers switched class (Fig. 3A). This phenomenon was associated with a variation in the transcriptional level of neighboring genes: The gene set associated with enhancers switching from the intermediate class to class 1 had an increased expression in TAC cardiomyocytes (Fig. 3B). This set included markers of hypertrophy, such as Actn1, Hif1a, Fstl1, and Anxa2 (Fig. S6B). In contrast, genes associated with enhancers shifting in the opposite direction were less expressed in hypertrophic cardiomyocytes. This demonstrates that enhancers are involved in promoting the expression of genes in hypertrophy through an epigenetic signature that regulates their activity.
Fig. 3.
Activity of enhancers in cardiac hypertrophy. (A) Distribution map illustrating the shift occurring between class 1 and intermediated class enhancers on induction of hypertrophy. Sham, cardiomyocytes in a basal condition; TAC, cardiomyocytes isolated from mice subjected to pressure overload. Enhancers with a peak score of MACS for H3K27ac higher than 50 were classified class 1, whereas enhancers with a peak score of less than 50 were classified as intermediate. (B) Box plots of gene expression values for all genes (bars in gray shades) and those associated with enhancers that change class on cardiac hypertrophy (yellow bar, intermediate class enhancers changing into class 1; blue bar, class 1 enhancers changing into intermediate class). Box plots represent interquartile ranges, with the bottom and the top of the boxes indicating the 25th and 75th percentiles, respectively; the internal bands correspond to the 50th percentile. P values were determined by unpaired Wilcoxon test. *P < 0.01. RPKM, reads per kilo base per million; Int., intermediate class; I, class 1.
Finally, to identify the transcription network within which the epigenetically regulated promoters and enhancers acted to govern gene expression in TAC cardiomyocytes, we searched for the presence of transcription factor-binding motifs within these genetic elements. Hypergeometric optimization of motif enrichment (HOMER) (27) analysis revealed that motifs for myocyte enhancer factor (MEF)2C and MEF2A were significantly enriched at enhancers, whereas motifs for specificity protein 1 (SP1), nuclear factor Y (NFY), and kruppel-like factor 4 (KLF4) were found at promoters (Fig. 4 A and B and Fig. S7A). To validate these predictions, we performed ChIP for MEF2C and MEF2A on cardiomyocytes isolated from sham and TAC mice. We found that MEF2C and MEF2A bound to 56.5% of the enhancers having binding motifs for these transcription factors and that were associated with genes modulated in cardiac hypertrophy, such as Grk5, Irs1, and Gpx3 (Fig. S7B). These results suggest that MEF2C and MEF2A, the two transcription factors that fundamentally promote gene expression in cardiac hypertrophy (12, 28), regulate the activity of enhancers.
Fig. 4.
Transcription networks of genes regulated epigenetically in cardiac hypertrophy. (A) The transcription binding motifs identified for promoters regulated epigenetically in cardiac hypertrophy (Left) and for putative, active enhancers (Right). Also see SI Materials and Methods. (B) Predicted genes regulated in cardiac hypertrophy through the binding of SP1, NFY, and KLF4 with promoters modulated epigenetically, and MEF2C and MEF2A with enhancers (Right). The colors of the nodes represent fold change of expression in cardiac hypertrophy, as indicated by the scale. (C) Model of how the transcription factors could act in regulating the expression of genes affected epigenetically in cardiac hypertrophy. On the basis of this model, MEFs govern the activity of enhancers, whereas SP1, NFY, and/or KLF4 affect the activity of promoters in triggering gene expression changes in cardiac hypertrophy. MEF, myocyte enhancer factor; SP1, transacting transcription factor 1; NFY, nuclear transcription factor-Y; KLF4, Kruppel-like factor 4; HDAC, histone deacetylase.
Discussion
In this study, we identified a specific epigenetic signature at promoters and discovered enhancers acting as regulatory elements associated with cardiac hypertrophy. So far, research on the epigenetics of heart failure has focused on histone acetylation mainly by means of mouse knockout models of genes encoding HDACs and HATs (12, 29). The studies revealed a key role of those enzymes in triggering gene expression changes in cardiac hypertrophy. Moreover, the chromatin remodeling factors Brg1, HDAC, and poly (ADP-ribose) polymerase (PARP) were demonstrated to cooperate in regulating gene expression of α- and β-myosin heavy chains during cardiac hypertrophy (30). However, little is still known about the genes regulated by histone acetylation, and the role of histone methylation remains largely uninvestigated. Indeed, although previous work has suggested an involvement of histone methylation in cardiac hypertrophy (31, 32), those researchers did not provide evidence for a role of this histone mark in regulating gene expression. Our study reveals that a specific epigenetic signature, defined by histone acetylation and methylation, regulates gene expression of hypertrophy by governing the activity of promoters. In fact, the expression of a large set of genes (325 of 1,109 genes) is regulated through a specific epigenetic signature. This signature is characterized by the mutual exclusion of activating (H3K9ac, H3K27ac, H3K4me3, and H3K79me2) and repressive (H3K9me2, H3K9me3, and H3K27me3) histone marks. Thus, these activating and repressive epigenetic modifications do not act cooperatively in regulating the activity of promoters but, instead, regulate distinct gene sets.
It is noteworthy that not all genes showed epigenetic changes consistent with epigenetic code theory. The expression of these genes could be regulated by mechanisms that go beyond the histone modifications studied in this work. These could include mechanisms that influence the activity of transcription factors (e.g., their expression, nuclear localization, activation by posttranslational modifications, presence of cofactors) and other epigenetic mechanisms that were not examined (e.g., DNA methylation, long noncoding RNAs, other histone modifications).
In addition, we have identified more than 9,000 putative active enhancers associated with cardiac hypertrophy. Surprisingly, the identified enhancers are largely unrelated to those previously described in heart development and cardiomyocyte differentiation (9, 10). Our results support the notion that during cardiac hypertrophy, the underlying gene expression reprogramming is regulated both by enhancers involved in heart development and by cis regulatory elements specifically active in this pathology (Fig. 4 and Fig. S6). Moreover, transcription network analysis of these genetic elements has revealed a role for MEF2C and MEF2A in regulating the activity of enhancers.
Although our findings do not elucidate the molecular pathways upstream of the histone marks, the fact that hypertrophy is associated with altered epigenetic profiles at enhancers and promoters does provide clues to the mechanisms regulating its pathogenesis.
Materials and Methods
For full details, see Transverse Aortic Constriction and Cardiomyocte Isolation and Transcription Network Analysis. Briefly, 2-mo-old male C57BL/6J mice were subjected, under anesthesia, to TAC, as described previously (33). All of the experiments were performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (publication 85–23, revised 1996) and approved by the local ethical committee of the Italian Ministry of Health. ChIP-seq and RNA-seq were carried out as described previously (33–36). MACS (18) and SICER (19) were used to identify the genomic regions bound to histones, whereas DiffBind (20) was used to identify the differential histone modification sites between sham and TAC cardiomyocytes.
Supplementary Material
Acknowledgments
We thank Rory Stark and Gordon Brown of Cancer Research UK and the Cambridge Research Institute for help with data analysis with DiffBind. This work was supported by an Advanced Grant (CardioEpigen; 294609) from the European Research Council, grants from Fondation LeDucq and Fondazione CARIPLO (12-4-5157157-31; to G.C.), Grant EPIGEN (Progetto Bandiera Epigenomica), and Ageing Project (Progetto Bandiera Invecchiamento; to R.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315155110/-/DCSupplemental.
References
- 1.Lompre AM, et al. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282(5734):105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- 2.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci USA. 1988;85(2):339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papait R, Greco C, Kunderfranco P, Latronico MV, Condorelli G. Epigenetics: A new mechanism of regulation of heart failure? Basic Res Cardiol. 2013;108(4):361. doi: 10.1007/s00395-013-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.May D, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44(1):89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110(4):479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, et al. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem. 2007;282(48):35078–35087. doi: 10.1074/jbc.M707083200. [DOI] [PubMed] [Google Scholar]
- 14.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 15.Kemi OJ, et al. Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. J Cell Physiol. 2008;214(2):316–321. doi: 10.1002/jcp.21197. [DOI] [PubMed] [Google Scholar]
- 16.Toischer K, et al. Differential cardiac remodeling in preload versus afterload. Circulation. 2010;122(10):993–1003. doi: 10.1161/CIRCULATIONAHA.110.943431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong X, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13(9):R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng C, et al. A statistical framework for modeling gene expression using chromatin features and application to modENCODE datasets. Genome Biol. 2011;12(2):R15. doi: 10.1186/gb-2011-12-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9(1):2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 24.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42(9):806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, et al. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281(14):9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto S, et al. Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation. 2006;113(5):679–690. doi: 10.1161/CIRCULATIONAHA.105.585182. [DOI] [PubMed] [Google Scholar]
- 30.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneda R, et al. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14(1):69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 32.Stein AB, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121(7):2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockman HA, et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci USA. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 35.Thomas-Chollier M, et al. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc. 2012;7(8):1551–1568. doi: 10.1038/nprot.2012.088. [DOI] [PubMed] [Google Scholar]
- 36.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.