A leading cause of antibiotic-resistant hospital infection (1), Enterococcus faecalis, decorates its surface with proteins that contribute to its ability to colonize and survive. Several proteins, including Esp (2), aggregation substance (3), a collagen adhesin (4), and a pilin (5), correlate with a particular disease or exacerbate the course of infection in a model system. In PNAS, Kandaswamy et al. (6) describe the targeting by human β-defensin 2 (hBD2) and resulting disorganization of the cellular machinery used by E. faecalis to secrete and anchor these proteins to the cell surface.
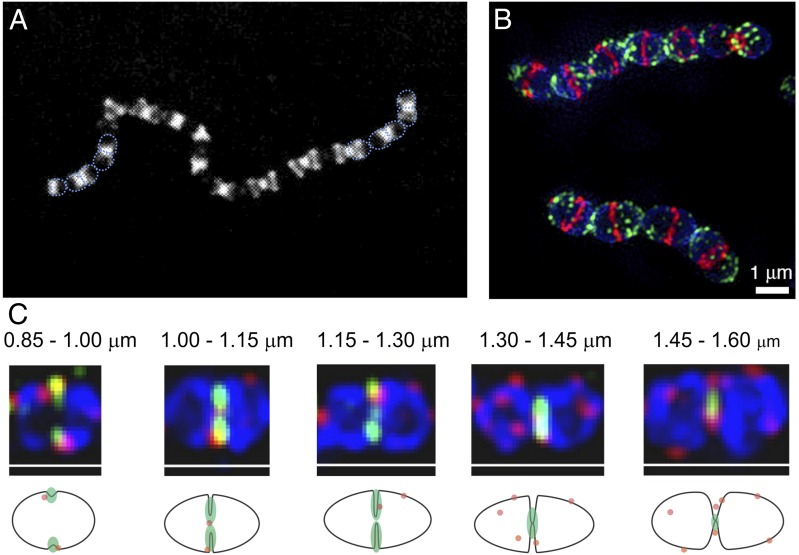
In 1962, Cole and Hahn (7) showed that in dividing cells, new cell wall was deposited by the ovoid Gram-positive Streptococcus pyogenes exclusively near the growing septum, and not by intercalation into existing cell wall (Fig. 1A). Rather than directly labeling peptidoglycan, however, these investigators used fluorescent antibodies to label surface-localized antiphagocytic M protein, or the group C cell wall carbohydrate. The authors observed that new M protein (or group C carbohydrate) was inserted at or near the developing septum, and spread from there toward the midcell as new cell wall accrued (Fig. 1A).
Fig. 1.
(A) New M protein deposited on the cell wall of M type 19 group A Streptococcus. Preexisting M protein was blocked using unlabeled antibody. After 30-min additional growth, fluorescein-labeled anti-M19 antibody stains the growing chain mainly near the septa of dividing cells. Blue-dashed cartoon lines depict approximate inferred cell boundaries, added for illustration (adapted from ref. 7). (Magnification, 2,100×.) (B) S. pyogenes, grown first in the presence of trypsin and pronase to removed preformed surface proteins, then subcultured for an additional 2 min before fixing and staining to label newly integrated cell wall proteins. Cells were stained with fluorescent antibody to M protein (red) and protein F (green), highlighting the differential initial localization of the YSIRK-signal peptide containing M protein at the septum (adapted from ref. 11). (C) Transient focal location of sortase (red) at or near the septum (green) at early stages in cell division of S. pyogenes (adapted from ref. 15). In early phases of S. pyogenes growth, sortase largely localizes to an area coincident with the septum. At later stages in cell division, SrtA appears to distribute more evenly around the cell, essentially as found in the accompanying examination of sortase distribution in E. faecalis (6).
With the appearance of nucleotide sequence data, the steps between translation of a gene product and its occurrence attached to the Gram-positive bacterial surface could begin to be inferred. Schneewind et al. (8) identified a C-terminal sequence motif common to many protein precursors destined for attachment to the Gram-positive cell surface. The C-terminal amino acid sequence motif, LPXTG, together with a positively charged C-terminal tail, were found to be required to efficiently anchor protein A from Staphylococcus aureus to the cell surface. The enzyme that catalyzes this transpeptidation reaction was subsequently identified and termed “sortase” (9).
In follow-up work, the laboratories of Lindahl and colleagues (10), Fischetti and colleagues (11), and Schneewind and colleagues (12) separately probed the additional role of an N-terminal secretion signal sequence with the motif YSIRK/GS in directing proteins to the septum of S. pyogenes (10, 11) and S. aureus (12). As shown in Fig. 1B, primary translation products with the YSIRK/GS motif, such as the M protein precursor of S. pyogenes (10,11), or ClfA of S. aureus (12), are secreted selectively at or near the growing septum. In contrast, streptococcal protein F (SfbI) (10, 11) or S. aureus proteins including SasF (12), all of which lack the motif, are deposited into old cell wall (Fig. 1B). Interestingly, because the YSIRK/GS motif can be mutated (10) or scrambled (12) without affecting the pattern of surface distribution, the specific feature of YSIRK-containing signal peptides responsible for the localization pattern remains obscure. S. aureus mutants defective in expression of a group of proteins containing abortive infectivity (ABI) domains are also aberrant in surface localization of YSIRK-motif proteins (13), but the mechanism for this association remains to be determined.
In parallel studies, Rosch and Caparon (14) dissected the process of secretion of the S. pyogenes exotoxin SpeB. During secretion, SpeB localizes to discrete foci near the septum, suggesting that protein secretion occurs through localized cell wall domains. The authors further found that the translocase that catalyzes transport through the membrane, SecA, colocalized with SpeB, which provided a new understanding of protein secretion by ovoid Gram-positive bacteria. This cell wall microdomain was termed the “ExPortal” (14). However, in electron micrographs of immunogold antibody-labeled cells, Lindahl and colleagues (10) did not find SecA localized to the septum of S. pyogenes, but rather distributed around its surface. In examining M protein deposition on the cell surface, Raz and Fischetti (15) used minimally treated whole cells and localized the sortase A (SrtA) of S. pyogenes to focal domains near the septum, similar to the findings of Rosch and Caparon (14). However, in later stages of growth, SrtA was found still in focal domains, but at points away from the septum (Fig. 1C). The localization of SrtA on the surface of cells appears to be a dynamic process, with early localization at or near the septum, and subsequent localization in focal patterns elsewhere around the cell surface (perhaps concomitant with dissolution of other higher-order structures related to peptidoglycan biosynthesis at the septum). Determination of the extent to which this is also true for the SecA translocase will require further study.
Because enterococci have emerged as leading causes of often multiple antibiotic-resistant hospital infection (1), there is substantial interest in developing new treatment modalities and understanding (and perhaps, augmenting) the activities of innate immunity, as well as current antimicrobial therapies. Although they are rugged microbes with intrinsic resistance to many antimicrobials (including detergents such as bile), enterococci are ordinarily held in check by factors of the innate immune system, including phagocytic cells and antimicrobial peptides. To understand how antimicrobial peptides target enterococci, with implications for the design of new therapeutics, Kandaswamy et al. (6) investigated the early interactions between human β-defensins and the E. faecalis cell. By fluorescently labeling minimally perturbed whole cells of E. faecalis, the investigators show that both the SecA translocase and the cellular sortase SrtA localize to foci near the septum of dividing cells. Interestingly, this focal localization at the septum is observed to hold true for SecA throughout the cell cycle. However, as observed by Raz and Fischetti (15), sortase appears to move away from the septum later in growth, suggesting a looser or more transient connection to the septum than for SecA.
Interestingly, Kandaswamy et al. (6) observed that human β-defensins, at levels below those required for inhibition of growth, bind the cell wall of E. faecalis in early growth in a focal pattern at the septum, analogous to the localization of both SecA and SrtA. Moreover, this deposition pattern was even more discrete in a multiple peptide resistance factor 2 (mprF2) mutant, which is defective in an enzyme that aminoacylates and reduces the negative charge of anionic membrane lipids (6). It therefore was of interest to know whether this pattern was congruent with the focal localization of SrtA and SecA. Surprisingly, these investigators found that hBD2 disrupted the focal localization of both SrtA and SecA, leading to an even distribution of these proteins around the enterococcal surface. The authors propose a model for the higher-order organization of the SecA translocase, the Sec YEG translocon, and sortases, whereby these proteins cluster in domains of anionic membrane lipids, which are then targeted by cationic peptides. The implication is that as these anionic lipid microdomains are modified by aminoacylation (or association with cationic peptides and possibly other cationic factors), this organization breaks down. The extent to which anionic microdomains in the bacterial membrane undergo flux during growth is not well understood, but may provide the basis for the variation observed in the localization of proteins associated with secretion and anchoring. It is currently not clear how the choice of insertion at the septum, versus insertion at the poles, relates to the timing of gene expression [e.g., S. aureus adhesins expressed early in growth phase before the quorum-induced Agr-mediated switch to expression of secreted exotoxins (16)]; or how an inducible protein, such as E. faecalis aggregation substance, is effectively distributed on the surface of pheromone-responsive donor cells following pheromone induction of potentially nondividing cells (17, 18).
The work by Kandaswamy et al. (6) suggests that sublethal levels of defense peptides may have evolved to disrupt or impede the surface localization of proteins that enhance colonization or exacerbate the course of infection. Antimicrobial compounds with dual activities, such as disrupting colonization and virulence at low levels, and being bacteriostatic or bactericidal by another mechanism at higher levels, may have an advantage in reducing the probability of the occurrence and outgrowth of resistant strains. This multiple targeting strategy may be the reason cationic antimicrobial peptides are so widely distributed in nature (19).
Acknowledgments
Production of this work was made possible by support from Public Health Service Grant AI083214 “Harvard-wide Program on Antibiotic Resistance” and fellowship support from Grant EY007145 (to D.V.T.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 20230.
References
- 1.Gilmore MS, Lebreton F, van Schaik W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol. 2013;16(1):10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67(1):193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow JW, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37(11):2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rich RL, et al. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274(38):26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 5.Nallapareddy SR, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandaswamy K, et al. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci USA. 2013;110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole RM, Hahn JJ. Cell wall replication in Streptococcus pyogenes. Science. 1962;135(3505):722–724. doi: 10.1126/science.135.3505.722. [DOI] [PubMed] [Google Scholar]
- 8.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70(2):267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson F, et al. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442(7105):943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 11.Raz A, Talay SR, Fischetti VA. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes. Mol Microbiol. 2012;84(4):631–647. doi: 10.1111/j.1365-2958.2012.08047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27(20):2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol. 2010;78(1):238–252. doi: 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304(5676):1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 15.Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci USA. 2008;105(47):18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 17.Wanner G, Formanek H, Galli D, Wirth R. Localization of aggregation substances of Enterococcus faecalis after induction by sex pheromones. An ultrastructural comparison using immuno labelling, transmission and high resolution scanning electron microscopic techniques. Arch Microbiol. 1989;151(6):491–497. doi: 10.1007/BF00454864. [DOI] [PubMed] [Google Scholar]
- 18.Olmsted SB, Erlandsen SL, Dunny GM, Wells CL. High-resolution visualization by field emission scanning electron microscopy of Enterococcus faecalis surface proteins encoded by the pheromone-inducible conjugative plasmid pCF10. J Bacteriol. 1993;175(19):6229–6237. doi: 10.1128/jb.175.19.6229-6237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4(7):529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]